Abstract
Poly(lactic-co-glycolic acid) (PLGA) is a popular material used to prepare nanoparticles for drug delivery. However, PLGA nanoparticles lack desirable attributes including active targeting abilities, resistance to aggregation during lyophilization, and the ability to respond to dynamic environmental stimuli. To overcome these issues, we fabricated a nanoparticle consisting of a PLGA core encapsulated within a shell of poly(N-isopropylacrylamide). Dynamic light scattering and transmission electron microscope imaging were used to characterize the nanoparticles, while an MTT assay and ELISA suggested biocompatibility in THP1 cells. Finally, a collagen type II binding assay showed successful modification of these nanoparticles with an active targeting moiety.
Introduction
Poly(lactic-co-glycolic acid) (PLGA) is a biocompatible, biodegradable synthetic polymer that is easy to fabricate into size-specific nanoparticles and has a well-documented ability for sustained therapeutic release.Citation1-Citation3 Although many investigators and commercial entities have embraced these desirable attributes to construct PLGA nanoparticles for use in research and commercial products,Citation1,Citation3-Citation5 PLGA nanoparticles still have significant limitations when it comes to using them to deliver therapeutics to a specific disease site. One such limitation is that PLGA nanoparticles, when delivered intravenously, have no active targeting capabilities and are restricted to passive targeting via the enhanced permeability and retention (EPR) effect seen in cancerous and inflamed tissues.Citation6-Citation8 Tissue accumulation via the EPR effect can provide potential benefits for cancer drug delivery,Citation6 but it renders drug delivery for other disease states that lack the EPR effect problematic. Other investigators have attempted to overcome this challenge by modifying PLGA nanoparticles for “active” targeting with the addition of monoclonal antibodies.Citation9 Another challenge is the long-term storage of aqueous suspensions of PLGA nanoparticle systems, due to hydrolytic degradation of the polymer and subsequent release of the encapsulated therapeutic.Citation10,Citation11 To overcome this issue, researchers have used lyophilization to prepare therapeutic-containing PLGA nanoparticles for long-term storage.Citation10,Citation11However, this simple solution introduces another problem: lyophilization causes the PLGA nanoparticles to aggregate into clumps that upon rehydration, readily fall out of solution.Citation10,Citation11 Finally, PLGA nanoparticles are limited in their responses to environmental stimuli like temperature or pH. Although environmental sensitivity is not a requirement for nanoparticle systems, it does provide advantages, such as the ability to control therapeutic release from nanoparticle carriers only when they are exposed to environments inherent to particular disease states.Citation12
In an effort to overcome the limitations associated with PLGA nanoparticles, as well as increase their functionality, we developed a method to encapsulate PLGA nanoparticles within a shell of poly(N-isopropylacrylamide) (pNIPAM). pNIPAM was chosen because it is a versatile material with some inherently unique properties. The pNIPAM shell can be synthesized as a homopolymer, or as a copolymer with incorporation of a variety of chemical moieties with defined concentrations that can then be further modified with active targeting functional groups.Citation13-Citation15 Additionally, pNIPAM can tolerate lyophilization;Citation16 we hypothesized that a pNIPAM shell would protect encapsulated PLGA nanoparticles from aggregation during lyophilization. Finally, pNIPAM is a temperature responsive polymer that can undergo a phase transition at a lower critical solution temperature (LCST) around 31–32°C.Citation17 At temperatures below its LCST, pNIPAM nanoparticles are swollen and hydrophilic.Citation13 However, once the temperature increases above the LCST, pNIPAM nanoparticles collapse as the polymer becomes hydrophobic.Citation13 Thus, by encapsulating PLGA nanoparticles in a pNIPAM shell, the nanoparticles should have the ability to respond to environmental stimuli, such as temperature. This response could then be used for loading and/or controlling the release of therapeutics from the pNIPAM layer.Citation18 In this paper, we characterize this novel PLGA core + pNIPAM shell nanoparticle system.
Results and Discussion
Physical characterization of the core + shell nanoparticles
Nanoparticles were formed through a two-step process that included: the formation of a PLGA core followed by the synthesis of a pNIPAM shell around these pre-formed PLGA cores (). Verification of successful encapsulation of the PLGA cores with the pNIPAM shells was initially achieved using dynamic light scattering to measure the size of the nanoparticles before and after the addition of the shell to determine the change in nanoparticle diameter. The diameter of the core + shell nanoparticles () increased post addition of the pNIPAM shell, suggesting successful encapsulation. pNIPAM shell thickness ranged from ~100 nm to ~200 nm at 25°C, where an increase in shell thickness corresponded to an increase in the mole percent of acrylic acid, and therefore charge density. This phenomenon has been shown previously in the literature.Citation19
Figure 1. Schematic of the fabrication of PLGA core + pNIPAM shell nanoparticles (A and B) and the subsequent chemistry (C) used to append the collagen type II binding peptide targeting ligand.
