Abstract
Data are accumulating to support a role for adipose-derived mesenchymal stem cells (MSCs) in breast cancer progression; however, to date most studies have relied on adipose MSCs from non-breast sources. There is a particular need to investigate the role of adipose MSCs in the pathogenesis of basal-like breast cancer, which develops at a disproportionate rate in pre-menopausal African-American women with a gain in adiposity. The aim of this study was to better understand how breast adipose MSCs (bMSCs) contribute to the progression of basal-like breast cancers by relying on isogenic HMT-3255 S3 (pre-invasive) and T4-2 (invasive) human cells that upon transplantation into nude mice resemble this tumor subtype. In vitro results suggested that bMSCs may contribute to breast cancer progression in multiple ways. bMSCs readily penetrate extracellular matrix components in part through their expression of matrix metalloproteinases 1 and 3, promote the invasion of T4-2 cells and efficiently chemoattract endothelial cells via a bFGF-independent, VEGF-A-dependent manner. As mixed xenografts, bMSCs stimulated the growth, invasion and desmoplasia of T4-2 tumors, yet these resident stem cells showed no observable effect on the progression of pre-invasive S3 cells. While bMSCs form vessel-like structures within Matrigel both in vitro and in vivo and chemoattract endothelial cells, there appeared to be no difference between T4-2/bMSC mixed xenografts and T4-2 xenografts with regard to intra- or peri-tumoral vascularity. Collectively, our data suggest that bMSCs may contribute to the progression of basal-like breast cancers by stimulating growth and invasion but not vasculogenesis or angiogenesis.
Introduction
It has long been appreciated that stromal cells within the breast tumor microenvironment contribute to mammary carcinogenesis.Citation1,Citation2 Even though adipose tissue is the most abundant stromal constituent in the breast, little is known about the involvement of resident adipose-derived cells in the development of breast cancer.Citation3 Increased adiposity and obesity are associated with an elevated risk of developing the most prevalent form of breast cancer (luminal type A) in post-menopausal women,Citation4-Citation6 an association assumed to be largely due to adipose tissue being a primary extra-gonadal source of estrogen.Citation4 Basal-like breast cancers are unique in that both pre- and post-menopausal women with elevated waist-hip ratios and body mass indices are at increased risk, with the prevalence being highest among premenopausal African-American women.Citation7,Citation8
Since the majority of basal-like breast cancers do not express estrogen receptor, progesterone receptor or Her-2, targeted therapeutic options are limited and prognosis is usually poor.Citation7,Citation8 This absence of hormone receptors suggests that adiposity contributes to the development of basal-like breast cancer via a hormone-independent mechanism(s). Data supporting a role of adipokines (cytokines secreted by adipose tissue) in promoting breast cancer cell growth and angiogenesisCitation5,Citation9 is certainly consistent with this idea. Moreover, recent studiesCitation10 suggest that mesenchymal stem cells from mammary fat depots differentiate into carcinoma-associated fibroblasts in response to breast cancer-derived soluble mediators and promote cancer cell invasion. The observations that (1) breast cancers often develop in close association with fat,Citation11 (2) during the aging process mammary tissue tends to become fattier and less dense,Citation12 and (3) age is a risk factor for breast cancerCitation13 make the study of adipose-derived cells as they relate to mammary carcinogenesis of particular interest.
Stromal cells within adipose tissue have traditionally been called “pre-adipocytes”, based on observations that these cells can accumulate lipid droplets in vitro as well as revert to a more fibroblastic appearance.Citation14 It was first reported in 2001 that these adipose stromal cells (ASCs) are not limited to adipocytic differentiation; they can also differentiate along osteogenic, chondrogenic, endothelial and perhaps even neural cell lineages, thus making them indistinguishable in this regard from MSCs from bone marrow.Citation15,Citation16 Consequently, ASCs are also referred to as MSCs, multipotent mesenchymal stromal cells and mesenchymal stem-like cells.Citation17-Citation19 Here, we use the MSC designation or bMSC if derived from breast adipose tissue.
Adipose tissue throughout the body, including within the breast, is an abundant source of MSCs.Citation3,Citation19 Experimental data demonstrate that MSCs from bone marrow and adipose tissue can efficiently home to tumors, including breast cancer,Citation20-Citation22 where they can stimulate breast cancer growth and invasion,Citation20,Citation23-Citation25 modulate inflammatory cells within the tumor microenvironment,Citation26 and give rise to carcinoma-associated fibroblasts.Citation10,Citation27,Citation28 But only recently have investigations focused on the role of local adipose MSCs on the progression of breast cancer.Citation3,Citation29 And, no study has specifically evaluated the effects of adipose MSCs on the progression of breast cancer resembling the basal-type, which arguably has the most intriguing and complex association with adiposity. Here, we generate experimental data to support the mid-late stage involvement of resident adipose MSCs in the progression of basal-type breast cancer and, in doing so, we establish a mixed xenograft model for future elucidation of molecular mechanisms contributing to this promoting effect.
Results
bMSC immunophenotyping
Within 7 population doublings of isolation, bMSCs were immunophenotyped in order assess cell purity using BM-MSCs as a comparison, as recently described.Citation3 We observed intense and widespread expression of all MSC markers tested (CD29, CD73, CD90 and CD105) yet undectable immunolabeling for CD31, an endothelial-specific marker and CD45 and CD14, which would identify lymphocytes and monocytes, respectively (data not shown). This was the case for the 4 strains of bMSCs and the two strains of BM-MSCs.
bMSCs express multiple matrix metalloproteinases and pro-angiogenic mediators
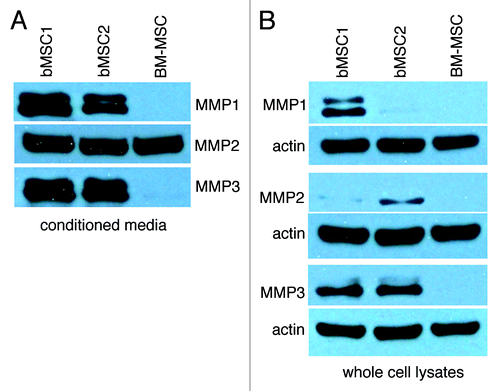
Since few studies have used breast adipose-derived MSCs to study mammary cancer progression, we performed gene expression profiling to begin to appreciate similarities and differences between MSCs from local fat depots and bone marrow. A comparison of the gene expression profiles of 3 bMSC and 2 BM-MSC cell strains identified 79 unique probe sets (GEO Accession Number: GSE24433).Citation3 Seventeen of the differentially expressed genes were expressed at significantly higher levels in bMSCs relative to BM-MSCs with MMP3 (stromelysin 1) being at the top of the list with a geometric fold change of +147.7 (Table S1). To validate our global gene expression analysis data, we selected 3 genes (MMP3 {+147.7}; IL1RA {+6.0}; and IGF2 {-48.8}) and confirmed their differential expression by qRT-PCR (Fig. S1). 254 highly expressed probe sets were identified for both bMSCs and BM-MSCs and overlayed onto a global molecular network (Ingenuity software, IPKB) (Table S2). Western blotting confirmed that bMSCs express the cleaved forms of MMP1, MMP2 and MMP3 proteins and secrete these proteases into culture media, whereas only MMP2 was detectable in whole cell lysates or conditioned media from BM-MSC cultures (). Casein- and gelatin-based zymography further demonstrated that MMP3 and MMP2, detected in abundance in bMSC conditioned media, were enzymatically active (data not shown). Through our gene expression analysis and confirmatory assays, it also became apparent that bMSCs express numerous pro-angiogenic mediators including MMPs, bFGF, VEGF-A (; Fig. S1 and Table S3) and multiple integrins (α5β1, α2β1 and α5β5) (qRT-PCR data not shown).
