Abstract
Maintaining high gene expression level during long-term culture is critical when producing therapeutic recombinant proteins using mammalian cells. Transcriptional silencing of promoters, most likely due to epigenetic events such as DNA methylation and histone modifications, is one of the major mechanisms causing production instability. Previous studies demonstrated that the core CpG island element (IE) from the hamster adenine phosphoribosyltransferase gene is effective to prevent DNA methylation. We generated one set of modified human cytomegalovirus (hCMV) promoters by insertion of one or two copies of IE in either forward or reverse orientations into different locations of the hCMV promoter. The modified hCMV with one copy of IE inserted between the hCMV enhancer and core promoter in reverse orientation (MR1) was most effective at enhancing expression stability in CHO cells without comprising expression level when compared with the wild type hCMV. We also found that insertion of IE into a chimeric murine CMV (mCMV) enhancer and human elongation factor-1α core (hEF) promoter in reverse orientation did not enhance expression stability, indicating that the effect of IE on expression stability is possibly promoter specific.
Introduction
Chinese hamster ovary (CHO) cells are the predominant mammalian hosts for producing recombinant therapeutics due to their capacity to perform proper folding, assembly, and correct post-translational modifications.Citation1 One major issue faced when using CHO cells for therapeutic protein production is production instability, where a production cell line loses productivity during long-term culture.Citation2 A typical industrial process scale up takes about 2–3 months. A substantial loss of productivity during the process affects both product yield and quality and compromises regulatory approval of the product.Citation2 Extensive studies have indicated that production instability is caused by two major mechanisms: (1) loss of transgene copies, and (2) transcriptional silencing of promoters.Citation3-Citation8 The molecular mechanism for the loss in transgene copies is not clear. Transcriptional silencing of promoters is linked to epigenetic events such as DNA methylation and histone modifications.Citation6-Citation9 DNA methylation is a biochemical process where a methyl group is added to the cytosine of CpG dinucleotide in mammalian cells. Removal of CpGs in the promoter is one approach to prevent promoter silencing caused by DNA methylation.Citation10 Another commonly used approach to overcome transcriptional silencing is to use epigenetic regulatory DNA elements such as insulators, locus control region, matrix attachment region (MAR), stabilizing anti-repressor element (STAR), and ubiquitous chromatin region opening element (UCOE).Citation11-Citation19
The core CpG island element (IE) isolated from the hamster adenine phosphoribosyltransferase (APRT) gene is an alternative DNA element that is shown to be effective at preventing DNA methylation.Citation20 The smaller size of IE (~120 base pairs) makes it easier to use than other DNA elements which have several thousand base pairs. A previous study demonstrated that insertion of IE into the retroviral long-terminal repeat (LTR) protected the viral vector from silencing in stably transfected NIL-2, HEK293, and QT6 cells.Citation21 The function of DNA elements could be dependent on the vector context and cells.Citation22 Plasmid vectors are preferred for therapeutic protein production in mammalian cells due to regulatory requirements. It is unclear whether IE can prevent silencing of recombinant genes expressed from plasmid vectors in CHO cells. In a recent work, we studied the effect of inserting IE into the human cytomegalovirus (hCMV) promoter in plasmid vectors on the expression level and stability in CHO cells during long-term culture.Citation23 In this work, we further evaluated whether the effect of IE on expression level and stability is promoter specific by extending its use to a chimeric viral-mammalian promoter. Chimeric promoters have the potential to provide better expression levels and stability than naturally occurring promoters.Citation24,Citation25
Inserting IE into the hCMV Promoter Enhances Expression Stability
In our recent work, 12 modified hCMV promoters were generated by inserting either one or two copies of IE in both forward and reverse orientations upstream of the hCMV enhancer, between the hCMV enhancer and core promoter (CP), or downstream of CP (). They were compared with the wild type (WT) hCMV for expression level and stability in stably transfected clones using enhanced green fluorescence protein (EGFP) as a reporter protein. Each clone was passaged for 8 wk in the absence of selection reagents as part of stability testing. Cell lines that maintain stable production without selection reagents are preferred for mass production of therapeutic proteins as selection reagents are toxic and expensive.Citation2 The percentage of EGFP expressing cells and EGFP geometric mean fluorescence intensity of each clone were quantified before and after stability testing using FACS Calibur.