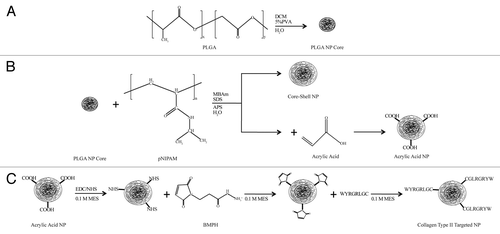
Table 1. Characterization of Various Nanoparticle Samples
Additional verification of successful encapsulation of the pNIPAM shell was provided by confirming a phase transition when the temperature was raised above its LCST. Because pNIPAM is a temperature sensitive polymer, we expected to see a difference in nanoparticle size, with the nanoparticle diameter being smaller at 37°C (above pNIPAM LCST) compared with 25°C (below pNIPAM LCST). All core + shell nanoparticles showed reduced diameters at 37°C compared with 25°C (). This trend is most pronounced with shell thicknesses that at 37°C range only from ~10 nm to ~100 nm with an increase in shell thickness again corresponding to an increase in the mole percent of acrylic acid incorporated into the pNIPAM.
To assess colloidal stability of our core + shell nanoparticles above and below the phase transition temperature, we measured their ζ-potentials. According to the literature, nanoparticle ζ-potentials above 30 mV or below -30 mV are considered stable.Citation20 As shown in , all nanoparticles, are near -30 mV indicating that they are stable.
Effects of lyophilization on core + shell nanoparticles
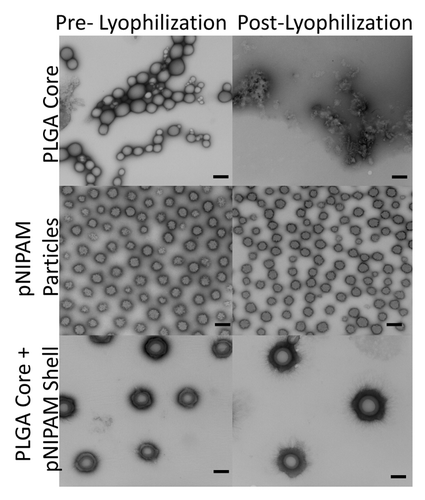
Lyophilization is an effective way to prevent the release of therapeutics loaded into PLGA nanoparticles during long-term storage.Citation10,Citation11 However, if not performed correctly, lyophilization can result in aggregation of the PLGA nanoparticles that prevents resolubilization of the clumped nanoparticles ().Citation10,Citation11 We used TEM imaging to visually confirm that well-defined spherical PLGA and PLGA core + pNIPAM shell nanoparticles were successfully fabricated (). However, as also observed by others,Citation10,Citation11 TEM imaging showed that lyophilization caused the PLGA nanoparticles to form aggregates (). This aggregation resulted in an inability to measure diameter and ζ-potential of post-lyophilized PLGA nanoparticles. In contrast, encapsulation of PLGA nanoparticles with pNIPAM shells prevented aggregation of the PLGA nanoparticles following lyophilization, further confirming that the pNIPAM shell fully encapsulated the PLGA core. Additionally, we found that lyophilization of the core + shell nanoparticles does not affect their size or ζ-potential at 25°C or 37°C ().