Figure 1. Breast MSCs secrete MMP1, MMP2 and MMP3. (A and B) Conditioned media was collected and concentrated from the respective MSC cultures, monolayers were lysed and samples were analyzed by SDS-PAGE. Here we show bMSC donor strains 1 and 2 as representative data. Two additional bMSC strains (including donor #3) yielded similar high levels of the respective MMPs.
bMSCs readily degrade extracellular matrix components and promote the invasion of HMT-3522 T4-2 breast cancer cells in vitro
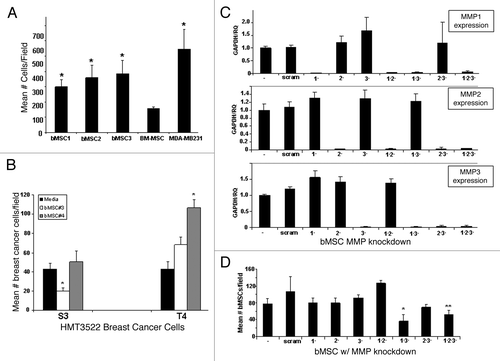
The expression of multiple MMPs by bMSCs prompted us to test whether these local stem cells may contribute to mammary carcinogenesis, in part by degrading extracellular matrix and facilitating cancer cell invasion. Using a transwell assay approach, we demonstrated that bMSCs readily penetrate basement membrane Matrigel® at levels significantly greater than BM-MSCs and at levels more comparable to the highly invasive breast cancer cell line, MDA-MB231 (). bMSCs also penetrated a polymerized type I collagen barrier, the major component of interstitial matrix (data not shown). siRNA-based technology was used to transiently knock-down the expression of MMP1, MMP2 and MMP3 in various combinations in bMSCs. All siRNA constructs efficiently and selectively reduced mRNA expression of the respective MMP genes (); yet single MMP gene knock-down did not affect the ability of bMSCs to penetrate Matrigel®-coated transwell inserts (). The only combinations of MMP knock-down to significantly reduce the number of MSCs invading through the Matrigel®-coated barrier were double knock-down of MMP1 and MMP3 and triple knock-down (). Having demonstrated that bMSCs possess potent matrix degrading potential, we employed an alternative transwell assay configuration to test whether bMSCs are able to promote the invasion of breast cancer cells of the basal-type. HMG-3522 S3 cells and the isogenic derivative, T4-2, were seeded on top of Matrigel®-coated inserts with tissue culture media or bMSCs in the lower well. The mean number of breast cancer cells penetrating the Matrigel® barrier was greater for two different bMSC cell strains, but only bMSC4 resulted in a statistically significant difference (). In contrast, bMSCs did not potentiate the invasion of S3 cells, but rather one strain caused a statistically significant reduction in invasion frequency while the other had no effect. Since bMSC4 had the most pronounced effect on promoting the invasion of T4-2 cells, this strain was used for MMP knock-down experiments. Unlike the matrix degrading potential of bMSCs, which was dependent on expression of MMP 1 and 3, their ability to potentiate the invasion of T4-2 cells through Matrigel® was independent of MMP function (Fig. S2).
Figure 2. Breast MSCs readily penetrate extracellular matrix via a MMP 1- and 3-dependent mechanism. (A) A standard transwell invasion assay was performed to compare the ability of MSCs (BM: bone marrow; b: breast) to penetrate a Matrigel® barrier relative to a highly invasive breast cancer cell line (MDA-MB231). Values reflect mean ± SD from triplicate wells with statistical comparisons to BM-MSC (* p < 0.05). (B) A standard transwell invasion assay was performed to test the ability of bMSCs (in the lower chambers) to facilitate the invasion of S3 and T4-2 breast cancer cells. (C) qRT-PCR demonstrates that all MMP siRNA constructs efficiently and specifically knock-down the respective genes. (D) Transient MMP knock-downs reveal that the ability of bMSCs to degrade Matrigel® is dependent in part on MMP1 and MMP3. * p value < 0.05 based on a 2-tailed student t test compared with untransfected controls. ** p value < 0.05 when compared with the scrambled control. Values represent the mean ± SD based on three random fields from triplicate inserts per culture. * p value < 0.05 based on a 2-tailed student t test compared with the respective media only controls.
bMSCs chemoattract human endothelial cells and exhibit branching morphogenesis in vitro and in vivo
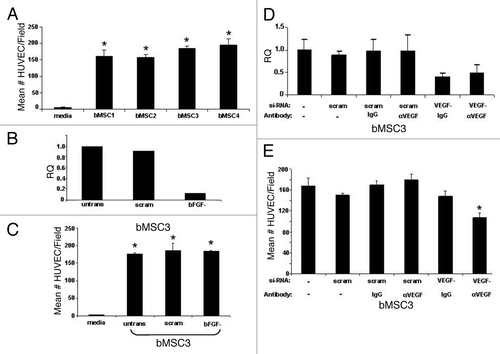
Since bMSCs express numerous pro-angiogeneic mediators, we hypothesized that these local adult stem cells may contribute to the vascularization of breast cancers. Using a transwell migration assay, we demonstrate that bMSCs consistently and efficiently chemoattract human endothelial cells (), suggesting that bMSCs may play a role in angiogenesis. Based on high levels of expression of bFGF and VEGF-A by bMSCs (Table S3), we selected these two factors as possible mediators of endothelial cell chemotaxis. While efficient knock-down of bFGF in bMSCs was consistently achieved (), there was no significant difference between the number of endothelial cells migrating to the lower surface of the transwell insert whether bMSCs in the lower wells were untransfected, transfected with a scrambled si-RNA, or bFGF si-RNA (). At best, we were only able to consistently knockdown VEGF-A via si-RNA by ~60% in bMSCs (), necessitating a dual inhibition strategy of si-RNA plus a neutralizing antibody. Our finding that dual inhibition of VEGF-A in bMSCs correlates with a significant reduction in the number of migrating endothelial cells implicates VEGF-A as a positive mediator of endothelial cell recruitment by bMSC ().