Figure 1. Schematic representation of vectors for evaluation of promoters with IE inserted at different locations. IE, core CpG island element; Enhancer, hCMV or mCMV enhancer; CP, hCMV or human elongation factor-1α (hEF) core promoter; IRESatt, mutated encephalomycocarditis virus (EMCV) internal ribosomal entry site (IRES) with attenuated translation efficiency; SpA, simian virus 40 early polyadenylation signal; EGFP, enhanced green fluorescence protein cDNA. mNPT, mutated neomycin phosphotransferase cDNA with amino acid D at 261 changed to G.
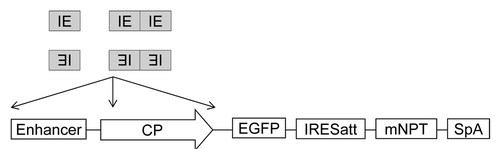
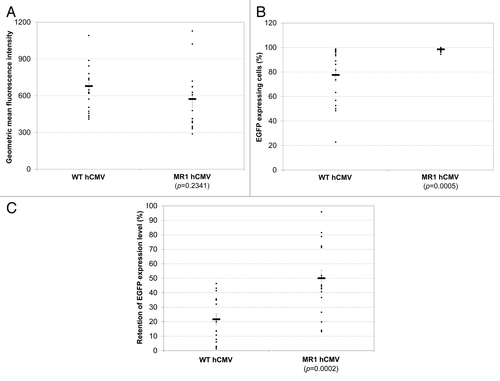
We observed that the effect of IE on expression level and stability involved complex interactions between the location, orientation, and copy numbers of IE within the hCMV promoter. Inserting IE downstream of the hCMV CP decreased EGFP expression regardless of IE copy numbers and orientations, while the effect on expression level at other locations was varied. Only six of the modified hCMV promoters (two copies of IE inserted upstream of hCMV enhancer in reverse orientation, one copy of IE inserted between the hCMV enhancer and CP in forward orientation, one copy of IE inserted between the hCMV enhancer and CP in reverse orientation, two copies of IE inserted between the hCMV enhancer and CP in reverse orientation, and two copies of IE inserted downstream of the hCMV CP in reverse orientation) enhanced expression stability. Among all the modifications, inserting IE between the hCMV enhancer and CP in reverse orientation, which was designated as MR1 hCMV, was most effective at enhancing expression stability without compromising expression levels (). All cells were verified to be EGFP expressing before stability testing. After 8 wk of culture, the percentage of EGFP expressing cells in all clones generated using the WT hCMV declined (). The average percentage of EGFP expressing cells of the 18 clones was 78% with the worst clone only having 23% of the population still expressing EGFP. In contrast, all clones generated using the MR1 hCMV had close to 100% EGFP expressing cells at the end of stability testing. Maintaining EGFP expression in cells did not guarantee that EGFP expression level would not decline (). EGFP expression level in all 18 clones generated using the WT hCMV dramatically declined after 8 wk of culture. On average, the 18 clones retained 22% of their original expression with the best clone retaining 46% of EGFP expression. While EGFP expression levels in all MR1 hCMV clones also declined after 8 wk of culture, one third of the clones retained over 70% of their start EGFP expression and could be considered stable.Citation26 In contrast to the WT hCMV, MR1 hCMV increased the average retention of EGFP expression level more than 2-fold, reaching 50%. MR1 hCMV also improved antibody expression stability of methotrexate (MTX) amplified CHO cell lines. Stably transfected pools generated using MR1 hCMV maintained over 60% of their original monoclonal antibody titer after 8 wk of culture in the absence of MTX, while the WT hCMV generated pools only retained 37% (results not shown).
