Abstract
Autophagy is a membrane-trafficking process whereby double-membrane vesicles called autophagosomes engulf and deliver intracellular material to the vacuole for degradation. Atg4 is a cysteine protease with an essential function in autophagosome formation. Mounting evidence suggests that reactive oxygen species may play a role in the control of autophagy and could regulate Atg4 activity but the precise mechanisms remain unclear. In this study, we showed that reactive oxygen species activate autophagy in the model yeast Saccharomyces cerevisiae and unraveled the molecular mechanism by which redox balance controls Atg4 activity. A combination of biochemical assays, redox titrations, and site-directed mutagenesis revealed that Atg4 is regulated by oxidoreduction of a single disulfide bond between Cys338 and Cys394. This disulfide has a low redox potential and is very efficiently reduced by thioredoxin, suggesting that this oxidoreductase plays an important role in Atg4 regulation. Accordingly, we found that autophagy activation by rapamycin was more pronounced in a thioredoxin mutant compared with wild-type cells. Moreover, in vivo studies indicated that Cys338 and Cys394 are required for the proper regulation of autophagosome biogenesis, since mutation of these cysteines resulted in increased recruitment of Atg8 to the phagophore assembly site. Thus, we propose that the fine-tuning of Atg4 activity depending on the intracellular redox state may regulate autophagosome formation.
Abbreviations:
- Ape1, aminopeptidase I
- Asc, ascorbate
- ATG, autophagy-related
- Cvt, cytoplasm-to-vacuole targeting
- Eh, redox potential
- Em, midpoint redox potential
- DTNB, 5, 5′-dithiobis (2-nitro-benzoic acid)
- DTT, dithiothreitol
- DTTox, oxidized DTT
- DTTred, reduced DTT
- GSH, reduced glutathione
- GSNO, S-nitrosoglutathione
- GSSG, oxidized glutathione
- Gsr, glutathione reductase
- IAM, iodoacetamide
- NEM, N-ethylmaleimide
- PAS, phagophore assembly site
- PE, phosphatidylethanolamine
- PTM, post-translational modification
- rap, rapamycin
- ROS, reactive oxygen species
- SD, synthetic minimal medium
- Trr1, thioredoxin reductase 1
- Trx1, thioredoxin 1
- YPD, yeast peptone dextrose
Introduction
Macroautophagy (hereafter autophagy) is a major catabolic process by which eukaryotic cells degrade intracellular material. During autophagy, organelles and protein complexes are engulfed in bulk within a double-membrane vesicle known as the autophagosome and delivered to the vacuole and lysosome for their degradation and recycling.Citation1,2 Under specific conditions, organelles and other cytoplasmic contents can also be targeted to the vacuole via selective autophagy pathways.Citation3,4
Autophagy is widely conserved through evolution, and accordingly, autophagy-related (ATG) genes have been identified from yeasts to mammals. The autophagy machinery is composed of conserved Atg proteins that mediate the formation of the autophagosome membrane. Core Atg proteins localize to a site for autophagosome formation called the phagophore assembly site (PAS), in which precursor membranes organize to generate a complete autophagosome.Citation2 Among these core Atg proteins, Atg8 plays an essential role in the formation of the autophagosome. Atg8 associates with both inner and outer membranes of the autophagosome by covalent binding to phosphatidylethanolamine (PE) at a highly conserved glycine residue exposed at the C terminus.Citation5 Conjugation of Atg8 to PE is catalyzed by the sequential action of a set of Atg proteins in a ubiquitin-like system.Citation6 First, the cysteine protease Atg4 processes the C terminus of nascent Atg8 to expose the conserved glycine residue. Atg8 is then activated by the E1-like enzyme Atg7, transferred to the E2-like enzyme Atg3, and finally conjugated to PE in a reaction that requires the participation of the E3-like ligase Atg12–Atg5-Atg16.Citation5-9 In addition to processing the C terminus of Atg8, Atg4 also functions as a deconjugating enzyme that cleaves the amide bond between Atg8 and PE.Citation5 This deconjugating activity of Atg4 releases Atg8 from PE in membranes and plays an important role in autophagy regulation by allowing Atg8 recycling.Citation5,Citation10-12 Consistent with the essential role of Atg4 in autophagosome formation, yeast cells lacking this protease are unable to assemble the PAS and exhibit null autophagic activity.Citation5,8
Mounting evidence suggests that reactive oxygen species (ROS) may play a role in the control of autophagy.Citation13-17 In mammals, nutrient starvation stimulates ROS formation that appears to be necessary for autophagosome biogenesis and hence autophagy activation.Citation18 In photosynthetic organisms, autophagy is induced under oxidative stress conditions,Citation19,20 especially in the light during ROS production in chloroplasts.Citation21 ROS are well-established signal molecules regulating diverse cellular pathways but their exact role in the control of autophagy and the associated molecular mechanisms remain to be established. At present, Atg4 is the only Atg protein whose activity has been shown to be redox regulated.Citation18,22 In human ATG4A and ATG4B, this regulation was proposed to involve Cys81, possibly by interfering with the activity of the nearby catalytic Cys77.Citation18 However, since Cys81 is not conserved in most eukaryotes including yeast and plants (Fig. S1), if Atg4 is also redox regulated in these organisms, the underlying molecular mechanism must be distinct. In this study, we investigated the redox regulation of yeast Atg4 and unraveled a molecular mechanism by which redox signals control Atg4 activity and autophagy.
Results
Autophagy is linked to the intracellular redox state in Saccharomyces cerevisiae
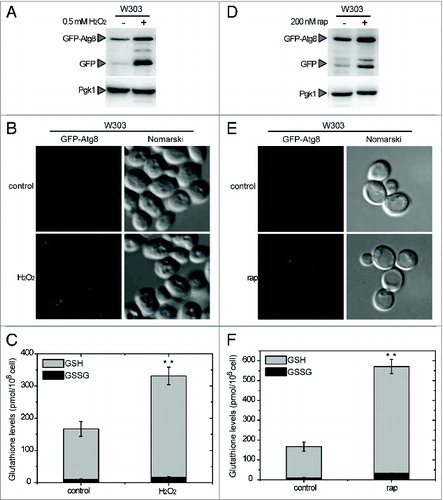
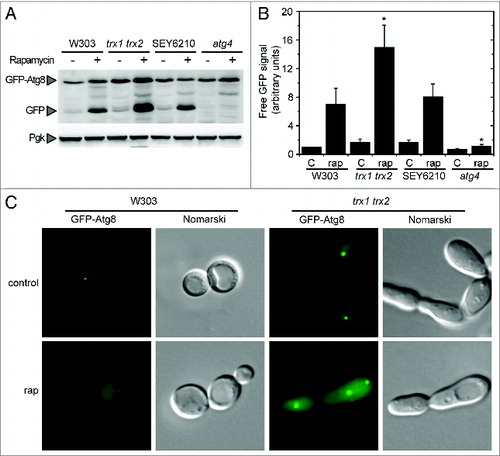
To investigate the connection between ROS and autophagy in yeasts, we analyzed autophagy induction when cells are subjected to oxidative stress triggered by hydrogen peroxide. The analysis of GFP-Atg8, indicated that autophagy was induced when yeast cells were treated with H2O2 (). In these conditions, the total glutathione pool, comprising reduced (GSH) and oxidized (GSSG) glutathione, was markedly increased (). This increase is a marker of oxidative stressCitation23 and reflects the alterations of the intracellular redox state triggered by H2O2. To further explore a connection between autophagy and oxidative stress, glutathione levels were also determined in yeast cells subjected to autophagy activation. For this purpose, cells were treated with rapamycin, which triggers autophagy via TOR inhibition.Citation24 Our results indicated that rapamycin treatment resulted not only in autophagy activation () but also in a pronounced increase of the total glutathione pool (). Consistently, an increase of the glutathione pool was previously reported in yeast during the first hours of nitrogen starvation, conditions that also trigger autophagy activation.Citation25 Taken together, our results suggest the existence of a link between autophagy and the intracellular redox state in yeasts. This indicates that, as in mammals, ROS may play a role in the control of autophagy in yeasts and prompted us to investigate the regulation of Atg4, the only Atg protein subject to redox regulation reported so far in other systems.Citation18,22
Figure 1. Connection between autophagy and the intracellular redox state in Saccharomyces. Analysis by western-blot of GFP-Atg8 cleavage assay in wild-type (WT) yeast cells growing exponentially in SD and treated with 0.5 mM H2O2 for 4 h (A) or 200 nM rapamycin for 2 h (D). Thirty micrograms of total extracts from untreated or treated cells were resolved by 12% SDS-PAGE followed by western blotting with anti-GFP and anti-Pgk1. The GFP-Atg8 fusion or free GFP proteins are marked with arrowheads. (B and E) Cells described in (A and D), respectively, were collected and processed for fluorescence microscopy analysis. The signal corresponds to GFP-Atg8. (C and F) Total gluthatione levels were measured in cells treated as described in (A and D), respectively. The data are represented as mean ± standard deviation from 5 independent experiments. “**,” Differences were significant at P < 0.01 according to the Student t test between untreated (control) and H2O2-treated cells (C), and between untreated (control) and rapamycin-treated cells (F).