Figure 3. Breast MSCs chemoattract human endothelial cells via a bFGF independent, VEGF-A dependent pathway. (A) Transwell migration data demonstrate that bMSCs from 4 individual donor strains chemoattract endothelial cells at a similar frequency. In all cases, the chemoattraction efficiency was statistically different than the media controls (* p < 0.05). (B) Efficient knock-down of bFGF in bMSCs as determined by qRT-PCR. Untransfected cells and a scrambled si-RNA construct were included as baseline controls. (C) bFGF knock-down has no significant effect on the ability of bMSCs to chemoattract endothelial cells. * p values < 0.05 when compared with media only controls. (D) Knock-down of VEGF-A in bMSCs as determined by qRT-PCR. As expected, the presence of neutralizing antibody or IgG had no affect on relative quantitation (RQ) of VEGF-A mRNA. (E) Inhibition of VEGF-A by siRNA and neutralizing antibody results in a statistically significant (* p < 0.05) reduction in the ability of bMSCs to chemoattract human endothelial cells. IgG1 served as a negative control for VEGF-neutralization. All values represent the mean number of cells per microscopic field based on 3 fields/insert and 3 inserts per condition.
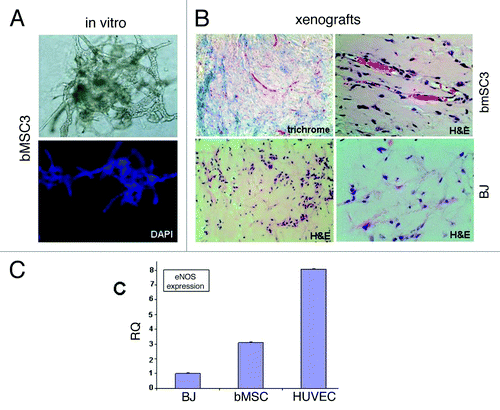
Recognizing that MSCs from non-breast sites have been reported to undergo endothelial differentiation, we tested whether bMSC could directly differentiate into vascular networks (vasculogenesis), thus potentially providing an additional mechanism by which bMSCs could contribute to breast cancer vascularity. When cultured within growth factor-reduced Matrigel®, bMSCs assembled into branching structures as early as 2 d in culture (data not shown) with an extensive vascular-like network forming by day 4 (). Following subcutaneous injection within Matrigel® into athymic nude mice, bMSCs arranged into numerous small vessel-like structures with many containing red blood cells within the lumen, suggesting a connection to the existing murine blood supply (, top panels). BJ fibroblasts were similarly inoculated for comparison, however, retrieval of these xenografts proved challenging due to their small size. Upon microscopic evaluation, there was no evidence of vessel-like structures within the BJ xenografts (, bottom panels). Expression levels of endothelial-specific nitric oxide synthase (eNOS) was ~3 fold higher in bMSC xenografts compared with BJ xenografts (), further suggesting a possible role for bMSCs in promoting vascularity.
Figure 4. Evidence supporting breast MSC endothelial differentiation. (A) bMSCs form extensive vascular-like structures 4 d after culturing within Matrigel® (20x light and fluorescent images). DAPI (lower panel) allowed the visualization of nuclei. (B) bMSCs also form vessel-like structures when subcutaneously injected within Matrigel® into athymic nude mice. Masson Trichrome and H&E histochemical staining permits the identification of numerous small vessel-like structures with red blood cells within lumens (upper panels), suggesting a connection to the host’s vascular system. Shown are specimens obtained 2 weeks after subcutaneous inoculation, however vascular networks were observed as early as 4 d (data not shown). No evidence of vascular-like structures in xenografts following the subcutaneous injection of BJ fibroblasts within Matrigel®. Shown is a 6-d xenograft, however, similar negative findings were observed at days 2, 4, 9 and 14 post-injection. (C) qRT-PCR data comparing eNOS expression levels in subcutaneous xenografts 6 d after inoculation with BJ fibroblasts, bMSCs or HUVECs all within Matrigel®.
bMSCs promote the growth and invasion but not the vascularity of basal-like breast cancers in vivo
Having generated in vitro data suggesting the involvement of bMSCs in degrading extracellular matrix components, facilitating the invasion of basal-like T4-2 breast cancer cells, recruiting endothelial cells and perhaps forming blood vessels de novo, we next tested the effects of bMSCs on breast cancer progression as mixed xenografts with S3 or T4-2 cells. We transplanted pre-invasive S3 cells and T4-2 cells, which are invasive with the ability to form slow growing tumors with squamous and basal metaplastic histology,Citation30 in the absence vs. presence of bMSCs into cleared mammary fat pads. Since the individual donor strains were virtually indistinguishable regarding their immunophenotypes, gene expression profiles and differentiation potential,Citation3 we selected one (bMSC3) for the animal studies. Pure S3 and S3/bMSC mixed xenograft tumors were undetectable at necropsy 8 weeks after mammary fat pad transplantation (). Only upon careful microscopic evaluation did we observe focal/residual tumor cells in 2 of 6 S3 xenografts and 4 of 6 S3/bMSC mixed xenografts (data not shown).
Table 1. Summary of fat pad xenograft tumor frequencies and volumes
Inoculation of T4-2 cells into cleared mammary fat pads resulted in a high (10/10) tumor incidence ( and ), as reported previously.Citation30 These pure xenografts were all slow growing, moderate-to-well differentiated carcinomas with squamous metaplasia (). For some T4-2 pure xenografts, we observed histologic evidence of invasion, but it was predominantly limited to small, focal areas (, middle 3 panels). When T4-2 cells were orthotopically coinjected with bMSCs the effects on breast cancer progression were striking. Mean tumor volumes were significantly larger than the pure T4-2 xenografts (p = 0.019) (, ), the desmoplastic response, as assessed by Masson Trichrome histochemical staining, was much more pronounced (, bottom panels) and the Ki-67 labeling indices were significantly higher (). Areas of marked proliferation were observed exclusively in the mixed xenografts and typically adjacent to stromal cells and the associated fibrosis (). In contrast, Ki-67-positive cells within the pure T4-2 xenografts were rare and sporadic in their distribution. Moreover, the mixed xenografts were not as well-circumscribed as T4-2 xenografts, typically displaying more extensive invasion (, bottom panels). Of note, the areas of T4-2 invasion in the mixed xenografts were generally in close association with desmoplasia (, middle and right lower panels) and readily evident, whereas additional sectioning was often necessary to detect evidence of invasion in the pure T4-2 xenografts. While we observed a trend toward increased intra-tumoral vascularity in the bMSC/T4-2 mixed xenografts relative to pure T4-2 specimens, the differences were not statistically significant (, patterned bars). There was also no significant difference in the peri-tumoral vascularity between the pure vs. mixed orthotopic xenografts. In order to more closely recapitulate our Matrigel® branching morphogenesis assays (shown in ), we subcutaneously injected T4-2 cells ± bMSC#3 within Matrigel® into the flanks of athymic nude mice and determined the mean number of vessels per low power field. We found that both the pure and mixed subcutaneous xenografts were more vascular within and around the tumors than the respective orthotopic tumors (, solid bars). However, despite testing a second injection site and the addition of Matrigel®, there was still no statistically significant difference in the vascularity of T4-2 and T4-2/bMSC mixed xenografts.