Figure 2. Comparison of the WT hCMV and MR1 hCMV for EGFP expression level and stability. (A) EGFP expression level in stably transfected clones before stability testing. (B) Percentage of EGFP expressing cells in different clones at the end of stability testing. (C) Retention of EGFP expression level in different clones at the end of stability testing. Six clones each were isolated from three separately transfected pools for a total of 18 clones for each promoter. The percentage of EGFP expressing cells and EGFP geometric mean fluorescence intensity were quantified before and after stability testing for each clone using FACS Calibur. The retention of EGFP expression level for each clone was calculated as the EGFP geometric mean fluorescence intensity measured at the end of stability testing divided by that before stability testing for the same clone. Each dot represents values measured for one clone. The horizontal bar and error bars represent the average and standard error of values of 18 clones. P value for statistical difference between the WT hCMV and MR1 CMV was calculated using two-tailed Student’s t test.
Effect of IE on Expression Stability is Promoter Specific
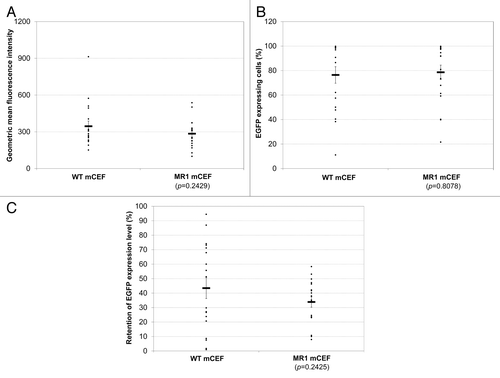
We next tested if IE could improve the expression stability of a chimeric promoter by inserting one copy of IE between the murine CMV (mCMV) enhancer and the core promoter from the human elongation factor-1α gene (hEF) in reverse orientation to generate MR1 mCEF. WT (no IE inserted) and MR1 mCEF were compared for expression level and stability in stably transfected clones using EGFP as a reporter protein as described above for the hCMV promoters. Consistent with the previous results (), the 18 MR1 mCEF clones had comparable EGFP geometric mean fluorescence intensity with those generated using the WT mCEF before stability testing (). However, MR1 mCEF did not enhance expression stability as determined by both the percentage of EGFP expression cells and retention of EGFP expression levels (). All clones generated using the WT and MR1 mCEF had 100% of EGFP expressing cells before stability testing. After 8 wk of culture, the majority of clones generated using both promoters had less than 100% of EGFP expressing cells. The average percentage of EGFP expressing cells in the 18 clones from each promoter was similar, dropping to 76% for the WT mCEF and 79% for the MR1 mCEF. Similarly, the difference in average retention of EGFP expression level was not statistically significant between clones generated using the WT and MR1 mCEF. However, five WT mCEF clones retained over 70% of their starting EGFP expression level while none of 18 MR1 mCEF clones retained expression over 60%.
Figure 3. Comparison of the WT mCEF and MR1 mCEF for EGFP expression level and stability. (A) EGFP expression in stably transfected clones before stability testing. (B) Percentage of EGFP expressing cells in different clones at the end of stability testing. (C) Retention of EGFP expression level in different clones at the end of stability testing. Eighteen clones, six from each pool, were isolated from three pools generated by each promoter. Each dot represents values measured for one clone. The percentage of EGFP expressing cells and EGFP geometric mean fluorescence intensity were quantified before and after stability testing for each clone using FACS Calibur. The retention of EGFP expression level for each clone was calculated as the EGFP geometric mean fluorescence intensity measured at the end of stability testing divided by that before stability testing for the same clone. Each dot represents values measured for one clone. The horizontal bar and error bars represent the average and standard error of values of 18 clones. P value for statistical difference between the WT mCEF and MR1 mCEF was calculated using two-tailed Student’s t test.