Atg4 activity is redox-regulated
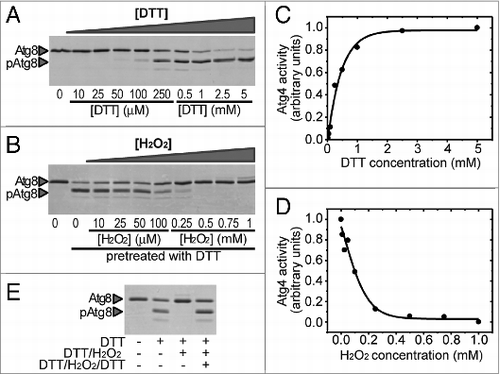
In order to analyze the redox regulation of yeast Atg4, the corresponding recombinant protein was purified and its activity was monitored. Atg4 processes Atg8 at a conserved glycine residue located at the C terminus of Atg8. For Atg4 activity assays we employed Atg8 from Chlamydomonas reinhardtii as previously described.Citation19 In contrast with yeast Atg8 that only possesses a single amino acid after the glycine cleavage site, Chlamydomonas Atg8 harbors a 15-amino acid extension after the glycine allowing detection and quantification of the unprocessed (Atg8) and processed (pAtg8) forms of Atg8 in Coomassie Blue-stained SDS-PAGE gels. Purified Atg4 was found to be functional as its activity increased in a concentration- and time-dependent manner (Fig. S2). This activity was determined in a standard Atg4 activity buffer containing the reducing agent DTT (dithiothreitol) at 1 mM. In order to determine whether Atg4 activity might be influenced by the redox environment, we incubated the protein in the presence of varying concentrations of reducing (DTT) or oxidizing (H2O2) compounds. Atg8 processing by Atg4 was found to increase with increasing DTT concentrations (). DTT is probably required for 2 reasons. Indeed, as classically observed for many purified proteins,Citation26 we observed that after purification, nonreduced Atg4 forms inactive high molecular weight oligomers that are disrupted by low concentrations of DTT (25 μM) that allow monomerization of Atg4. However, under these conditions the monomer was inactive and higher DTT concentrations were required to yield a significant Atg4 activity. This suggests that Atg4 is activated by reduction. Moreover, reduced Atg4 was inhibited by H2O2 in a concentration-dependent manner () indicating that the protein can be inhibited by oxidation. Both Atg4 reduction and oxidation were found to be fully reversible (), strongly suggesting the involvement of a redox post-translational modification (PTM) in the regulation of Atg4 activity.
Figure 2. Atg4 is activated by reduction and reversibly inhibited by oxidation. Atg4 (4.5 μM) activity was monitored by following the cleavage of Atg8 (5 μM) from the unprocessed (Atg8) to the processed (pAtg8) form (indicated by arrowheads) by SDS-PAGE and Coomassie brilliant blue staining. (A) Effect of DTT on Atg4 activity. Atg4 was incubated for 2 h in the presence of DTT concentrations ranging from 10 μM to 5 mM. (B) Effect of H2O2 on the activity of prereduced Atg4. After pretreatment with 0.1 mM DTT for 1 h, Atg4 was incubated in the presence of increasing H2O2 concentrations for 2 h. Quantification of Atg4 activity from (A and B) is shown in (C and D), respectively. The reference sample for quantification (Atg4 activity = 1 in arbitrary units) was 5 mM DTT and 0 mM H2O2 (prereduced with DTT) in (A and B), respectively. (E) Inhibition of Atg4 activity by H2O2 is reversed by DTT. Atg4 protein was pretreated with 1 mM DTT for 2 h (lane 2); then, Atg4 was incubated with 2 mM H2O2 for 30 min (lane 3); finally, Atg4 was newly treated with 10 mM DTT for 30 min (lane 4). Control (lane 1): incubation with no addition. For all lanes, Atg8 was added to the reaction mixture for 1 h after each specific treatment to assess Atg4 activity.
Atg4 activity is dependent on a single disulfide with a very low redox potential
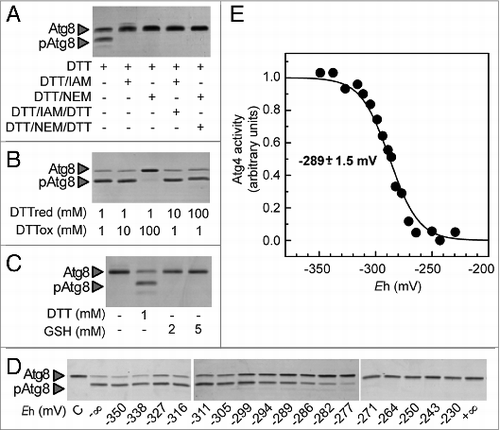
Cysteine alkylating agents such as iodoacetamide (IAM) or N-ethylmaleimide (NEM) are widely used to characterize redox PTMs. However, they were useless in the case of Atg4 since this enzyme is a cysteine protease whose activity is strictly dependent on a catalytic cysteine. Consistently, we found that reduced and active Atg4 is irreversibly inactivated by treatment with IAM or NEM (). To get further insight into Atg4 redox regulation, we analyzed Atg8 processing in the presence of varying amounts of reduced and oxidized DTT (DTTred and DTTox, respectively) (). The enzyme appeared to be activated at a DTTred/DTTox ratio between 10−1 and 10−2 suggesting that a very low redox potential is required for Atg4 activation. Consistently, while Atg4 was activated in the presence of 1 mM DTTred (Em,7 = -330 mV), GSH (Em,7 = -240 mV) proved inefficient even at 5 mM (). Finally, we performed a full DTT redox titration by varying the ambient redox potential (Eh) between -230 mV and -350 mV. After incubation at each potential, the extent of Atg4 activation was determined with the Atg8 proteolytic assay (). Atg4 activation strongly depended on the redox potential and no significant activity was detected when the Eh was higher than –277 mV. The experimental data of this in-gel redox titration gave excellent fits to the Nernst equation for the reduction of a single 2-electron component with a midpoint redox potential (Em), at pH 7.0, of -289 mV ± 1.5 mV (). Taken together, these data strongly suggest that the redox regulation of Atg4 activity is linked to the oxidoreduction of a single regulatory disulfide bond with a very low redox potential.