Responses of core + shell nanoparticles to dynamic environmental stimuli
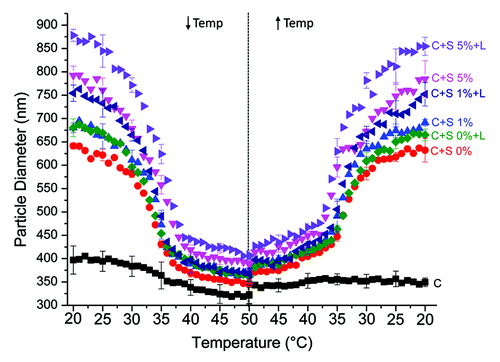
To further assess the response of our core + shell nanoparticles to temperature-based environmental stimuli, we used dynamic light scattering to measure the diameter of our core + shell nanoparticles as they were exposed to a dynamic range of temperatures from 20°C to 50°C to 20°C. We found that the core + shell nanoparticles readily respond within this temperature range with all core + shell nanoparticle types decreasing in diameter as the temperature was raised above their LCST (). This response was reversible, as the nanoparticles returned to their original diameter when the temperature was lowered back below their LCST. Additionally, the LCST of the pNIPAM shell was tuned by modifying the amount of acrylic acid that was incorporated as a co-monomer. As more acrylic acid was incorporated, the LCST of pNIPAM increased (). The core + shell nanoparticles with 0 mol% acrylic acid exhibited an LCST at ~31–32°C, while the 1 mol% acrylic acid had an LCST at ~33–34°C, and the core + shell nanoparticles with 5 mol% acrylic acid had an LCST at ~35°C (). This trend has previously been established in the literature.Citation19,Citation21 In the future, these core + shell nanoparticles could be engineered to respond to different environmentally based stimuli in addition to temperature by changing the co-monomer composition of the pNIPAM.Citation14
Core + shell nanoparticle biocompatibility
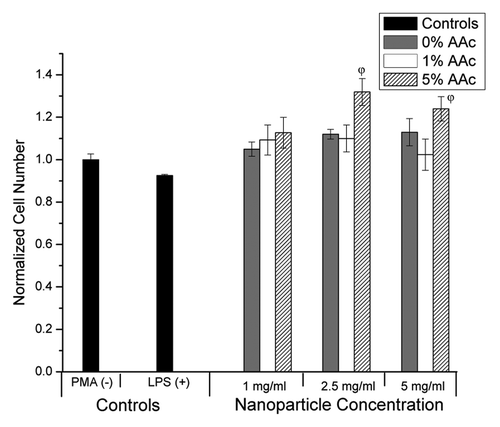
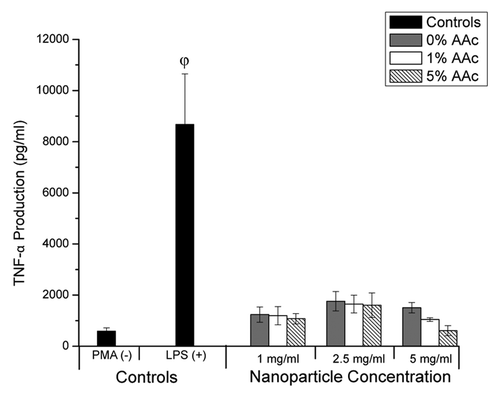
The biocompatibility of the core + shell nanoparticles was assessed by evaluating the toxicity and inflammatory response in an immortalized human monocyte cell line. This cell line was chosen because, in our experience, it shows increased sensitivity to toxicity compared with other cell lines. Additionally, monocytes have a significant role in the perpetuation of osteo- and rheumatoid arthritis,Citation22,Citation23an inflammatory disease model that will be used to evaluate the ability of these core + shell nanoparticles to deliver desired therapeutics in the future. The core + shell nanoparticles were not toxic at any of the concentrations tested, as we saw no significant reduction in THP1 cell number (). However, the core + shell nanoparticles with 5 mol% acrylic acid induced a significant increase in proliferation of the THP1 cells at concentrations of 2.5 and 5 mg/mL compared with all other treatments (p < 0.05; one-way ANOVA + Tukey post-hoc test). Next, the ability of the core + shell nanoparticles to elicit an inflammatory response was determined by measuring TNF-α production by THP1 cells using an ELISA. A significant inflammatory response, characterized by an increase in TNF-α production, is not desirable and would indicate non-biocompatibility. Similar to the PBS control, the core + shell nanoparticles did not elicit TNF-α production (; p > 0.05; one-way ANOVA), suggesting these nanoparticles are biocompatible though further testing will be conducted in the future to verify this preliminary work. The positive control, THP1 cells treated with lipopolysacharide, did induce TNF-α production as expected.
Targeting core + shell nanoparticles
In order to increase the targeting capabilities of our core + shell nanoparticles beyond the passive EPR effect,Citation6,Citation7 we modified them with an active targeting moiety—a peptide that binds collagen type II ().Citation24 This targeting moiety was chosen because our future work will utilize these nanoparticles to deliver therapeutics in an arthritic-based disease model, and collagen type II is readily found in the cartilage of the joints.Citation25 However, this chemistry could easily be applied to attach other targeting moieties that contain a free thiol functional group.