Discussion
Using MR1 hCMV ensured all cells still expressed EGFP although the expression level still declined over 30% in two thirds of the clones. We had recently observed that the decreased expression in these clones was not due to loss in EGFP gene copies.Citation23 DNA methylation was observed in both stable and unstable clones generated using the MR1 hCMV promoter, suggesting that IE enhanced expression stability by mechanisms other than preventing DNA methylation. Histone modifications are also associated with transcription silencing. A recent study indicated that different epigenetic regulatory elements associated with specific histone modifications prevented gene silencing.Citation27 For example, MARs were associated to histone marks usually linked to actively expressed genes, while an UCOE was found to act by preventing deposition of repressive chromatin marks. Analysis and comparison of histone modifications between clones generated using the WT and MR1 hCMV and between stable and unstable MR1 hCMV clones may help us understand how IE enhances expression stability and why transcriptional silencing still happened in some clones. This information will also define whether IE is a new epigenetic regulator or acts by mechanisms similar to existing epigenetic regulatory elements. As the length of IE is short, it would also be of practical interest to test whether combinations of IE with other epigenetic regulatory elements on the same plasmid vector could further enhance expression stability.
Inserting one copy of IE between the LTR and hCMV enhancer and CP in reverse orientation in a previous study and our recent work both enhanced expression stability.Citation21, Citation23 As both of the promoters originate from viral sources, we were interested to study if IE worked with other types of promoters. We extended the application of IE to a chimeric viral-mammalian promoter consisting of the mCMV enhancer and hEF CP, mCEF. A promoter of the CHO EF-1α gene was reported to provide more stable expression than the hCMV promoter.Citation28,Citation29 Similarly, we observed that expression from the chimeric WT mCEF using a hEF CP was more stable than the WT hCMV ( and ). As MR1 mCEF did not further enhance expression stability compared with the WT mCEF (), it is possible that IE works better with viral CP than mammalian CP and more enhancer- CP combinations should be evaluated. We also observed that the effect of IE was different when inserted into various locations on hCMV and in different orientations.Citation23 Use of IE did not enhance expression stability for mCEF possibly due to non-optimal location and insufficient IE copies. Besides enhancer and/or promoter source and the IE location and copy number, it is would also be interesting to look at the effect of enhancer and CP lengths on IE’s function. Previous studies observed that IE prevented methylation on DNA sequences within a limited range.Citation20,Citation30 hCMV and mCMV enhancers used in this work have 376 and 422 base pairs, respectively. hCMV and hEF CP have 203 and 250 base pairs, respectively. hCMV is 100 base pairs shorter than mCEF in total and could be another reason for the different effects IE had on expression stability.
A wider range of promoters and augmenting elements would be useful not only for recombinant protein production in CHO cells, but also for areas like synthetic biology and cell engineering. The IE element is relatively short compared with other common elements like MAR and UCOE and could be more easily applied during vector designs to further expand any existing promoter libraries.
Disclosure of Potential Conflicts of Interest
No potential conflict of interest was disclosed.
Acknowledgments
This work was supported by the Biomedical Research Council and the Science and Engineering Research Council of A*STAR (Agency for Science, Technology. and Research), Singapore.