Figure 5. Breast MSCs promote the growth but not vascularity of HMT-3522 T4-2 cells. (A) 8-week HMT-3522 T4-2 cells (T4-2) ± bMSC3 fat pad xenografts (scale bar: 3mm). (B) Determination of the percent of Ki-67 positive cells per microscopic field revealed a statistically significant difference (p ≤ 0.011) between the pure and mixed xenografts. Representative immunohistochemical staining reveal that pure T4-2 xenografts contained only rare Ki-67 positive cells (arrows in top panels), whereas T4-2/bMSC xenografts often contained focal areas of active proliferation in close association with fibrosis (f = fibrosis). Primary antibody was omitted as a negative control for all specimens (left panels). Values indicated in the right panels represent mean percent Ki-67 positivity and were calculated based on counting the number of immunolabeled cells and then dividing by the total number of cells in 3 random fields per xenograft. (C) Intra- and peri-tumoral vascularity was not significantly different in T4-2 vs. T4-2/bMSC mixed xenografts whether injected orthotopically (patterned bars) or subcutaneously within Matrigel® (solid bars). Vascularity was determined on H&E stained tissue sections as described in Materials and Methods. LPF: low power field.
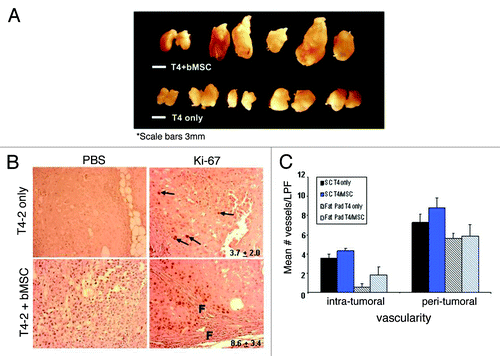
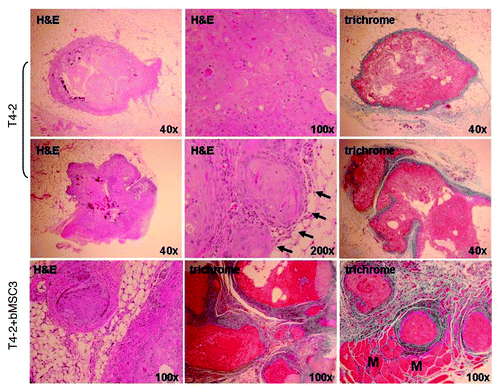
Figure 6. Representative histologic features of pure HMT-3522 T4-2 vs. mixed T4-2/bMSC fat pad xenografts. H&E staining demonstrated that pure T4-2 and T4-2/bMSC xenograft tumors formed well-differentiated mammary carcinomas displaying squamous and basal-like features. Masson’s trichrome staining revealed that the desmoplastic response was much more pronounced in the mixed xenografts. Pure T4-2 xenografts tended to be relatively small lesions encased in a minimal amount of fibrotic stroma (upper panels), with additional tissue sectioning often being required to obtain evidence of invasion (middle panels). In contrast, invasion through and beyond the cyst wall or pseudocapsule was often observed in a single H&E tissue section of T4-2/bMSC mixed xenografts (lower panels). Arrows indicate areas of superficial invasion into fat. M: muscle.
Discussion
Here, we provide data that are consistent with MSCs from breast adipose tissue contributing to the progression of basal-like breast cancer. Using a human pre-invasion (S3) to invasion (T4-2) progression model allowed us to elucidate when during the multi-step process of carcinogenesis resident MSCs could have their promoting influence. Both in vitro and mammary fat pad xenograft data suggest that bMSCs are unable to progress HMT-3522 S3, even though these cells display a partial loss of tissue polarity and a higher potential for acquiring invasive properties than its original cell population.Citation30 Since S3 cells adopt an invasive phenotype when exposed to conditioned media containing multiple T4-2 derived MMPs,Citation30 we were somewhat surprised that bMSCs, which express high levels of multiple, active MMPs, did not lead to the in vivo progression of S3 cells. Recognizing that S3 cells are very sensitive to their microenvironment, we tested the ability of bMSC to progress these pre-invasive cells at a second inoculation site: under the skin and together with Matrigel®. Again, there was no evidence to support disease progression (data not shown). Therefore, we conclude that bMSCs are unable to complement the pathways that are still deficient in S3 cells for conferring a stable malignant phenotype.
Once HMT-3522 cells transition to an invasive phenotype (as represented by T4-2 cells in this progression model), bMSCs appear to contribute to basal-like breast cancer progression by stimulating tumor growth, triggering desmoplasia and potentiating invasion. Microarray data revealed that bMSCs are high expresser of several members of a cell growth and proliferation network and they express, numerous matrix proteins and multiple MMPs, hinting that MSC from local fat depots may contribute to breast cancer progression via multiple mechanisms. Precisely how bMSCs promote the growth of T4-2 xenografts is an area of current investigation. In the mixed xenografts, we observed focal areas of T4-2 proliferation in close proximity to prominent desmoplasia suggesting that bMSCs and their associated fibrogenic response exert a growth stimulatory effect on the cancer cells. Since bMSCs express high levels of multiple MMPs, it is reasonable to speculate that these resident stem cells could indirectly contribute to breast cancer cell growth by triggering the proteolytic release of growth factors tethered to extracellular matrix.Citation31
In vitro, bMSCs readily degrade extracellular matrix (Matrigel®) and potentiate the invasion of T4-2 breast cancer cells. Relying on multiple transwell assay configurations, we have examined potential mediators of these activities, focusing on two MMPs (MMP1 and MMP3), that are expressed at high levels by bMSCs but not BM-MSCs and MMP2, which is expressed at high levels in both bMSCs and BM-MSCs. Using si-RNA technology, we demonstrate that while expression of MMP1 and MMP3 is critical for bestowing bMSCs with their potent matrix degrading potential, this alone does not explain how local MSCs facilitate T4-2 invasion, as this potentiation effect is not dependent on bMSCs expressing MMP 1, 2 and/or 3. Transwell invasion assays, while a simple and rapid approach for assessing invasive potential of cells, are unable to faithfully recapitulated the dynamic interactions among cells of the tumor microenvironment. Therefore, we extended our studies into immunodeficient nude mice and have presented histologic evidence that T4-2 cells when co-injected with bMSCs into mammary fat pads tend to invade more frequently and to a greater depth compared with T4-2 only xenografts, further suggesting that bMSCs contribute to the progression of basal-like breast cancers in part by facilitating invasion.