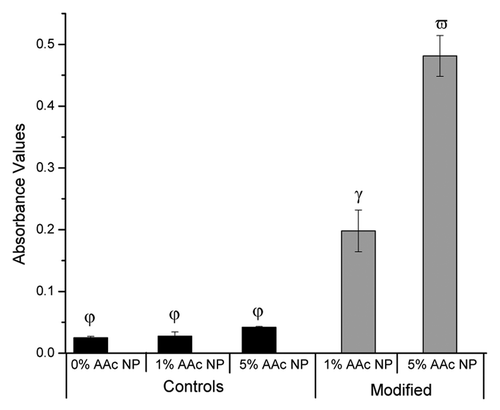
To assess whether incorporation of the collagen type II binding peptide allowed the core + shell nanoparticles to bind to collagen type II, we utilized a microplate assay. This assay involved coating a 96-well plate with collagen type II, incubating it with collagen type II targeting enabled core + shell nanoparticles, and then probing for the presence of the biotin labeled collagen type II peptide. The results from this assay indicate that core + shell nanoparticles modified with collagen type II binding peptide bound collagen type II as compared with unmodified core + shell nanoparticle controls (; p < 0.05, one-way ANOVA + Tukey post hoc test). Furthermore, the number of collagen type II binding peptide modified core + shell nanoparticles able to bind to collagen type II was directly related to the concentration of acrylic acid that was incorporated into the pNIPAM shell.
Figure 6. Collagen Type II binding assay for core + shell nanoparticles. 1% and 5% AAc modified NPs are statistically significant from each other and the controls as indicated by the different Greek letters (p < 0.05; one-way ANOVA + Tukey Post-hoc test). Data presented as mean ± standard deviation (n = 4).
Materials and Methods
Encapsulating pre-formed PLGA cores within synthesized pNIPAM shells
Core + shell nanoparticle preparation occurred in two steps: fabrication of the PLGA cores followed by the addition of the pNIPAM shell (). First, PLGA cores were prepared using a single emulsion technique.Citation26 Briefly, 5 g of poly(dl-lactide/glycolide) 50:50 (PLGA; Polysciences Inc.) were dissolved in 5 mL of dichloromethane (DCM; Sigma-Aldrich), added to 20 mL of 5% polyvinyl alcohol (PVA; Alfa Aesar; Avg. MW = 11,000–31,000), and homogenized for 30 s using a probe sonicator (Branson Sonifier 450) to generate a single emulsion. The emulsion was then added to 100 mL of rapidly stirred distilled water and left overnight to allow for full evaporation of the DCM. The PLGA nanoparticles were further purified via 4 × 30 min centrifugation washes at 10,000 g and 4°C with distilled water. Any clumps of PLGA nanoparticles that remained after centrifugation were disrupted using brief sonication. The PLGA cores were then encapsulated in pNIPAM shells using aqueous free radical precipitation polymerization under a nitrogen atmosphere.Citation27-Citation29 Briefly, 0.27 g N-isopropylacrylamide (2.385 mmol) (Polysciences Inc.), 0.021 g N-N'-methylene bisacrylamide (0.136 mmol) (Fluka), 0.012 g sodium dodecyl sulfate (0.042 mmol) (Sigma Aldrich), and 0.015 g ammonium persulfate (0.066 mmol) (Sigma Aldrich) were dissolved in 30 mL of distilled water and purged of oxygen by nitrogen bubbling. For nanoparticle targeting, either 1.67 µL (0.024 mmol or 1 mol%) or 8.35 µL (0.121 mmol or 5 mol%) of acrylic acid (AAc; Alfa Aesar) were included. Meanwhile, 20 mL of the PLGA nanoparticle cores were added to a 250 mL three-neck round bottom flask and equilibrated to 70°C for 20 min under nitrogen with stirring. Then, 10 mL of the shell solution was added to the 70°C equilibrated PLGA nanoparticle cores and allowed to polymerize. Additional 5 mL aliquots of shell solution were added 30, 50, 70 and 90 min after the initial polymerization. Polymerization continued for 6 h after the final addition of shell solution. Purification was achieved through dialysis of the PLGA core + pNIPAM shell nanoparticles against distilled water for 7 d in 15,000 MWCO dialysis tubing (Spectrum Laboratories, Inc.). Then centrifugation washes were performed to further isolate the core + shell nanoparticles from any pNIPAM homoparticles that may have formed. Any clumps of core + shell nanoparticles post centrifugation purification were dispersed using brief sonication. Finally, a portion of the samples were lyophilized and then rehydrated in distilled water.