Figure 3. Atg4 activity is dependent on the redox potential. Atg4 activity was monitored as in . (A) Atg4 activity is irreversibly inhibited by alkylating agents. Atg4 was pretreated with 1 mM DTT for 2 h (lane 1); then, incubated for 1 h with 10 mM iodoacetamide (IAM, lane 2) or 10 mM N-ethylmaleimide (NEM, lane 3); next, Atg4 was again incubated with 50 mM DTT for 30 min (lanes 4 and 5). (B) Atg4 activity is dependent on the DTTred/DTTox ratio. Atg4 activity was determined after incubation during 2 h in the presence of various DTTred/DTTox ratio (1/1, 1/10, 1/100, 10/1, 100/1 in mM). (C) Atg4 is not activated by GSH. Atg4 activity was analyzed after incubation for 2 h in the presence of 1 mM DTT (lane 2), 2 mM GSH (lane 3) or 5 mM GSH (lane 4), or in the absence of reducing agent (lane 1). (D) Redox titration of Atg4 activity. Atg4 activity was determined after incubation for 2 h at indicated Eh poised by 20 mM DTT in various dithiol/disulfide ratios. C: control untreated. The −∞ sample was used as reference for quantification. (E) Atg4 activities monitored as in (D) were interpolated by nonlinear regression of the data using a Nernst equation for 2 electrons exchanged (n = 2) and one redox component. The average midpoint redox potential (Em,7) of 4 independent experiments is reported in the figure as mean ± standard deviation.
Moreover, to investigate the possible involvement of other Cys modifications, Atg4 activity was analyzed after treatment with the NO donor S-nitrosoglutathione (GSNO) (Fig. S3). The incubation of prereduced Atg4 protein with GSNO resulted in Atg4 activity inhibition since no Atg8 processing was visualized. Inhibition by GSNO was totally reversed by DTT but not by other SNO specific electron donor as ascorbate, again supporting the formation of a disulfide bond (Fig. S3).
Atg4 is activated by thioredoxin reduction in vitro
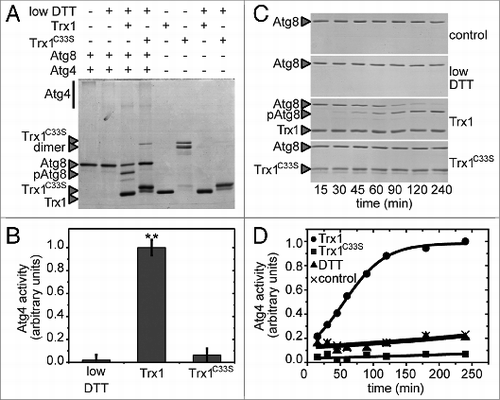
In the cytoplasm, regulatory disulfides with very low redox potentials, as measured for Atg4, are most generally reduced by small oxidoreductases named thioredoxins (Trx). S. cerevisiae contains 2 equivalent and redundant cytoplasmic thioredoxins, Trx1 and Trx2, which are reduced by the NADPH-dependent thioredoxin reductase Trr1. In order to test the ability of the Trx system to reduce Atg4, Trx1, and Trr1 from S. cerevisiae were cloned, expressed in E. coli and purified. A classical method employed in numerous biochemical assays to test the ability of Trx to reduce a target protein is based on the use of low concentrations of DTT that are able to efficiently reduce Trx in vitro but react very slowly with the disulfide of the target protein. In the presence of 25 μM DTT, Trx1 was clearly able to activate Atg8 cleavage by Atg4, whereas no processing was detected when the catalytic cysteine mutant Trx1C33S or only DTT were used (). As classically observed for many monocysteinic Trx active site mutants from diverse species, Trx1C33S is mainly a dimer in the absence of DTT, due to the formation of an intermolecular disulfide bond between the single reactive active site cysteine of 2 Trx1C33S monomers (). Although both Trr1 and Trx1 appeared functional based on control DTNB assays, Atg4 activation by Trx1 in the presence of NADPH and Trr1 could not be measured, most probably because Trr1 interferes with the Atg8 cleavage assay. To further characterize Atg4 reduction by Trx, we analyzed the time-course of Atg8 processing by Atg4 in the presence of Trx1 and low DTT concentrations (25 μM). Atg4 was efficiently reduced and activated by Trx1 compared with DTT alone or in the presence of Trx1C33S (). Therefore, our results strongly suggest a thioredoxin-dependent regulation of Atg4 activity due to the oxidoreduction of a regulatory disulfide bond.
Figure 4. Activation of Atg4 by thioredoxin. Atg4 activity was monitored as in . (A) Atg4 activity was measured after incubation for 1 h in the presence (+) or absence (−) of 25 μM DTT, 5 μM Trx1 and 5 μM Trx1C33S as indicated. (B) Quantification of Atg4 activity analyzed as in (A). The data are represented as mean ± standard deviation (n = 3). “**,” Differences were significant at P < 0.01 according to the Student t test between DTT and Trx1 samples, and between Trx1 and Trx1C33S samples. (C) Kinetics of Atg4 activation in the presence of diverse reductants as indicated using the same concentrations as in (A); control, no electron donor was added. (D) Quantification of Atg4 activity analyzed as in (C). The unprocessed (Atg8) and processed (pAtg8) forms of Atg8, Trx1 and monomeric and dimeric Trx1C33S are marked with arrowheads. The reference sample for quantification was Trx1+DTT and Trx1+DTT, 240 min in (B and D), respectively.
The trx1 trx2 mutant shows increased autophagy upon inhibition of TOR signaling
Our results described above indicated that Trx plays a role in Atg4 activity regulation in vitro. To investigate the biological significance of this finding, we analyzed autophagy induction by treatment with rapamycin in a yeast mutant strain lacking Trx1 and Trx2, the only cytoplasmic thioredoxins present in S. cerevisiae. It has been reported that the trx1 trx2 double mutant shows cell cycle defects leading to a significant increase in cell size and a greater proportion of large budded cells.Citation27 Regarding autophagy regulation, we found that the trx1 trx2 mutant displayed a higher autophagic activity upon rapamycin treatment compared with wild-type cells as determined by the GFP-Atg8 processing assay () or by fluorescence microscopy (). Although these observations might indicate that Trx plays a role in the redox control of autophagy in yeast cells, we cannot rule out an indirect effect of Trx deficiency. Indeed, the trx1 trx2 mutant exhibits a pleitropic phenotype including a high sensitivity to oxidative stressCitation28 that could affect autophagy independently of the role of Trx in the regulation of Atg4 activity. Therefore, we further investigated the molecular determinants of Atg4 redox regulation in vitro, in order to be subsequently able to establish more directly the importance of Trx in the regulation of autophagy in vivo.
Figure 5. Autophagy activation in the trx1 trx2 mutant. Autophagy induction was analyzed in wild-type (W303 and SEY6210), trx1 trx2 mutant and atg4 mutant strains treated with 200 nM rapamycin for 2 h. (A) Thirty micrograms of total extracts from the different strains grown in SD in the absence (−) or presence (+) of rapamycin were resolved by 12% SDS-PAGE followed by western blotting with anti-GFP and anti-Pgk1. The GFP-Atg8 fusion or free GFP proteins are marked with arrowheads. (B) Quantification of the signal corresponding to free GFP from (A). The reference sample for quantification was W303 control. The data are represented as mean ± standard deviation (n = 4). “*,” Differences were significant at P < 0.05 according to the Student t test between W303-rap and trx1 trx2 mutant-rap or atg4 mutant-rap. (C) Control or rapamycin-treated W303 and trx1 trx2 mutant cells were collected and processed for fluorescence microscopy analysis. The signal corresponds to GFP-Atg8.