References
- Wurm FM. Production of recombinant protein therapeutics in cultivated mammalian cells. Nat Biotechnol 2004; 22:1393 - 8; http://dx.doi.org/10.1038/nbt1026; PMID: 15529164
- Barnes LM, Bentley CM, Dickson AJ. Stability of protein production from recombinant mammalian cells. Biotechnol Bioeng 2003; 81:631 - 9; http://dx.doi.org/10.1002/bit.10517; PMID: 12529877
- Chusainow J, Yang YS, Yeo JH, Toh PC, Asvadi P, Wong NS, Yap MG. A study of monoclonal antibody-producing CHO cell lines: what makes a stable high producer?. Biotechnol Bioeng 2009; 102:1182 - 96; http://dx.doi.org/10.1002/bit.22158; PMID: 18979540
- He L, Winterrowd C, Kadura I, Frye C. Transgene copy number distribution profiles in recombinant CHO cell lines revealed by single cell analyses. Biotechnol Bioeng 2012; 109:1713 - 22; http://dx.doi.org/10.1002/bit.24428; PMID: 22234778
- Jun SC, Kim MS, Hong HJ, Lee GM. Limitations to the development of humanized antibody producing Chinese hamster ovary cells using glutamine synthetase-mediated gene amplification. Biotechnol Prog 2006; 22:770 - 80; http://dx.doi.org/10.1021/bp060004t; PMID: 16739961
- Kim M, O’Callaghan PM, Droms KA, James DC. A mechanistic understanding of production instability in CHO cell lines expressing recombinant monoclonal antibodies. Biotechnol Bioeng 2011; 108:2434 - 46; http://dx.doi.org/10.1002/bit.23189; PMID: 21538334
- Osterlehner A, Simmeth S, Göpfert U. Promoter methylation and transgene copy numbers predict unstable protein production in recombinant Chinese hamster ovary cell lines. Biotechnol Bioeng 2011; 108:2670 - 81; http://dx.doi.org/10.1002/bit.23216; PMID: 21618470
- Yang Y, Mariati, Chusainow J, Yap MG. DNA methylation contributes to loss in productivity of monoclonal antibody-producing CHO cell lines. J Biotechnol 2010; 147:180 - 5; http://dx.doi.org/10.1016/j.jbiotec.2010.04.004; PMID: 20430058
- Paredes V, Park JS, Jeong Y, Yoon J, Baek K. Unstable expression of recombinant antibody during long-term culture of CHO cells is accompanied by histone H3 hypoacetylation. Biotechnol Lett 2013; 35:987 - 93; http://dx.doi.org/10.1007/s10529-013-1168-8; PMID: 23468139
- Swindle CS, Kim HG, Klug CA. Mutation of CpGs in the murine stem cell virus retroviral vector long terminal repeat represses silencing in embryonic stem cells. J Biol Chem 2004; 279:34 - 41; http://dx.doi.org/10.1074/jbc.M309128200; PMID: 14559924
- Lindahl Allen M, Antoniou M. Correlation of DNA methylation with histone modifications across the HNRPA2B1-CBX3 ubiquitously-acting chromatin open element (UCOE). Epigenetics 2007; 2:227 - 36; http://dx.doi.org/10.4161/epi.2.4.5231; PMID: 18032920
- Araki Y, Hamafuji T, Noguchi C, Shimizu N. Efficient recombinant production in mammalian cells using a novel IR/MAR gene amplification method. PLoS One 2012; 7:e41787; http://dx.doi.org/10.1371/journal.pone.0041787; PMID: 22844523
- Dang Q, Auten J, Plavec I. Human beta interferon scaffold attachment region inhibits de novo methylation and confers long-term, copy number-dependent expression to a retroviral vector. J Virol 2000; 74:2671 - 8; http://dx.doi.org/10.1128/JVI.74.6.2671-2678.2000; PMID: 10684282
- Festenstein R, Tolaini M, Corbella P, Mamalaki C, Parrington J, Fox M, Miliou A, Jones M, Kioussis D. Locus control region function and heterochromatin-induced position effect variegation. Science 1996; 271:1123 - 5; http://dx.doi.org/10.1126/science.271.5252.1123; PMID: 8599090
- Girod PA, Nguyen DQ, Calabrese D, Puttini S, Grandjean M, Martinet D, Regamey A, Saugy D, Beckmann JS, Bucher P, et al. Genome-wide prediction of matrix attachment regions that increase gene expression in mammalian cells. Nat Methods 2007; 4:747 - 53; http://dx.doi.org/10.1038/nmeth1076; PMID: 17676049