Transmission electron microscopy (TEM) characterization
Images were taken of the pre- and post- lyophilized nanoparticle samples stained with 2% uranyl acetate. The stained nanoparticles were then placed on a glow-discharged 400 mesh coated with formvar + carbon film, and then placed in a Philips CM-100 TEM where images were captured on Kodak SO-163 electron image film.
Nanoparticle sizing and zeta potential
Measurements were taken with a Malvern Zetasizer Nano ZS90. Pre- and post-lyophilized nanoparticles were suspended in distilled water and analyzed for particle size in polystyrene cuvettes at 25°C and 37°C. Temperature sweeps were performed by varying temperature from 20°C to 50°C to 20°C in 1° increments with measurement of particle size with each change in degree. Disposable Malvern ζ-potential cuvettes were used to measure the ζ-potential of our particles at 25°C and 37°C. After making any change in temperature, nanoparticle samples were allowed to equilibrate for five minutes before any sizing or zeta measurements were made.
Peptide synthesis and purification
The collagen type II binding peptide single amino acid sequence consisting of WYRGRLGC was identified from the literature.Citation24 It was synthesized at a 0.4 mmol scale on Knorr-amide resin (Synbiosci Corp.) using standard FMOC (9-fluorenylmethyloxycarbonyl) chemistry. Two different chemistries were used to couple each amino acid (Synbiosci Corp). The first coupling reagents consisted of N-hydroxybenzotriazole (HOBt; Synbiosci) and N,N' diisopropylcarbodiimide (DIC; Sigma-Aldrich) and the second coupling reagents were O-Benzotriazole-N,N,N',N'-tetramethyl-uronium-hexafluoro-phosphate (HBTU; Synbiosci) and lutidine (Sigma-Aldrich). Following synthesis, the peptide was cleaved from the resin with 95% trifluoroacetic acid (Sigma-Aldrich), 2.5% water, 1.25% triisopropylsilane (Sigma-Aldrich), and 1.25% ethanedithiol (Sigma-Aldrich), precipitated in cold ether, and recovered by centrifugation. It was then purified with an acetonitrile gradient on an AKTA Explorer FPLC (GE Healthcare) equipped with a 22/250 C18 reversed phase column (Grace Davidson). Molecular weight was confirmed by time of flight MALDI mass spectrometry using a 4800 Plus MALDI TOF/TOF Analyzer (Applied Biosystems). Theoretical molecular weight of WYRGRLGC was calculated to be 1,009.1 while the actual molecular weight was found to be 1,009.58. A biotinylated version of the peptide (biotin-WYRGRLC) was purchased from Genscript and its theoretical molecular weight was calculated to be 1,179.42 while its actual molecular weight was found to be 1,179.8.
Modifying core + shell nanoparticle with targeting moiety
Nanoparticle targeting was achieved through the addition of a collagen type II binding peptide to the AAc groups on our core + shell nanoparticles using a heterobifunctional crosslinker (). Briefly, 0.4 mg of 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide hydrochloride (EDC; Thermo-Scientific) and 1.1 mg of N-hydroxylsulfosuccinimide (sulfo-NHS; Thermo-Scientific) were added to 1 mg of core + shell nanoparticles for 15 min in activation buffer (0.1 M 2-(N-morpholino)ethanesulfonic acid (MES; Amresco, pH 6.0). Excess EDC and sulfo-NHS was removed by a centrifuge wash. The heterobifunctional crosslinker, N-(β-maleimidopropionic acid) hydrazide (BMPH; Thermo-Scientific) was added to the activated nanoparticles (0.1 mg for 1 mol% AAc nanoparticles or 0.3 mg for 5 mol% AAc nanoparticles) for 2 h in coupling buffer (0.1 M MES, pH 7.2). Excess BMPH was removed using gel filtration chromatography through an ÄKTA Purifier FPLC (GE Healthcare) with Bio-Scale Mini Bio-Gel columns packed with polyacrylamide beads (Bio-Rad Laboratories). The collagen type II binding peptide (15% biotinylated) was added to the nanoparticles for 2 h in coupling buffer. Excess peptide was removed via gel filtration chromatography. Confirmation of peptide addition was performed using a flouraldehyde assay (Pierce), which reacts with free amines, and a streptavidin color development assay, which confirmed the presence of the biotinylated peptide on the nanoparticle surface (data not shown).