Cysteines 338 and 394 form a disulfide bond involved in the regulation of Atg4 activity
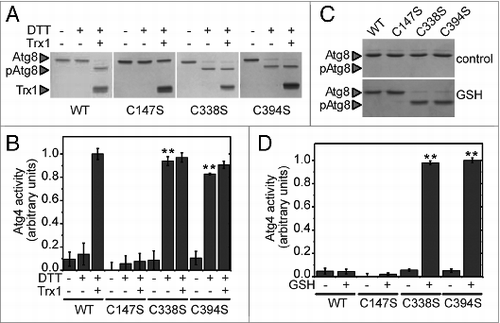
All our results point to the existence of a single disulfide bond responsible for redox regulation of S. cerevisiae Atg4. As MALDI-TOF mass spectrometry failed to detect all cysteine containing peptides, the identity of the cysteines involved in the disulfide was investigated by site-directed mutagenesis. Yeast Atg4 contains 12 cysteine residues among which only catalytic Cys147 is strictly conserved in all organisms (Fig. S1). Each of the 12 cysteines was individually mutated to serine and the corresponding recombinant proteins were produced and purified. The Atg8 cleavage activity of all Atg4 variants was analyzed in the presence of a low concentration of DTT with or without Trx1. As shown above, in these conditions, Atg4 WT was activated by reduction only in the presence of Trx1 (). Among the 12 variants, only 3 were distinguishable since all other mutants behaved as the WT enzyme (Fig. S4). As expected, the Atg4C147S mutant lacking the catalytic cysteine was inactive in all conditions tested (). Moreover, the Atg4C338S and Atg4C394S mutants were found to be independent of Trx activation since they were readily active in the presence of a low DTT concentration (). Similarly, these 2 mutants were active in the presence of GSH whereas Atg4 WT was not (). As mentioned previously, a mildly reducing environment provided by a low concentration of DTT or by GSH is required to keep the proteins in a disaggregated form but not for their redox activation (). Therefore, our results indicate that the monomeric forms of Atg4C338S and Atg4C394S mutants are not subjected to redox regulation and, consequently, are permanently active. Finally, these results indicate that Atg4 is activated through Trx-dependent reduction of a regulatory disulfide bond formed between Cys338 and Cys394.
Figure 6. The Cys-to-Ser mutant proteins Atg4C338S and Atg4C394S are not redox-regulated. Atg4 activity was monitored as in . (A) The activity of Atg4 WT, Atg4C147S, Atg4C338S, or Atg4C394S was determined after incubation for 2 h in the presence (+) or absence (−) of 25 μM DTT and 5 μM Trx1 as indicated. (C) Same as in (A) but after incubation for 2 h in the presence of 5 mM GSH. (B) and (D) Quantification of Atg4 activity determined as in (A and C), respectively. The reference sample for quantification was WT-Trx1+DTT and Atg4C394S+GSH in (B and D), respectively. The data are represented as mean ± standard deviation (n = 3). “**,” Differences were significant at P < 0.01 according to the Student t test between the DTT-WT and DTT-C338S or DTT-C394S mutants. The Atg8 and pAtg8 forms, and Trx1 are marked with arrowheads.
Expression of Atg4C338S or Atg4C394S stimulates PAS formation
In order to investigate the role of Atg4 redox regulation in vivo, we studied the functionality of Atg4 WT, Atg4C147S, Atg4,C338S and Atg4C394S in yeast mutant cells lacking endogenous Atg4. Yeast Atg8 processing by Atg4 is essential in both cytoplasmic-to-vacuole targeting (Cvt) and autophagy pathways. The atg4 mutant fails to deliver the Cvt pathway marker precursor aminopeptidase I (prApe1) from the cytoplasm to the vacuole, where it is processed to its mature form (mApe1).Citation5,8 Atg4 WT, Atg4,C338S and Atg4C394S were able to functionally replace endogenous Atg4 in the Cvt pathway, based on the ability of the complemented strains to restore maturation of prApe1 to mApe1 (). By contrast, the inactive Atg4C147S was not able to functionally complement the atg4 mutant. These data are consistent with our in vitro analyses () and indicate that Cys147, but not Cys338 and Cys394, is essential for Atg4 activity in vivo.
Figure 7. Complementation of an atg4 mutant strain with WT or mutant forms of Atg4. (A) atg4 mutant strain was transformed with empty vector (−) or a plasmid encoding Atg4 WT, Atg4C147S, Atg4C338S, or Atg4C394S, to express a HA-tagged version of the corresponding protein under the control of the ATG4 promoter. The wild-type strain (SEY6210) was used as a positive control. Thirty micrograms of total extracts from stationary phase cells grown in SD were resolved by 12% SDS-PAGE followed by western blotting with anti-Ape1, anti-Pgk1, anti-HA and anti-GFP. The precursor (prApe1) and mature (mApe1) forms of Ape1, Pgk1, HA-Atg4, GFP-Atg8 fusion or free GFP proteins are marked with arrowheads. (B) Cells growing exponentially in SD were collected and processed for fluorescence microscopy analysis. The signal corresponds to GFP-Atg8. (C) The percentage of cells from (B) displaying GFP-Atg8 puncta were quantified. The data are represented as mean ± standard deviation from 3 independent experiments. “*,” Differences were significant at P < 0.05 according to the Student t test between atg4 (WT) and atg4 (C338S) or atg4 (C394S). Number of cells quantified: n = 620 for atg4 (−), n = 700 for atg4 (WT), n = 600 for atg4 (C147S), n = 1275 for atg4 (C338S) and n = 1465 for atg4 (C394S).
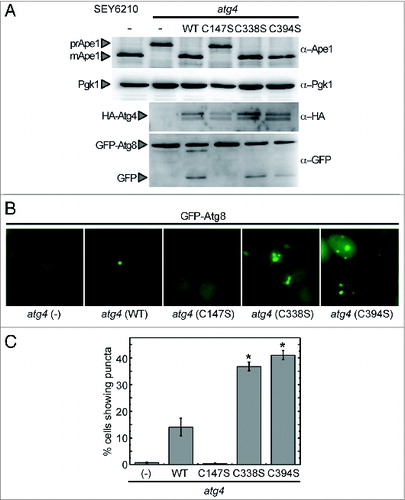
The functionality of WT and Atg4 mutant forms was also examined through the analysis of GFP-Atg8 processing, a biochemical assay widely used to monitor autophagic activity.Citation29 No significant difference was observed in the autophagic activity of cells expressing Atg4 WT and the 2 Atg4C338S and Atg4C394S mutants. By contrast, Atg4C147S was unable to promote autophagic activity (). To further analyze the role of Cys338 and Cys394 in the control of Atg4 activity, we examined the cellular distribution of GFP-Atg8 in atg4-deficient cells expressing Atg4 cysteine mutants. As previously shown,Citation8 no GFP-Atg8 was detected as punctate signal in atg4-deficient cells (), since Atg4 is fully required for the proper assembly of PAS and the presence of Atg8 at this structure. Expression of Atg4 WT rescued PAS formation and the GFP-Atg8 signal could be observed as dots in some cells (). In agreement with the null activity displayed by the Atg4C147S mutant both in vitro and in vivo, no PAS was detected in atg4-deficient cells expressing this mutant (). Interestingly, expression of Atg4C338S or Atg4C394S resulted in a significant increase in the number of cells showing puncta compared with cells expressing Atg4 WT under optimal growth (). Moreover, more than one puncta per cell could be observed in cells expressing Atg4C338S or Atg4C394S (). These data suggest that the redox regulation of yeast Atg4 may be required to control PAS initiation and/or formation.