Our Affymetrix data and confirmatory assays also revealed that bMSCs express a number of pro-angiogenic mediators, as has been reported previously for MSCs from multiple tissue sites.Citation32,Citation33 We demonstrate that bMSCs consistently and efficiently chemoattract human endothelial cells in vitro, suggesting their potential involvement in promoting tumor angiogenesis. We also show that expression of VEGF-A, but not bFGF, by bMSCs is necessary for chemoattracting endothelial cells. Our observation that bMSCs assemble into vessel-like structures within Matrigel® in vitro and in vivo, suggest that perhaps bMSC promote tumor vascularity by a second (and not necessarily mutually exclusive) mechanism: rather than recruiting endothelial cells to the tumor microenvironment, they incorporate into vessels within the tumor by directly differentiating into endothelial cells, as previously reported for adipose MSCs in an ischemic injury modelCitation34-Citation36 and the 4T1 murine model of breast cancer progression.Citation23 Despite our in vitro data favoring the potential involvement of bMSCs in promoting breast cancer vascularity, we did not observe a significant difference in capillary density either within or around the tumor when comparing T4-2 vs. T4-2/bMSC xenografts at two (fat pad and subcutaneous) inoculation sites. The precise molecular mechanism triggering the formation of cancer-associated blood vessels is unclear; however, hypoxia appears to be a critical mediator.Citation37 Since T4-2 and T4-2/bMSC xenografts are relatively slow growing it is possible that following an 8-week latency period these tumors had not yet become sufficiently hypoxic to trigger neo-angiogenesis. In this study, not a single xenograft displayed evidence of necrosis at necropsy or upon histological analysis, a finding that is at least consistent with this possibility.
Here, we have established a mixed xenograft model that will provide a useful tool for better understanding how MSCs within local fat depots contribute to the progression of basal-like breast cancers, with the ultimate goal of developing targeted therapeutic options for this aggressive variant. In this study, bMSCs were isolated from adipose tissue from residual reduction mammoplasty specimens, which were deemed cancer-free following gross and microscopic evaluations. Since most individuals undergoing this surgical procedure have an elevated body-mass index, adipose tissue may not be “normal” per se. Obesity is associated with systemic inflammation and pro-inflammatory proteins are secreted by hypertrophic adipocytes and stromal cells within adipose tissue,Citation38 which could contribute to bMSC-mediated breast cancer progression. It is important to now use this orthotopic mixed xenograft model to test adipose MSCs (breast vs. abdominal) isolated from individuals with elevated as well as normal body-mass indices.
Materials and Methods
Cell isolation and monolayer culturing
Breast adipose tissue was obtained from patients undergoing elective reduction mammoplasty procedures in the Department of Plastic Surgery, Virginia Commonwealth University. 500–700 g of adipose tissue was obtained per case, processed essentially as described previously,Citation39 and then cultured in low glucose DMEM medium supplemented with 10% FBS, 10 ng/ml epidermal growth factor and 1.0% antibiotic-antimycotic solution. Bone marrow-derived MSCs (BM-MSCs) were obtained from the iliac crest of normal donors in accordance with our institutional Internal Review Board using a standard isolation procedure. BM-MSCs were maintained in high glucose DMEM supplemented with 10% FBS, 1.0 ng/ml basic fibroblast growth factor (bFGF) and 1.0% penicillin/streptomycin. HMT-3522 S3 and T4-2 cells were maintained in chemically-defined media on type I collagen-coated tissue culture dishes (PureColCollagen, Advanced BioMatrix), as described previously.Citation30 MDA-MB231 cells were propagated in RPMI media 1640 (+ L-glutamine) supplemented with 10% FBS and 1.0% antibiotic-antimycotic solution. Human umbilical vein endothelial cells (HUVEC) were purchased from Lifeline Cell Technology and cultured in Vasculife VEGF complete media (Lifeline Cell Technology), as recommended by the manufacturer. BJ fibroblasts were cultured as previously described.Citation40 All cultures were kept in a humidified 37°C incubator in 5.0% CO2.
Flow cytometry
Cells were immunolabeled according to a standard protocol using the following panel of monoclonal antibodies (Millipore, Billerica, MA): CD14 (macrophage/monocyte marker); CD29 (marker of MSCs, endothelial cells, monocytes and platelets); CD31 (marker of endothelial cells); CD45 (general leukocyte marker); CD73 (expressed on MSCs, epithelia and endothelial cells and mature lymphocytes); CD90 (MSC marker); and CD105 (marker of MSCs, endothelial cells and presumably some stem/progenitors). A no primary antibody control was included for each cell strain tested. Alexa 488 anti-mouse (Molecular Probes) was used at a 1:400 dilution as a secondary antibody for each sample. The following cells provided positive controls: HUVEC (CD31 and CD105); a lymphoblastic cell line (CD45); and HL-60 (CD14). A standard cytometric analysis was performed using a Coulter Epics XL-MCL (Beckman Coulter, Brea, CA) in our institution’s core facility.
Expression analysis using oligonucleotide (affymetrix) microarrays
Total cellular RNA was isolated using Trizol reagent (Life Technologies, Inc.) from 3 bMSC and 2 BM-MSC strains (all PD 8–9) representing 5 different donors. Four μg of total RNA from each sample was used to generate double-stranded cDNA using a 24-mer oligodeoxythymidylic acid primer with a T7 RNA polymerase promoter site added to the 3′ end (Superscript cDNA Synthesis System; Life Technologies, Inc.). Second strand cDNA synthesis, cleanup and biotinylation were conducted according to our standard protocol.Citation41 Fragmented cRNA (10 µg) was hybridized on the GeneChip® Human Genome U133A 2.0 (HG-U133A 2.0) array and analyzed as described previously.Citation41
RT-PCR
For Affymetrix validation studies, qRT-PCR was used to assess expression levels of selected genes using TaqMan® chemistry. Probes and primer sets for detection of MMP1, MMP3, IGF2 and IL1RA transcripts were obtained from inventoried assays (Applied Biosystems, Foster City, CA). Gene-specific probes labeled in the 5′ end with FAM (6-carboxyfluorescein) and in the 3′ end with a dark quencher were used for all the target genes of interest. For all of the samples, cyclophilin A (PPIA) from the Predeveloped TaqMan® Assay reagents (Applied Biosystems) was used as an endogenous control. The experiments were performed in the ABI Prism 7500 Sequence Detection System (Applied Biosystems) using the TaqMan® One-Step PCR Master Mix Reagents Kit and analyzed as described previously.Citation41 SYRB® green-based qRT-PCR was used to assess the efficiency of si-RNA knock-down for the respective genes: MMP1 (forward ACACATCTGACCTACAGGATTGA, reverse GTGTGACATTACTCCAGAGTTGG; MMP2 (forward CTTCCAAGTCTGGAGCGATGT, reverse TACCGTCAAAGGGGTATCCAT); MMP3 (forward ATGGACAAAGGA-TACAACAGGGA, reverse TGTGAGTGAGTGATAGAGTGGG); bFGF (forward ATCAAAGGAGTGTGTGCTAACC, reverse ACTGCCCAGTTCGTTTCAGTG); and VEGF-A (forward CAACATCACCATGCAGATTATGC, reverse CCCACAGGGATTTTC-TTGTCTT).