Collagen type II binding assay
Modified nanoparticles were tested for their ability to bind to collagen type II. A 96-well plate (Greiner) was coated with collagen type II from chicken sternum (Sigma) in 0.25% acetic acid at a concentration of 0.5 mg/ml overnight. Following three washes, the plate was blocked with 1% bovine serum albumin (BSA; SeraCare Life Systems) for 1 h. After three more washes, the collagen type II binding peptide modified core + shell nanoparticles and unmodified controls were incubated in the collagen type II coated plate for 1 h. Following three more washes, streptavidin (R&D Systems) was diluted 200× in 1% BSA and incubated for 20 min in the plate. After more washing to remove unbound streptavidin, a color solution (R&D Systems) was added for 20 min. Sulfuric acid (Mallinckrodt Chemicals) was then used to stop the reaction and absorbance was read at 450 nm with a correction at 540 nm.
Cell culture
Immortalized human monocytes (THP1, ATCC) were grown in RPMI 1640 with L-glutamine (Mediatech Inc.) supplemented with 0.05 mM mercaptoethanol (Sigma-Aldrich), 10 mM HEPES (Mediatech Inc.), 1 mM sodium pyruvate (Mediatech Inc.), 10% fetal bovine serum (Hyclone), and 1% penicillin/streptomycin (Mediatech Inc.). Cells were used between passage number 4 and 12 for all assays and maintained at 37°C with 5% CO2.
Nanoparticle biocompatibility
The biocompatibility of the nanoparticles was assessed by measuring toxicity and pro-inflammatory cytokine expression in THP1 cells. Cells were seeded at a density of 250,000 cells/mL in 96-well plates (Corning) and treated with 10 ng/mL phorbol 12-myristate 13-acetate (PMA, Sigma-Aldrich) for 48 h to induce differentiation, which was confirmed by the monocytes becoming adherent. Following a change of media, cells were treated with various concentrations of core + shell nanoparticles. Control samples received PBS (negative control) or 50 ng/mL lipopolysaccharide (LPS, Sigma-Aldrich) (positive control). After 24 h, the media was collected for cytokine analysis and an MTT-based assay was performed to determine cell toxicity using the Aqueous One Proliferation Kit (Promega) according to manufacturer’s instructions. Briefly, 20 µL of reagent was added directly to 100 µL of cells and media. After two hours of incubation in the cell culture incubator, the absorbance was read at 490 nm with a correction at 650 nm.
The ability of the particles to cause an inflammatory response was determined by running conditioned cell media on a TNF-α ELISA (PeproTech) according to manufacturer instructions. Briefly, Nunc MaxiSorp 96-well plates were coated with capture antibody overnight. After blocking for one hour with 1% bovine serum albumin (Sera Lifesciences) in PBS, samples and standards were incubated for two hours with gentle rotation. Following incubation with a detection antibody and an avidin-horse radish peroxidase conjugate, the samples were developed with the addition of 2,2'-azino-bis(3-ethylbenzthiazoline-6-sulphonic acid) (ABTS) liquid substrate (Sigma-Aldrich) and monitored at 405 nm with a correction at 650 nm.
Statistical Analysis
Data was analyzed for differences using a single factor ANOVA with a Tukey post-hoc test. A value of p < 0.05 was used for all analyses. Graphs are depicted as mean ± standard deviation.
Conclusions
In closing, we successfully encapsulated PLGA core nanoparticles with a pNIPAM shell. The addition of this pNIPAM shell conferred many benefits including the ability to protect the PLGA cores from aggregating during lyophilization, the ability to modify the nanoparticles with active targeting moieties in a concentration-dependent manner, and the ability to change size based on external environmental-based stimuli. These core + shell nanoparticles are non-toxic and do not elicit an inflammatory response in THP1 cells suggesting biocompatibility. All of these abilities will prove important in future in vivo studies focused on examining the ability of these nanoparticles to deliver therapeutics in an arthritic-based disease model.