Discussion
Recent lines of evidence indicate that ROS may play an important role in the control of autophagy but the molecular mechanisms underlying this regulation remain to be established.Citation13-16 This suggests that the activity of one or several key enzymes of the autophagy machinery, such as Atg proteins, may be regulated in response to changes of intracellular redox conditions. In the present study, we provide evidence linking autophagy and ROS in S. cerevisiae. On the one hand, we confirmed that manipulation of the cellular redox state, by hydrogen peroxide treatment, results in autophagy activation in yeasts. On the other hand, we showed that induction of autophagy by rapamycin treatment is accompanied by an alteration of the intracellular redox state as indicated by an increase of the cellular glutathione pool. In addition, we unraveled that a redox regulatory mechanism controls the activity of the key autophagy protein Atg4 in yeasts. These results indicate that yeast Atg4 is active in the reduced form and inactive in the oxidized form and this regulation is reversible suggesting the involvement of redox post-translational modifications. A similar effect of redox conditions was reported for Atg4 homologs in human and Arabidopsis suggesting that the redox regulation of Atg4 may be broadly conserved in eukaryotes.Citation18,22 Our analyses further revealed that yeast Atg4 activation depends on the ambient redox potential and that the transfer of 2 electrons is involved in this reaction indicating a regulation implicating a single disulfide bond. Moreover, the low midpoint redox potential of this putative disulfide, -289 mV at pH 7.0, suggested that Atg4 activity might be controlled through dithiol/disulfide exchange involving strong reducing oxidoreductases. Consistently, Trx was found to be a very efficient activator of Atg4, suggesting that this oxidoreductase may be the physiological electron donor for Atg4 activation in S. cerevisiae. In close agreement, in vivo analysis showed that autophagy is altered in the trx1 trx2 double mutant. Trx is involved in the control of numerous cellular processes including DNA synthesis, sulfate assimilation, transcription, cell division, apoptosis, stress responses, and cell signalingCitation28,30 through the reduction and/or oxidation of target proteins. Indeed, the trx1 trx2 mutant displays a pleiotropic phenotype, including prolonged cell cycle S phase, due to inefficient DNA synthesis, defective sulfate assimilation, and a pronounced sensitivity to peroxides.Citation27,28,31 Furthermore, due to the main role of Trx in the antioxidant defense by providing electrons to peroxiredoxins, glutathione peroxidases, and methionine sulfoxide reductases, the intracellular redox state of this mutant is notably modified.Citation28 Therefore, whether the effect of trx1 and trx2 mutations on autophagy is direct, caused by impaired redox regulation of Atg4, or an indirect consequence of the multiple cellular roles of thioredoxins and especially their function in peroxide metabolic pathways cannot be distinguished. Nevertheless, our in vitro and in vivo results suggest that Trx may have a function in the redox control of autophagy and this role may be conserved in other eukaryotes since Trx is present in all organisms. Accordingly, a quantitative proteomic approach has recently identified Trx and Trx reductase as autophagosome-associated proteins.Citation32
Site-directed mutagenesis revealed that the single regulatory disulfide bond controlling Atg4 activity is formed between Cys338 and Cys394. In vivo, this disulfide could be formed by reaction of Atg4 with oxidized Trx or by direct interaction with ROS. In the latter case, one of the 2 cysteines would be oxidized, e.g., to sulfenic acid in the presence of H2O2 or to nitrosothiols after reaction with an NO-donor. This oxidized cysteine would then react with the other proximal cysteine leading to formation of the Cys338-Cys394 disulfide, as demonstrated in the presence of an NO-donor (Fig. S3). Based on multiple sequence alignments, the catalytic Cys147 is the only cysteine strictly conserved and Atg4 sequences appear to contain numerous insertions specific to some species in different portions of the protein and, thereby illustrating the high diversity of this protease family in eukaryotes (Fig. S1B and S1C). Cys394 is located in a sequence insertion specific to a limited number of ascomycete species and is therefore not present in most organisms. However, in all Atg4 sequences including mammals and plants, a cysteine is always present in the region corresponding to S. cerevisiae Cys338, within a stretch of 3 amino acids (Fig. S1A). We can also observe that most Atg4 sequences harbor one or several additional cysteines down- stream of Cys338 that could fulfill the same functional role as Cys394 in S. cerevisiae. Therefore, we hypothesize that a disulfide bond formed between Cys338 and Cys394 in yeast, or Cys338 and a C-terminal cysteine in other eukaryotes, might be formed to control Atg4 activity. The redox regulation may be more complex in mammals since Cys81 of human Atg4A and Atg4B, which is only conserved in tetrapod homologs of these 2 proteins, has been shown to participate in the redox regulation of Atg4 activity.Citation18 In other redox-regulated enzymes, regulatory disulfides have appeared progressively throughout evolution leading to more sophisticated regulations in multicellular eukaryotes compared with unicellular eukaryotes.Citation33,34 Therefore, the redox control of autophagy, through redox post-translational modifications of Atg4, might be conserved in unicellular and multicellular eukaryotes, but the precise molecular mechanism might not be exactly the same. Further studies will be required to unravel the exact mechanism of Atg4 redox regulation and to especially investigate the role of the conserved Cys338 in diverse species.
Human ATG4B is the only Atg4 protein for which the 3-dimensional structure has been reported.Citation35,36 Due to sequence divergence with the human protein, it was not possible to build an accurate model of yeast Atg4 structure through homology modeling. However, considering the positions of the active site and of the regulatory Cys338 in the protein, formation of the disulfide bond between Cys338 and the C-terminal arm containing Cys394 is likely to interfere with the activity of yeast Atg4 by limiting the accessibility of Cys147 and/or by interfering with Atg8 binding. This mode of regulation is reminiscent of 2 previously characterized Trx-regulated enzymes, NADP-dependent malate dehydrogenase, and A2B2 glyceraldehyde-3-phosphate dehydrogenase.Citation37,38 In these enzymes, a C-terminal arm blocks access to the active site in the oxidized form and reduction of the enzyme by Trx leads to a conformational change that releases the C-terminal arm thereby providing full access of the substrates and cofactors to the active site.Citation34 Interestingly, as observed in the case of yeast Atg4, A2B2 glyceraldehyde-3-phosphate dehydrogenase is able to form redox dependent higher order AnBn oligomers.Citation39 Although we could not obtain evidence of the formation of Atg4 oligomers in vivo and we consider that they appear due to the purification procedure, we cannot rule out the possibility that such oligomers may have a functional role in vivo under specific conditions. Further studies will be required to elucidate the structural determinants of Atg4 redox regulation and to determine whether the C-terminal portion of the protein may act, as reported for other Trx targets, as a redox sensitive autoinhibitory domain.
Autophagosome formation is probably one of the most complex and regulated steps of autophagy. Many Atg proteins play a role in autophagosome biogenesis, but the cysteine protease Atg4 carries out a main function since it controls the steady-state of Atg8 lipidation. Atg8–PE localizes to all the autophagy structures, such as the PAS, the phagophore, the autophagosome, and the autophagic body.Citation8,40 Atg4 functions in immature Atg8 cleavage, free and processed Atg8 recycling from the autophagosome membranes and Atg8 releasing from inappropriate membranes,Citation5,8,Citation10-12 but the fine-tuning of Atg4 activity regulation remains unknown. Since Atg4 acts as both conjugating and deconjugating enzyme, its activity is expected to be tightly and finely regulated. In our study, we found that expression of Atg4 harboring mutations C338S or C394S did not impair Cvt and autophagy pathways but resulted in a stimulation of PAS formation under normal growth conditions (). These results suggest that the higher Atg4 activity caused by the absence of Cys338 or Cys394 might provoke an increased Atg8–PE concentration that might be directed toward the PAS. This finding is in close agreement with recent studies reporting that a defect in Atg4 deconjugating activity results in Atg8 mislocalization to the vacuolar rim and other membrane structures such as the endoplasmic reticulum, producing a higher percentage of cells displaying GFP-Atg8 puncta without increasing autophagic activity.Citation10-12 Redox regulation of yeast Atg4 may therefore be crucial to control the equilibrium between free Atg8 and Atg8–PE and thereby the initiation of PAS formation. This mechanism provides a first link between the activity of an Atg protein and the intracellular redox state that may allow local ROS concentrations to affect the autophagy process. This is certainly not the only link between ROS and autophagy and future studies will likely unravel additional redox mechanisms affecting Atg4 and other components of the autophagic machinery.
Materials and Methods
Chemicals and replications
Otherwise indicated, all chemicals were purchased from Sigma-Aldrich. The most relevant chemicals used in this study were DTTox (Sigma, D3511); DTT red (Sigma, 43815); GSH (Sigma, G4251); GSNO (Sigma, N4148); hydrogen peroxide (Sigma, H1009), IAM (Sigma, I1149); NEM (Sigma, E1271); rapamycin (LC Laboratories, R-5000).
Statistical methods
All experiments were performed at least in triplicate and data are presented as mean ± standard deviation. The statistical significance of data was verified by the Student t test whenever required.