Functional network analyses
A data set containing Affymetrix® probe set IDs as gene identifiers and corresponding normalized unlogged expression summaries was uploaded into the Ingenuity Pathways Analysis (Ingenuity® Systems, www.ingenuity.com) application. Each gene identifier was mapped to its corresponding gene object in the Ingenuity Pathways Knowledge Base (IPKB). These genes, called Focus Genes, were overlaid onto a global molecular network developed from information contained in the IPKB. Networks of these Focus Genes were then algorithmically generated based on their connectivity, or interactions between one another. Biological networks were ranked by score, where the score corresponds to the likelihood of a set of genes being found in the networks due to random chance. The 254 highly expressed probe sets (BM-MSCs and bMSCs) were overlaid onto a global molecular network developed from information contained in the IPKB. Networks of these genes were then algorithmically generated based on their connectivity. The 79 differentially expressed genes were similarly analyzed.
MMP Western Blotting
MSCs were cultured overnight in serum-free media. 500 μl of conditioned media was collected, concentrated 10-fold using Microcon® centrifugal filter devices (Millipore, Billerica, MA) and resolved on an 8% SDS polyacrylamide gel. A standard RIPA buffer was used to lyse the monolayer cultures and protein concentrations were measured by the Bio-Rad Dc protein assay (Hercules, CA), according to the manufacturer’s protocol. Twenty-five µg of each lysate was subjected to SDS-PAGE. Following electrotransfer, nitrocellulose membranes were probed with 1.0 µg/ml of antibodies generated against MMP1, MMP2, MMP3 (R&D Systems), or actin (Sigma Chemical Company) as a loading control and processed as described previously,Citation42 using goat anti-mouse horseradish peroxidase-conjugated secondary antibody (Bio-Rad) followed by chemoluminescent detection using SuperSignal Pico Chemiluminescent Substrate (ThermoScientific).
Gelatin and casein zymography
Conditioned media from one-day, near confluent bMSC cultures was collected and concentrated as described above for western blotting and then analyzed by SDS-PAGE zymography relying on precast 12% casein and 10% gelatin gels (Biorad) to demonstrate MMP3 and MMP2 activity, respectively.Citation43
Antibody arrays
48 h conditioned media was collected from near confluent bMSCs cultures and sent to the antibody array testing service at Ray Biotech. A customized Quantibody® array was designed to measure levels of 8 soluble proteins, including MMP1, MMP2, MMP3, bFGF and VEGF-A in the media samples.
si-RNA knock-down
Transient knock-down of MMP1, MMP2, MMP3, bFGF and/or VEGF-A in bMSCs was accomplished using small interfering RNA (siRNA) constructs from Ambion (MMP1: ID s8849; MMP2: s8851; MMP3: s8853; bFGF: S223531; VEGF-A: S460) and Lipofectamine RNAiMAX reagent (Invitrogen), according to the manufacter’s recommendation. Silencer Select negative control #1 siRNA (Ambion #4390843) was used as a negative control. 48 h after transfection, cells were harvested for qRT-PCR validation and transwell invasion assays.
Transwell assays
Three transwell assay configurations were used: (1) bMSCs seeded on top of either polymerized collagenCitation44 or Matrigel® 30 to assess the interstitial and basement membrane matrix degrading potential of these resident MSCs, respectively. As a reference, highly invasive MDA-MB231 human breast cancers cells were included in the assays; (2) HMT-3522 S3 or T4-2 breast epithelial cells seeded on top of an insert coated with basement membrane Matrigel® (1 h at room temperature with 7.0% growth factor-reduced Matrigel®, BD Biosciences, in a total media volume of 20 μl)Citation30 and either media or bMSCs in the bottom well to determine whether bMSCs promote the invasion of breast cancer cells of the basal-type; and (3) HUVEC seeded on top of uncoated transwell inserts with bMSCs in the bottom chamber to assess the ability of local MSCs to chemoattract endothelial cells. MSC cells with transient knock-down of candidate mediators (as described above) or scrambled controls were seeded on either the top or bottom chambers depending on the particular transwell assay configuration. In the case of VEGF-A, where the efficiency of knock-down was typically 65–70%, bMSCs were incubated with a neutralizing antibody for VEGF-A (R&D Systems, MAB293; 10 μg/35 mm dish) or IgG1 (R&D Systems, MAB002; 10 μg/35 mm dish) 24 h after transient transfection and subjected to an endothelial migration assay (described below) in the continued presence of either VEGF-A antibody or IgG1. For all transwell assays, samples were tested in triplicate transwell inserts (24-well format; 8-microns; BD Biosciences), seeding 100,000 cells in 200 μl media on the top of the inserts and 250,000 of the respective cells (or just media) in the lower wells of a multi-cluster dish. Inserts for endothelial migration assays were processed after 18–24 h and for invasion assays processing was at 44–48 h using a standard procedure.Citation30
Branching morphogenesis assay
bMSCs, HUVECs (positive control) or BJ fibroblasts (negative control) were cultured on a Matrigel® overlayCitation45 using their respective growth media. Cultures were monitored daily under light microscopy for vessel-like structures. At end points of 2, 4 and 6 d, cells were fixed for 10 min in 4.0% paraformaldehyde at room temperature and then nuclei were labeled with DAPI for fluorescent imaging. These 3 cell types were also subcutaneously injected within Matrigel® into athymic nude mice (3 million cells in a 150 μl volume of Matrigel® diluted 1:1 in media/injection × 2 injections per mouse). 2, 4, 6 and 14 d post-injection, mice were sacrificed, xenografts excised and then specimens were either processed for routine histology or used for RNA isolation. To confirm differentiation along an endothelial lineage, qRT-PCR was conducted to measure mRNA levels of e-NOS (forward primer: TGATGGCGAAGCGAGTGAAG; reverse primer: CTGCTGTGCGTAGCTCTGG), as described above.