| Abbreviations: | ||
| PLGA | = | poly(lactic-co-glycolic acid) |
| pNIPAM | = | poly(N-isopropylacrylamide) |
| EPR | = | enhanced permeability and retention |
| LCST | = | lower critical solution temperature |
| PVA | = | polyvinyl alcohol |
| AAc | = | acrylic acid |
| TEM | = | transmission electron microscopy |
| THP1 | = | human monocytes |
| DCM | = | dichloromethane |
| FMOC | = | 9-fluorenylmethyloxycarbonyl |
| HOBt | = | N-hydroxybenzotriazole |
| DIC | = | N,N' diisopropylcarbodiimide |
| HBTU | = | O-Benzotriazole-N,N,N',N'-tetramethyl-uronium-hexafluoro-phosphate |
| EDC | = | 1-ethyl-3-[3-dimethylaminopropyl]carbodiimide hydrochloride |
| sulfo-NHS | = | N-hydroxylsulfosuccinimide |
| MES | = | 2-(N-morpholino)ethanesulfonic acid |
| BMPH | = | N-[β-maleimidopropionic acid] hydrazide |
| BSA | = | bovine serum albumin |
| PMA | = | phorbol 12-myristate 13-acetate |
| LPS | = | lipopolysaccharide |
| PBS | = | phosphate buffered saline |
| TNF-α | = | tumor necrosis factor-α |
| ELISA | = | enzyme-linked immunosorbent assay |
| ABTS | = | 2,2'-azino-bis(3-ethylbenzthiazoline-6-sulphonic acid), N-methyl-bis-acrylamide, MBAm |
Acknowledgments
We thank Debra M. Sherman for her assistance in preparing and imaging samples in the Purdue University Life Science Microscope Facility. We also would like to thank Becca Scott for her expertise in producing . Additionally, this material is based upon work supported by the National Science Foundation Graduate Research Fellowship under Grant No. 103049 (J.L.B.).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- Shive MS, Anderson JM. Biodegradation and biocompatibility of PLA and PLGA microspheres. Adv Drug Deliv Rev 1997; 28:5 - 24; http://dx.doi.org/10.1016/S0169-409X(97)00048-3; PMID: 10837562
- Budhian A, Siegel SJ, Winey KI. Haloperidol-loaded PLGA nanoparticles: systematic study of particle size and drug content. Int J Pharm 2007; 336:367 - 75; http://dx.doi.org/10.1016/j.ijpharm.2006.11.061; PMID: 17207944
- Panyam J, Labhasetwar V. Biodegradable nanoparticles for drug and gene delivery to cells and tissue. Adv Drug Deliv Rev 2003; 55:329 - 47; http://dx.doi.org/10.1016/S0169-409X(02)00228-4; PMID: 12628320
- Hans ML, Lowman AM. Biodegradable nanoparticles for drug delivery and targeting. Curr Opin Solid State Mater Sci 2002; 6:319 - 27; http://dx.doi.org/10.1016/S1359-0286(02)00117-1
- Wischke C, Schwendeman SP. Principles of encapsulating hydrophobic drugs in PLA/PLGA microparticles. Int J Pharm 2008; 364:298 - 327; http://dx.doi.org/10.1016/j.ijpharm.2008.04.042; PMID: 18621492
- Acharya S, Sahoo SK. PLGA nanoparticles containing various anticancer agents and tumour delivery by EPR effect. Adv Drug Deliv Rev 2011; 63:170 - 83; http://dx.doi.org/10.1016/j.addr.2010.10.008; PMID: 20965219
- Maeda H, Wu J, Sawa T, Matsumura Y, Hori K. Tumor vascular permeability and the EPR effect in macromolecular therapeutics: a review. J Control Release 2000; 65:271 - 84; http://dx.doi.org/10.1016/S0168-3659(99)00248-5; PMID: 10699287
- Sandanaraj BS, Gremlich HU, Kneuer R, Dawson J, Wacha S. Fluorescent nanoprobes as a biomarker for increased vascular permeability: implications in diagnosis and treatment of cancer and inflammation. Bioconjug Chem 2010; 21:93 - 101; http://dx.doi.org/10.1021/bc900311h; PMID: 19958018
- Kocbek P, Obermajer N, Cegnar M, Kos J, Kristl J. Targeting cancer cells using PLGA nanoparticles surface modified with monoclonal antibody. J Control Release 2007; 120:18 - 26; http://dx.doi.org/10.1016/j.jconrel.2007.03.012; PMID: 17509712
- Chacón M, Molpeceres J, Berges L, Guzmán M, Aberturas MR. Stability and freeze-drying of cyclosporine loaded poly(D,L lactide-glycolide) carriers. Eur J Pharm Sci 1999; 8:99 - 107; http://dx.doi.org/10.1016/S0928-0987(98)00066-9; PMID: 10210732