Strains, media and growth conditions
The yeast strains used in this study are listed in Table S1. Yeast cells were grown in rich medium (YPD; 1% yeast extract [Difco, 10215203], 2% peptone [Difco, 211705], and 2% glucose [wt/vol]) (Sigma, G7021) or synthetic minimal medium (SD; 0.67% yeast nitrogen base [Difco, 211940], 2% glucose, supplemented with the appropriate amino acids and vitamins.) When indicated, yeast cells were grown in the presence of 0.5 mM hydrogen peroxide (H2O2; Sigma, H1009) or 200 nM rapamycin (rap; LC Laboratories, R-5000) for the indicated times.
Cloning and protein purification
The coding regions of TRX1 (312 bp) and TRR1 (960 bp) from Saccharomyces cerevisiae were amplified using the primers 5′TRX1 and 3′TRX1, or 5′TRR1 and 3′TRR1, respectively, and cloned into pET3c-His plasmid between NdeI and BamHI sites, to express the corresponding His-tagged proteins. The His-tagged Cys-to-Ser Trx1 mutant, denoted as Trx1C33S, was obtained by overlapping PCR using the wild-type version of the TRX1 gene and 5′TRX1C33S and 3′TRX1C33S mutagenic primers, and cloned into pET28a (Novagen, 69864-3) plasmid between NdeI and XhoI sites. The ATG4 coding region from S. cerevisiae was cloned into pET28a as described.Citation19 The His-tagged Cys-to-Ser Atg4 mutants, denoted as Atg4C55S, Atg4C59S, and Atg4C138S, were obtained by overlapping PCR using the wild-type version of the ATG4 gene and 5′ATG4C55S and 3′ATG4C55S, 5′ATG4C59S and 3′ATG4C59S or 5′ATG4C138S and 3′ATG4C138S mutagenic primers, respectively. Atg4C147S, Atg4C235S, Atg4C240S, Atg4C288S, Atg4C338S, Atg4C394S, Atg4C445S, Atg4C451S and Atg4C494S (see Table S3 for details). Cys-to-Ser mutant clones were synthesized by GeneCust Europe (http://www.genecust.com). The primers used in this study are detailed in Table S2. All clones were verified by DNA sequencing and used to transform E. coli BL21 (DE3) strain.
All recombinant proteins were expressed by induction at exponential growth phase (OD600nm ∼0.5) with 0.5 mM isopropyl-β-D-thiogalactopyranoside (Sigma, I6758) for 3 h at 37°C. The recombinant proteins Trx1, Trr1, and Trx1C33S were purified by affinity chromatography on a His-Select Nickel Affinity Gel (Sigma, P6611) following manufacturer's instructions. For Trx1 and Trr1 purifications, an ammonium sulfate (Sigma, A5132) precipitation step (40% to 90% and 40% to 80% saturation, respectively) was performed before affinity chromatography. Prior to purification, His-tagged Atg4 WT and the cysteine mutant proteins were solubilized with 4 M urea (Sigma, 51456) due to the presence of the recombinant proteins in inclusion bodies. Then, the proteins were purified from the soluble fraction by affinity chromatography as described above. The Atg8 protein from Chlamydomonas reinhardtii was purified as described.Citation19 The proteins used in this study are listed in Table S3.
Glutathione determination
Yeast cells from liquid cultures were collected by centrifugation (5000 g, 5 min), washed once in 50 mM sodium phosphate (pH 7.5) buffer, resuspended in 0.2 N HCl and vortexed 10 times for 30 s each time in the presence of glass beads (Sigma, G8772). Crude extracts were cleared by centrifugation at 15000 g for 20 min at 4°C. 500 μl of sample were neutralized by adding 50 μl of 50 mM NaH2PO4 (pH 7.5) and 0.2 N NaOH to a final pH between 5 and 6. The neutralized sample was directly used for measuring total glutathione (reduced [GSH] plus oxidized [GSSG] glutathione) by the recycling assay initially described by Tietze.Citation41 The method relies on the Gsr (glutathione reductase)-dependent reduction of 5,5′-dithiobis(2-nitro-benzoic acid) (DTNB; Sigma, D8130) at 412 nm. Oxidized glutathione was measured after treatment of neutralized sample with 10 mM 4-vinylpyridine (VPD, Sigma V320-4) for 30 min at 25°C. To remove excess VPD, the derivatized sample was centrifuged twice at 15000 g for 20 min at 4°C. To measure total glutathione or GSSG, sample was added to a mix containing 120 mM NaH2PO4 (pH 7.5), 300 μM DTNB, 500 μM NADPH, 1 mM EDTA (pH 8), 1 U/ml GR (Sigma, G3664), and DTNB reduction was measured at 412 nm. Different GSH (Sigma, G4251) concentrations ranging from 0 to 5 μM were used as standards.
Preparation and analysis of proteins
Saccharomyces cells from liquid cultures were collected by centrifugation (5000 g, 5 min), washed once in 50 mM TRIS-HCl (pH 7.5), resuspended in lysis buffer (50 mM TRIS-HCl pH 7.5, 1 mM Pefabloc (Sigma, 76307) and 0.1% Triton X100 (Sigma,T9284) and vortexed 10 times for 30 s each time in the presence of glass beads (Sigma, G8772). Crude extracts were cleared by centrifugation at 15000 g for 20 min at 4°C. Proteins were quantified with the BCA assay (Sigma, B9643) as described by the manufacturer.
In vitro Atg4 cleavage assays
The typical reaction mixture contained 4.5 μM Atg4, 5 μM Atg8, 1 mM 1,4-dithiothreitol (DTT; Sigma, 43815) and 1 mM EDTA (Sigma, E6511) in Tris-buffered saline (50 mM Trizma base, 138 mM NaCl, 27 mM KCl, pH 8). When indicated, Atg4 WT and cysteine variants were preincubated in the presence of reducing (DTT or GSH), oxidizing [DTTox (Sigma, D3511) or H2O2 (Sigma, H1009)], alkylating [IAM (Sigma, I1149) or NEM (Sigma, E1271)], or S-nitrosoglutathione (GSNO; Sigma, N4148), compounds alone or in combination at the indicated times and concentrations. For the analysis of thioredoxin activity, 25 μM DTT ± 5 μM Trx1 or Trx1C33S were used at the indicated incubation time. The reaction mixtures were incubated at 25°C for the indicated time and stopped by addition of β-mercaptoethanol-free Laemmli sample buffer followed by 5 min of boiling. Proteins were resolved on 15% SDS-PAGE gels and stained with Coomassie brilliant blue (Sigma, 27816). For Atg4 activity quantification, gels were scanned with Odyssey (LI-COR, Biosciences, Siemensstrasse, Germany) and the signals corresponding to unprocessed (Atg8) and processed (pAtg8) Atg8 were quantified with the software Image Studio (LI-COR, Biosciences, Siemensstrasse, Germany). The Atg4 activity was considered as the relation between pAtg8/(Atg8+pAtg8), and it was normalized between 0 and 1, with 0 corresponding to totally oxidized and inactive Atg4 and 1 corresponding to totally reduced and active Atg4. The Atg4 activities (in arbitrary units) were plotted using the software OriginPro 8 (OriginLab).
Redox titration
Redox titration of Atg4 activity was performed by incubating the Atg4 protein (4.5 μM) in 50 mM TRIS-HCl pH 7.5, 1 mM EDTA at defined Eh values imposed by reduced and oxidized DTT (DTTred and DTTox, respectively) in various dithiol/disulfide ratios (20 mM as final DTT concentration). After equilibrating Atg4 samples in 100 μl redox buffers for 2 h at 25°C, 10 μl were added to the reaction mixture containing 5 μM Atg8 and 1 mM EDTA in 50 mM TRIS-HCl buffer at pH 7.5 for 1 h at 25 °C. Atg8 cleavage was visualized by Coomassie blue-stained gel and Atg4 activity was quantified as described above. Titration data were fitted to Nernst equation (n = 2, one component) using nonlinear regression with the CoHort software as described.Citation42 The midpoint redox potential (Em) is reported as the mean ± standard deviation of 4 independent experiments. The redox potential values (Eh) are expressed at pH 7.0.