Analysis of tumor formation
Female Hsd athymic nude mice (strain: Foxnlnu) were purchased from Harlan Laboratories and cared for in accordance with international guidelines (Institutional Animal Care and Use Committee, Virginia Commonwealth University, Richmond, VA; protocol #AD20203). Athymic nude mice rather than SCID mice were selected since HMT-3522 S3 and T4-2 cells have been extensively studied in this host.Citation30 Number 4 inguinal fat pads were cleared at 3 weeks of age and then immediately inoculated with 2 million HMT-3522 S3 or T4-2 cells ± 2 million bMSCs in a total volume of 30 μl. For subcutaneous injections, 3 million S3 or T4-2 cells ± 2 million breast MSCs were injected in 50% Matrigel® in a total volume of 150μl into the rear flanks of 7–8 week old mice. Eight weeks after xenotransplantation, the animals were sacrificed, at which time tumors were excised, measured, fixed in 10% formaldehyde, paraffin-embedded and then sectioned for histological staining (H&E and Masson Trichrome) and Ki-67 immunohistochemistry (see below). Initially, two H&E sections per xenograft were analyzed by a board-certified pathologist (MI). Invasion was defined as extension/invasion of the tumor beyond the initial area of injection, the pseudocapsule, or cyst wall. For those tumors with no evidence of microscopic invasion, additional tissue sectioning was performed. This involved H&E staining every tenth tissue section for up to a total of 10 sections per specimen. The number of peri-tumoral and intra-tumoral vessels per low power field (LPF) was determined using H&E stained tissue sections. When possible, 10 fields of stroma and 10 fields of tumor tissue were evaluated for each xenograft. For many pure T4-2 xenografts, intra-tumoral and peri-tumoral vascularity was determined based on fewer than 10 microscopic fields of each due to the small specimen size and modest desmoplastic response.
Ki-67 immunohistochemistry
Tissue sections were processed according to a routine immunohistochemical staining procedure.Citation42 The Ki-67 primary antibody (1:50 dilution of H-300; Santa Cruz Biotechnologies, Santa Cruz, CA) was applied to the tissue sections for overnight incubation at 4°C. Omission of the primary antibody was included as a negative control for each specimen. All images of Ki-67 immunostained xenografts were magnified by Nikon Eclipse E600 and captured by Nikon Digital Camera DXM1200F, using the Nikon ACT-1 image-editing program. Proliferation indices were determined based on three random microscopic fields per xenograft.
Statistical analysis
Statistical analysis of Ki-67 proliferation indices, the mean number of cells per microscopic fields (i.e., transwell invasion and migration assays) and the mean number of blood vessels per low powered field involved pairwise comparison using a 2-tailed student t-test with a p value 0.05 deemed statistically significant.
| Abbreviations: | ||
| ASCs | = | adipose stromal cells |
| bFGF | = | basic fibroblast growth factor |
| BM-MSCs | = | bone marrow-derived mesenchymal stem cells |
| bMSCs | = | breast mesenchymal stem cells |
| EGF | = | epidermal growth factor |
| eNOS | = | endothelial-type nitric oxide synthase |
| FBS | = | fetal bovine serum |
| H&E | = | hematoxylin and eosin |
| HUVEC | = | human umbilical vein endothelial cells |
| IGF2 | = | insulin-like growth factor 2 |
| IL1RA | = | interleukin-1 receptor antagonist |
| IPKB | = | Ingenuity Pathways Knowledge Base |
| LPF | = | low power field |
| MMP | = | matrix metalloproteinase |
| MSC | = | mesenchymal stem cell |
| PPIA | = | peptidylprolyl isomerase A (cyclophilin A) |
| PD | = | population doubling |
| RQ | = | relative quantitation |
| siRNA | = | small interfering RNA |
| VEGF-A | = | vascular endothelial growth factor A |
Additional material
Download Zip (431.5 KB)Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Peter Ma for his assistance in the isolation of bMSCs, Rachel Shin and Robert Elmore for assisting with transwell assay cell counts, Dr. Mina Bissell (Lawrence Berkeley National Laboratory) for providing HMT-3522 T4-2 cells and Dr. Jennifer Wayne (Virginia Commonwealth University) for providing BM-MSC cultures. We are also appreciative of the excellent service provided by the Anatomic Pathology Research Service and histotechnology at Virginia Commonwealth University. Microscopy was performed at the Virginia Commonwealth University Dept. of Anatomy and Neurobiology Microscopy Facility, supported, in part, with funding from NIH-NINDS Center core grant 5P30NS047463. Services and products in support of the research project were generated by the VCU Massey Cancer Center Flow Cytometry Shared Resource, supported in part with funding from NIH-NCI Cancer Center Support Grant P30 CA016059.
This work was supported by a National Institutes of Health KO1 CA105050–01A131, a Department of Defense BCRP Concept Award (BC085416), a Department of Defense BCRP Idea Award (BC094734), the Jeffress Memorial Trust and the Department of Pathology at Virginia Commonwealth University (to LWE). Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the NIH or DOD.
References
- Radisky ES, Radisky DC. Stromal induction of breast cancer: inflammation and invasion. Rev Endocr Metab Disord 2007; 8:279 - 87; http://dx.doi.org/10.1007/s11154-007-9037-1; PMID: 17447144
- Egeblad M, Littlepage LE, Werb Z. The fibroblastic coconspirator in cancer progression. Cold Spring Harb Symp Quant Biol 2005; 70:383 - 8; http://dx.doi.org/10.1101/sqb.2005.70.007; PMID: 16869775
- Zhao M, Dumur CI, Holt SE, Beckman MJ, Elmore LW. Multipotent adipose stromal cells and breast cancer development: Think globally, act locally. Mol Carcinog 2010; 49:923 - 7; http://dx.doi.org/10.1002/mc.20675; PMID: 20842668
- Cleary MP, Grossmann ME. Minireview: Obesity and breast cancer: the estrogen connection. Endocrinology 2009; 150:2537 - 42; http://dx.doi.org/10.1210/en.2009-0070; PMID: 19372199
- Roberts DL, Dive C, Renehan AG. Biological mechanisms linking obesity and cancer risk: new perspectives. Annu Rev Med 2010; 61:301 - 16; http://dx.doi.org/10.1146/annurev.med.080708.082713; PMID: 19824817
- Boyd NF, Martin LJ, Chavez S, Gunasekara A, Salleh A, Melnichouk O, et al. Breast-tissue composition and other risk factors for breast cancer in young women: a cross-sectional study. Lancet Oncol 2009; 10:569 - 80; http://dx.doi.org/10.1016/S1470-2045(09)70078-6; PMID: 19409844