- Girod PA, Zahn-Zabal M, Mermod N. Use of the chicken lysozyme 5′ matrix attachment region to generate high producer CHO cell lines. Biotechnol Bioeng 2005; 91:1 - 11; http://dx.doi.org/10.1002/bit.20563; PMID: 15889435
- Kwaks TH, Barnett P, Hemrika W, Siersma T, Sewalt RG, Satijn DP, Brons JF, van Blokland R, Kwakman P, Kruckeberg AL, et al. Identification of anti-repressor elements that confer high and stable protein production in mammalian cells. Nat Biotechnol 2003; 21:553 - 8; http://dx.doi.org/10.1038/nbt814; PMID: 12679786
- Pikaart MJ, Recillas-Targa F, Felsenfeld G. Loss of transcriptional activity of a transgene is accompanied by DNA methylation and histone deacetylation and is prevented by insulators. Genes Dev 1998; 12:2852 - 62; http://dx.doi.org/10.1101/gad.12.18.2852; PMID: 9744862
- Zhang F, Frost AR, Blundell MP, Bales O, Antoniou MN, Thrasher AJ. A ubiquitous chromatin opening element (UCOE) confers resistance to DNA methylation-mediated silencing of lentiviral vectors. Mol Ther 2010; 18:1640 - 9; http://dx.doi.org/10.1038/mt.2010.132; PMID: 20588258
- Siegfried Z, Eden S, Mendelsohn M, Feng X, Tsuberi BZ, Cedar H. DNA methylation represses transcription in vivo. Nat Genet 1999; 22:203 - 6; http://dx.doi.org/10.1038/9727; PMID: 10369268
- Senigl F, Plachý J, Hejnar J. The core element of a CpG island protects avian sarcoma and leukosis virus-derived vectors from transcriptional silencing. J Virol 2008; 82:7818 - 27; http://dx.doi.org/10.1128/JVI.00419-08; PMID: 18550662
- Mariati, Ho SC, Yap MG, Yang Y. Evaluating post-transcriptional regulatory elements for enhancing transient gene expression levels in CHO K1 and HEK293 cells. Protein Expr Purif 2010; 69:9 - 15; http://dx.doi.org/10.1016/j.pep.2009.08.010; PMID: 19899222
- Mariati, Yeo JH, Koh EY, Ho SC, Yang Y. Insertion of core CpG island element into human CMV promoter for enhancing recombinant protein expression stability in CHO cells. Biotechnol Prog 2014; 30:523 - 34; http://dx.doi.org/10.1002/btpr.1919; PMID: 24789630
- Blazeck J, Alper HS. Promoter engineering: recent advances in controlling transcription at the most fundamental level. Biotechnol J 2013; 8:46 - 58; http://dx.doi.org/10.1002/biot.201200120; PMID: 22890821
- Ho SCL, Yang Y. Identifying and engineering promoters for high level and sustainable therapeutic recombinant protein production in cultured mammalian cells. Biotechnol Lett 2014; 36:1569 - 79; http://dx.doi.org/10.1007/s10529-014-1523-4; PMID: 24737078
- Bailey LA, Hatton D, Field R, Dickson AJ. Determination of Chinese hamster ovary cell line stability and recombinant antibody expression during long-term culture. Biotechnol Bioeng 2012; 109:2093 - 103; http://dx.doi.org/10.1002/bit.24485; PMID: 22896849
- Majocchi S, Aritonovska E, Mermod N. Epigenetic regulatory elements associate with specific histone modifications to prevent silencing of telomeric genes. Nucleic Acids Res 2014; 42:193 - 204; http://dx.doi.org/10.1093/nar/gkt880; PMID: 24071586
- Chan KKK, Wu SM, Nissom PM, Oh SKW, Choo ABH. Generation of high-level stable transgene expressing human embryonic stem cell lines using Chinese hamster elongation factor-1 alpha promoter system. Stem Cells Dev 2008; 17:825 - 36; http://dx.doi.org/10.1089/scd.2007.0233; PMID: 18788934
- Running Deer J, Allison DS. High-level expression of proteins in mammalian cells using transcription regulatory sequences from the Chinese hamster EF-1alpha gene. Biotechnol Prog 2004; 20:880 - 9; http://dx.doi.org/10.1021/bp034383r; PMID: 15176895
- Hejnar J, Hájková P, Plachy J, Elleder D, Stepanets V, Svoboda J. CpG island protects Rous sarcoma virus-derived vectors integrated into nonpermissive cells from DNA methylation and transcriptional suppression. Proc Natl Acad Sci U S A 2001; 98:565 - 9; http://dx.doi.org/10.1073/pnas.98.2.565; PMID: 11209056