Yeast complementation
For expression of HA-tagged Atg4 WT, Atg4C147S, Atg4C338S, or Atg4C394S in WT (SEY6210) or atg4 (UNY123) mutant cells expressing GFP-Atg8 from an integrative plasmid, the cDNA of the ATG4 gene and the mutant versions were cloned into SalI and SphI sites of centromeric pHAC22 vector under the control of the endogenous ATG4 promoter. The ATG4 promoter (300 bp upstream ATG4 cDNA) was cloned into EcoRI and SalI sites of pHAC22.
Immunoblot analysis
For immunoblot analyses, total protein extracts (30 μg) were subjected to SDS-PAGE and then transferred to nitrocellulose membrane (GE Healthcare, 10 401 396) for antibody binding. The anti-Ape1, anti-Pgk1 (Invitrogen, 459250), anti-HA (Santa Cruz Biotechnology, sc-7392) and anti-GFP (Roche, 11 814 460 001) antibodies were diluted to 1:3000, 1:40000, 1:1000 and 1:1000, respectively. The ECL prime chemiluminescent system (GE-Healthcare, RPN2232) was used to detect the proteins with horseradish peroxidase-conjugated secondary antibodies (Sigma, A6154).
Fluorescence microscopy
For fluorescence microscopy, yeast cells were grown to OD600 nm∼0.4 in SD medium and fixed with 4% formaldehyde (Merck, 1.04003.1000) in phosphate-buffered saline (10 mM Na2HPO4, 2 mM KH2PO4, 137 mM NaCl, 2.7 mM KCl) for 30 min prior to observation. Samples were examined on a DM6000B microscope (Leica, Wetzlar, Germany) with an ORCA-ER camera (Hamamatsu, Hamamatsu, Japan) and processed with the Leica Application Suite Advanced Fluorescence software package (Leica).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website.
Supplemental Material.pdf
Download PDF (3.3 MB)Acknowledgments
Yeast wild-type (SEY6210) and atg4 (UNY123) mutant strains, pGFP-Atg8 vector and anti-Ape1 antiserum was kindly provided by Dr D Klionsky. pHAC22 plasmid was kindly provided by Dr M Hall. Yeast wild-type (W303) and trx1 trx2 (EMY63) mutant strains were kindly provided by Dr Naima Belgareh_Touzé and Drs Emmanuelle Issakidis-Bourguet and Eric Muller, respectively. The authors would like to thank Dr Naïma Belgareh-Touzé for helpful discussions and technical advices.
Funding
This work was supported in part by Agence Nationale de la Recherche Grant 12-BSV5-0019 REDPRO2 and LABEX DYNAMO ANR-11-LABX-0011 (to CHM and SDL), and Ministerio de Economía y Competitividad Grant BFU2012-35913 and Junta de Andalucía Grant CVI-7336 (to JLC). MEPP was supported by an IEF EU Marie Curie Fellowship (PIEF-GA-2011-298652-REDOXDYNAMICS).
References
- He C, Klionsky DJ. Regulation mechanisms and signaling pathways of autophagy. Annu Rev Genet 2009; 43:67-93; PMID:19653858; http://dx.doi.org/10.1146/annurev-genet-102808-114910
- Mizushima N, Yoshimori T, Ohsumi Y. The role of Atg proteins in autophagosome formation. Annu Rev Cell Dev Biol 2011; 27:107-32; PMID:21801009; http://dx.doi.org/10.1146/annurev-cellbio-092910-154005
- Schreiber A, Peter M. Substrate recognition in selective autophagy and the ubiquitin-proteasome system. Biochim Biophys Acta 2014; 1843:163-81; PMID:23545414; http://dx.doi.org/10.1016/j.bbamcr.2013.03.019
- Weidberg H, Shvets E, Elazar Z. Biogenesis and cargo selectivity of autophagosomes. Annu Rev Biochem 2011; 80:125-56; PMID:21548784; http://dx.doi.org/10.1146/annurev-biochem-052709-094552
- Kirisako T, Ichimura Y, Okada H, Kabeya Y, Mizushima N, Yoshimori T, Ohsumi M, Takao T, Noda T, Ohsumi Y. The reversible modification regulates the membrane-binding state of Apg8/Aut7 essential for autophagy and the cytoplasm to vacuole targeting pathway. J Cell Biol 2000; 151:263-76; PMID:11038174; http://dx.doi.org/10.1083/jcb.151.2.263
- Ichimura Y, Kirisako T, Takao T, Satomi Y, Shimonishi Y, Ishihara N, Mizushima N, Tanida I, Kominami E, Ohsumi M, et al. A ubiquitin-like system mediates protein lipidation. Nature 2000; 408:488-92; PMID:11100732; http://dx.doi.org/10.1038/35044114
- Hanada T, Noda NN, Satomi Y, Ichimura Y, Fujioka Y, Takao T, Inagaki F, Ohsumi Y. The Atg12-Atg5 conjugate has a novel E3-like activity for protein lipidation in autophagy. J Biol Chem 2007; 282:37298-302; PMID:17986448; http://dx.doi.org/10.1074/jbc.C700195200
- Kim J, Huang WP, Klionsky DJ. Membrane recruitment of Aut7p in the autophagy and cytoplasm to vacuole targeting pathways requires Aut1p, Aut2p, and the autophagy conjugation complex. J Cell Biol 2001; 152:51-64; PMID:11149920; http://dx.doi.org/10.1083/jcb.152.1.51
- Sakoh-Nakatogawa M, Matoba K, Asai E, Kirisako H, Ishii J, Noda NN, Inagaki F, Nakatogawa H, Ohsumi Y. Atg12-Atg5 conjugate enhances E2 activity of Atg3 by rearranging its catalytic site. Nat Struct Mol Biol 2013; 20:433-9; PMID:23503366; http://dx.doi.org/10.1038/nsmb.2527
- Nair U, Yen WL, Mari M, Cao Y, Xie Z, Baba M, Reggiori F, Klionsky DJ. A role for Atg8-PE deconjugation in autophagosome biogenesis. Autophagy 2012; 8:780-93; PMID:22622160; http://dx.doi.org/10.4161/auto.19385
- Nakatogawa H, Ishii J, Asai E, Ohsumi Y. Atg4 recycles inappropriately lipidated Atg8 to promote autophagosome biogenesis. Autophagy 2012; 8:177-86; PMID:22240591; http://dx.doi.org/10.4161/auto.8.2.18373
- Yu ZQ, Ni T, Hong B, Wang HY, Jiang FJ, Zou S, Chen Y, Zheng XL, Klionsky DJ, Liang Y, et al. Dual roles of Atg8-PE deconjugation by Atg4 in autophagy. Autophagy 2012; 8:883-92; PMID:22652539; http://dx.doi.org/10.4161/auto.19652
- Filomeni G, Desideri E, Cardaci S, Rotilio G, Ciriolo MR. Under the ROS…thiol network is the principal suspect for autophagy commitment. Autophagy 2010; 6:999-1005; PMID:20639698; http://dx.doi.org/10.4161/auto.6.7.12754
- Huang J, Lam GY, Brumell JH. Autophagy signaling through reactive oxygen species. Antioxid Redox Signal 2011; 14:2215-31; PMID:20874258; http://dx.doi.org/10.1089/ars.2010.3554
- Pérez-Pérez ME, Lemaire SD, Crespo JL. Reactive oxygen species and autophagy in plants and algae. Plant Physiol 2012; 160:156-64; PMID:22744983; http://dx.doi.org/10.1104/pp.112.199992
- Scherz-Shouval R, Elazar Z. Regulation of autophagy by ROS: physiology and pathology. Trends Biochem Sci 2011; 36:30-8; PMID:20728362; http://dx.doi.org/10.1016/j.tibs.2010.07.007
- Navarro-Yepes J, Burns M, Anandhan A, Khalimonchuk O, del Razo LM, Quintanilla-Vega B, Pappa A, Panayiotidis MI, Franco R. Oxidative stress, redox signaling, and autophagy: cell death versus survival. Antioxid Redox Signal 2014; 21:66-85; PMID:24483238; http://dx.doi.org/10.1089/ars.2014.5837