- Holzer M, Vogel V, Mäntele W, Schwartz D, Haase W, Langer K. Physico-chemical characterisation of PLGA nanoparticles after freeze-drying and storage. Eur J Pharm Biopharm 2009; 72:428 - 37; http://dx.doi.org/10.1016/j.ejpb.2009.02.002; PMID: 19462479
- Petros RA, DeSimone JM. Strategies in the design of nanoparticles for therapeutic applications. Nat Rev Drug Discov 2010; 9:615 - 27; http://dx.doi.org/10.1038/nrd2591; PMID: 20616808
- Pelton RH, Chibante P. Preparation of aqueous latices with N-isopropylacrylamide. Colloids Surf 1986; 20:247 - 56; http://dx.doi.org/10.1016/0166-6622(86)80274-8
- Liu RX, Fraylich M, Saunders BR. Thermoresponsive copolymers: from fundamental studies to applications. Colloid Polym Sci 2009; 287:627 - 43; http://dx.doi.org/10.1007/s00396-009-2028-x
- Das M, Mardyani S, Chan WCW, Kumacheva E. Biofunctionalized pH-responsive microgels for cancer cell targeting: Rational design. Adv Mater 2006; 18:80 - 3; http://dx.doi.org/10.1002/adma.200501043
- Cheng CJ, Chu LY, Zhang J, Wang HD, Wei G. Effect of freeze-drying and rehydrating treatment on the thermo-responsive characteristics of poly(N-isopropylacrylamide) microspheres. Colloid Polym Sci 2008; 286:571 - 7; http://dx.doi.org/10.1007/s00396-007-1817-3
- Heskins M, Guillet JE. Solution Properties of Poly(N-isopropylacrylamide). J Macromol Sci -Chem 1968; 2:1441 - 55; http://dx.doi.org/10.1080/10601326808051910
- Afrassiabi A, Hoffman AS, Cadwell LA. Effect of temperature on the release rate of biomolecules from thermally reversible hydrogels. J Membr Sci 1987; 33:191 - 200; http://dx.doi.org/10.1016/S0376-7388(00)80377-4
- Kratz K, Hellweg T, Eimer W.. Influence of charge density on the swelling of colloidal poly(N-isopropylacrylamide-co-acrylic acid) microgels. . Colloids and Surfaces a-Physicochemical and Engineering Aspects 2000; 170:137 - 49; http://dx.doi.org/10.1016/S0927-7757(00)00490-8
- Somasundaran P. Encyclopedia of Surface and Colloid Science, Volume 2. New York / London: Taylor & Francis, 2006.
- Feil H, Bae YH, Feijen J, Kim SW. Effect of comonomer hydrophilicity and ionization on the lower critical solution temperature of N-isopropylacrylamide copolymers. Macromolecules 1993; 26:2496 - 500; http://dx.doi.org/10.1021/ma00062a016
- Westacott CI, Sharif M. Cytokines in osteoarthritis: mediators or markers of joint destruction?. Semin Arthritis Rheum 1996; 25:254 - 72; http://dx.doi.org/10.1016/S0049-0172(96)80036-9; PMID: 8834014
- Choy EHS, Panayi GS. Cytokine pathways and joint inflammation in rheumatoid arthritis. N Engl J Med 2001; 344:907 - 16; http://dx.doi.org/10.1056/NEJM200103223441207; PMID: 11259725
- Rothenfluh DA, Bermudez H, O’Neil CP, Hubbell JA. Biofunctional polymer nanoparticles for intra-articular targeting and retention in cartilage. Nat Mater 2008; 7:248 - 54; http://dx.doi.org/10.1038/nmat2116; PMID: 18246072
- Cremer MA, Rosloniec EF, Kang AH. The cartilage collagens: a review of their structure, organization, and role in the pathogenesis of experimental arthritis in animals and in human rheumatic disease. J Mol Med (Berl) 1998; 76:275 - 88; http://dx.doi.org/10.1007/s001090050217; PMID: 9535561
- Xu PS, Gullotti E, Tong L, Highley CB, Errabelli DR, Hasan T, et al. Intracellular drug delivery by poly(lactic-co-glycolic acid) nanoparticles, revisited. Mol Pharm 2009; 6:190 - 201; http://dx.doi.org/10.1021/mp800137z; PMID: 19035785
- Jones CD, Lyon LA. Synthesis and characterization of multiresponsive core-shell microgels. Macromolecules 2000; 33:8301 - 6; http://dx.doi.org/10.1021/ma001398m
- Jones CD, Lyon LA. Dependence of shell thickness on core compression in acrylic acid modified poly(N-isopropylacrylamide) core/shell microgels. Langmuir 2003; 19:4544 - 7; http://dx.doi.org/10.1021/la034392+
- Berndt I, Richtering W. Doubly temperature sensitive core-shell microgels. Macromolecules 2003; 36:8780 - 5; http://dx.doi.org/10.1021/ma034771+