- Scherz-Shouval R, Shvets E, Fass E, Shorer H, Gil L, Elazar Z. Reactive oxygen species are essential for autophagy and specifically regulate the activity of Atg4. EMBO J 2007; 26:1749-60; PMID:17347651; http://dx.doi.org/10.1038/sj.emboj.7601623
- Pérez-Pérez ME, Florencio FJ, Crespo JL. Inhibition of target of rapamycin signaling and stress activate autophagy in Chlamydomonas reinhardtii. Plant Physiol 2010; 152:1874-88; PMID:20107021; http://dx.doi.org/10.1104/pp.109.152520
- Xiong Y, Contento AL, Nguyen PQ, Bassham DC. Degradation of oxidized proteins by autophagy during oxidative stress in Arabidopsis. Plant Physiol 2007; 143:291-9; PMID:17098847; http://dx.doi.org/10.1104/pp.106.092106
- Pérez-Pérez ME, Couso I, Crespo JL. Carotenoid deficiency triggers autophagy in the model green alga Chlamydomonas reinhardtii. Autophagy 2012; 8:376-88; PMID:22302003; http://dx.doi.org/10.4161/auto.18864
- Woo J, Park E, Dinesh-Kumar SP. Differential processing of Arabidopsis ubiquitin-like Atg8 autophagy proteins by Atg4 cysteine proteases. Proc Natl Acad Sci U S A 2014; 111:863-8; PMID:24379391; http://dx.doi.org/10.1073/pnas.1318207111
- Auchère F, Santos R, Planamente S, Lesuisse E, Camadro JM. Glutathione-dependent redox status of frataxin-deficient cells in a yeast model of Friedreich's ataxia. Hum Mol Genet 2008; 17:2790-802; PMID:18562474; http://dx.doi.org/10.1093/hmg/ddn178
- Noda T, Ohsumi Y. Tor, a phosphatidylinositol kinase homologue, controls autophagy in yeast. J Biol Chem 1998; 273:3963-6; PMID:9461583; http://dx.doi.org/10.1074/jbc.273.7.3963
- Mehdi K, Penninckx MJ. An important role for glutathione and gamma-glutamyltranspeptidase in the supply of growth requirements during nitrogen starvation of the yeast Saccharomyces cerevisiae. Microbiology 1997; 143:1885-9; PMID:9202464; http://dx.doi.org/10.1099/00221287-143-6-1885
- Simpson RJ. Stabilization of proteins for storage. Cold Spring Harb Protoc 2010; 2010:top79; PMID:20439424; http://dx.doi.org/10.1101/pdb.top79
- Muller EG. Thioredoxin deficiency in yeast prolongs S phase and shortens the G1 interval of the cell cycle. J Biol Chem 1991; 266:9194-202; PMID:2026619
- Toledano MB, Delaunay-Moisan A, Outten CE, Igbaria A. Functions and cellular compartmentation of the thioredoxin and glutathione pathways in yeast. Antioxid Redox Signal 2013; 18:1699-711; PMID:23198979; http://dx.doi.org/10.1089/ars.2012.5033
- Shintani T, Huang WP, Stromhaug PE, Klionsky DJ. Mechanism of cargo selection in the cytoplasm to vacuole targeting pathway. Dev Cell 2002; 3:825-37; PMID:12479808; http://dx.doi.org/10.1016/S1534-5807(02)00373-8
- Lillig CH, Holmgren A. Thioredoxin and related molecules–from biology to health and disease. Antioxid Redox Signal 2007; 9:25-47; PMID:17115886; http://dx.doi.org/10.1089/ars.2007.9.25
- Garrido EO, Grant CM. Role of thioredoxins in the response of Saccharomyces cerevisiae to oxidative stress induced by hydroperoxides. Mol Microbiol 2002; 43:993-1003; PMID:11929546; http://dx.doi.org/10.1046/j.1365-2958.2002.02795.x
- Dengjel J, Høyer-Hansen M, Nielsen MO, Eisenberg T, Harder LM, Schandorff S, Farkas T, Kirkegaard T, Becker AC, Schroeder S, et al. Identification of autophagosome-associated proteins and regulators by quantitative proteomic analysis and genetic screens. Mol Cell Proteomics 2012; 11:014035; PMID:22311637; http://dx.doi.org/10.1074/mcp.M111.014035
- Lemaire SD, Quesada A, Merchan F, Corral JM, Igeno MI, Keryer E, Issakidis-Bourguet E, Hirasawa M, Knaff DB, Miginiac-Maslow M. NADP-malate dehydrogenase from unicellular green alga Chlamydomonas reinhardtii. A first step toward redox regulation? Plant Physiol 2005; 137:514-21; PMID:15579663; http://dx.doi.org/10.1104/pp.104.052670
- Michelet L, Zaffagnini M, Morisse S, Sparla F, Pérez-Pérez ME, Francia F, Danon A, Marchand CH, Fermani S, Trost P, et al. Redox regulation of the Calvin-Benson cycle: something old, something new. Front Plant Sci 2013; 4:470; PMID:24324475; http://dx.doi.org/10.3389/fpls.2013.00470
- Satoo K, Noda NN, Kumeta H, Fujioka Y, Mizushima N, Ohsumi Y, Inagaki F. The structure of Atg4B-LC3 complex reveals the mechanism of LC3 processing and delipidation during autophagy. EMBO J 2009; 28:1341-50; PMID:19322194; http://dx.doi.org/10.1038/emboj.2009.80
- Sugawara K, Suzuki NN, Fujioka Y, Mizushima N, Ohsumi Y, Inagaki F. Structural basis for the specificity and catalysis of human Atg4B responsible for mammalian autophagy. J Biol Chem 2005; 280:40058-65; PMID:16183633; http://dx.doi.org/10.1074/jbc.M509158200
- Fermani S, Sparla F, Falini G, Martelli PL, Casadio R, Pupillo P, Ripamonti A, Trost P. Molecular mechanism of thioredoxin regulation in photosynthetic A2B2-glyceraldehyde-3-phosphate dehydrogenase. Proc Natl Acad Sci U S A 2007; 104:11109-14; PMID:17573533; http://dx.doi.org/10.1073/pnas.0611636104
- Johansson K, Ramaswamy S, Saarinen M, Lemaire-Chamley M, Issakidis-Bourguet E, Miginiac-Maslow M, Eklund H. Structural basis for light activation of a chloroplast enzyme: the structure of sorghum NADP-malate dehydrogenase in its oxidized form. Biochemistry 1999; 38:4319-26; PMID:10194350; http://dx.doi.org/10.1021/bi982876c
- Trost P, Fermani S, Marri L, Zaffagnini M, Falini G, Scagliarini S, Pupillo P, Sparla F. Thioredoxin-dependent regulation of photosynthetic glyceraldehyde-3-phosphate dehydrogenase: autonomous vs. CP12-dependent mechanisms. Photosynth Res 2006; 89:263-75; PMID:17031544; http://dx.doi.org/10.1007/s11120-006-9099-z
- Suzuki K, Kirisako T, Kamada Y, Mizushima N, Noda T, Ohsumi Y. The pre-autophagosomal structure organized by concerted functions of APG genes is essential for autophagosome formation. EMBO J 2001; 20:5971-81; PMID:11689437; http://dx.doi.org/10.1093/emboj/20.21.5971
- Tietze F. Enzymic method for quantitative determination of nanogram amounts of total and oxidized glutathione: applications to mammalian blood and other tissues. Anal Biochem 1969; 27:502-22; PMID:4388022; http://dx.doi.org/10.1016/0003-2697(69)90064-5
- Zaffagnini M, Michelet L, Massot V, Trost P, Lemaire SD. Biochemical characterization of glutaredoxins from Chlamydomonas reinhardtii reveals the unique properties of a chloroplastic CGFS-type glutaredoxin. J Biol Chem 2008; 283:8868-76; PMID:18216016; http://dx.doi.org/10.1074/jbc.M709567200