- Millikan RC, Newman B, Tse CK, Moorman PG, Conway K, Dressler LG, et al. Epidemiology of basal-like breast cancer. Breast Cancer Res Treat 2008; 109:123 - 39; http://dx.doi.org/10.1007/s10549-007-9632-6; PMID: 17578664
- Rakha EA, Ellis IO. Triple-negative/basal-like breast cancer: review. [review] Pathology 2009; 41:40 - 7; http://dx.doi.org/10.1080/00313020802563510; PMID: 19089739
- Manabe Y, Toda S, Miyazaki K, Sugihara H. Mature adipocytes, but not preadipocytes, promote the growth of breast carcinoma cells in collagen gel matrix culture through cancer-stromal cell interactions. J Pathol 2003; 201:221 - 8; http://dx.doi.org/10.1002/path.1430; PMID: 14517839
- Jotzu C, Al E, Welte G, Li J, Hennessy BT, Devarajan E, et al. Adipose tissue-derived stem cells differentiate into carcinoma-associated fibroblasts-like cells under the influence of tumor-derived factors. Anal. Cell Path. (Amst) 2010; 33:61 - 79
- Bulun SE, Sharda G, Rink J, Sharma S, Simpson ER. Distribution of aromatase P450 transcripts and adipose fibroblasts in the human breast. J Clin Endocrinol Metab 1996; 81:1273 - 7; http://dx.doi.org/10.1210/jc.81.3.1273; PMID: 8772611
- Ginsburg OM, Martin LJ, Boyd NF. Mammographic density, lobular involution and risk of breast cancer. Br J Cancer 2008; 99:1369 - 74; http://dx.doi.org/10.1038/sj.bjc.6604635; PMID: 18781174
- Dumitrescu RG, Cotarla I. Understanding breast cancer risk-where do we stand in 2005?. J. Cell. Mol. Bio. 2005; 9:208 - 21
- Rahimi N, Tremblay E, McAdam L, Roberts A, Elliott B. Autocrine secretion of TGF-beta 1 and TGF-beta 2 by pre-adipocytes and adipocytes: a potent negative regulator of adipocyte differentiation and proliferation of mammary carcinoma cells. In Vitro Cell Dev Biol Anim 1998; 34:412 - 20; http://dx.doi.org/10.1007/s11626-998-0023-z; PMID: 9639104
- Zuk PA, Zhu M, Mizuno H, Huang J, Futrell JW, Katz AJ, et al. Multilineage cells from human adipose tissue: implications for cell-based therapies. Tissue Eng 2001; 7:211 - 28; http://dx.doi.org/10.1089/107632701300062859; PMID: 11304456
- Zuk PA, Zhu M, Ashjian P, De Ugarte DA, Huang JI, Mizuno H, et al. Human adipose tissue is a source of multipotent stem cells. Mol Biol Cell 2002; 13:4279 - 95; http://dx.doi.org/10.1091/mbc.E02-02-0105; PMID: 12475952
- Gimble JM, Nuttall ME. Adipose-derived stromal/stem cells (ASC) in regenerative medicine: pharmaceutical applications. Curr Pharm Des 2011; 17:332 - 9; http://dx.doi.org/10.2174/138161211795164220; PMID: 21375497
- Caimi PF, Reese J, Lee Z, Lazarus HM. Emerging therapeutic approaches for multipotent mesenchymal stromal cells. Curr Opin Hematol 2010; 17:505 - 13; http://dx.doi.org/10.1097/MOH.0b013e32833e5b18; PMID: 20729733
- Schäffler A, Büchler C. Concise review: adipose tissue-derived stromal cells--basic and clinical implications for novel cell-based therapies. Stem Cells 2007; 25:818 - 27; http://dx.doi.org/10.1634/stemcells.2006-0589; PMID: 17420225
- Karnoub AE, Dash AB, Vo AP, Sullivan A, Brooks MW, Bell GW, et al. Mesenchymal stem cells within tumour stroma promote breast cancer metastasis. Nature 2007; 449:557 - 63; http://dx.doi.org/10.1038/nature06188; PMID: 17914389
- Ishii G, Sangai T, Oda T, Aoyagi Y, Hasebe T, Kanomata N, et al. Bone-marrow-derived myofibroblasts contribute to the cancer-induced stromal reaction. Biochem Biophys Res Commun 2003; 309:232 - 40; http://dx.doi.org/10.1016/S0006-291X(03)01544-4; PMID: 12943687
- Goldstein RH, Reagan MR, Anderson K, Kaplan DL, Rosenblatt M. Human bone marrow-derived MSCs can home to orthotopic breast cancer tumors and promote bone metastasis. Cancer Res 2010; 70:10044 - 50; http://dx.doi.org/10.1158/0008-5472.CAN-10-1254; PMID: 21159629
- Muehlberg FL, Song YH, Krohn A, Pinilla SP, Droll LH, Leng X, et al. Tissue-resident stem cells promote breast cancer growth and metastasis. Carcinogenesis 2009; 30:589 - 97; http://dx.doi.org/10.1093/carcin/bgp036; PMID: 19181699
- Sun B, Roh KH, Park JR, Lee SR, Park SB, Jung JW, et al. Therapeutic potential of mesenchymal stromal cells in a mouse breast cancer metastasis model. Cytotherapy 2009; 11:289 - 98, 1, 298; http://dx.doi.org/10.1080/14653240902807026; PMID: 19308770
- Welte G, Alt E, Devarajan E, Krishnappa S, Jotzu C, Song YH. Interleukin-8 derived from the local tissue-resident stromals cells promotes cell invasion. Mol. Carcinogenesis 2011;
- Razmkhah M, Jaberipour M, Erfani N, Habibagahi M, Talei AR, Ghaderi A. Adipose derived stem cells (ASCs) isolated from breast cancer tissue express IL-4, IL-10 and TGF-β1 and upregulate expression of regulatory molecules on T cells: do they protect breast cancer cells from the immune response?. Cell Immunol 2011; 266:116 - 22; http://dx.doi.org/10.1016/j.cellimm.2010.09.005; PMID: 20970781
- Mishra PJ, Mishra PJ, Humeniuk R, Medina DJ, Alexe G, Mesirov JP, et al. Carcinoma-associated fibroblast-like differentiation of human mesenchymal stem cells. Cancer Res 2008; 68:4331 - 9; http://dx.doi.org/10.1158/0008-5472.CAN-08-0943; PMID: 18519693
- Haviv I, Polyak K, Qiu W, Hu M, Campbell I. Origin of carcinoma associated fibroblasts. Cell Cycle 2009; 8:589 - 95; http://dx.doi.org/10.4161/cc.8.4.7669; PMID: 19182519
- Zhang Y, Daquinag A, Traktuev DO, Amaya-Manzanares F, Simmons PJ, March KL, et al. White adipose tissue cells are recruited by experimental tumors and promote cancer progression in mouse models. Cancer Res 2009; 69:5259 - 66; http://dx.doi.org/10.1158/0008-5472.CAN-08-3444; PMID: 19491274
- Rizki A, Weaver VM, Lee SY, Rozenberg GI, Chin K, Myers CA, et al. A human breast cell model of preinvasive to invasive transition. Cancer Res 2008; 68:1378 - 87; http://dx.doi.org/10.1158/0008-5472.CAN-07-2225; PMID: 18316601
- Rundhaug JE. Matrix metalloproteinases and angiogenesis. J Cell Mol Med 2005; 9:267 - 85; http://dx.doi.org/10.1111/j.1582-4934.2005.tb00355.x; PMID: 15963249
- Blasi A, Martino C, Balducci L, Saldarelli M, Soleti A, Navone SE, et al. Dermal fibroblasts display similar phenotypic and differentiation capacaity to fat-derived mesenchymal stem cells, but differ in anti-inflammatory and angiogenic potential. Vacular Cell 2011; 3:5; http://dx.doi.org/10.1186/2045-824X-3-5
- Kinnaird T, Stabile E, Burnett MS, Lee CW, Barr S, Fuchs S, et al. Marrow-derived stromal cells express genes encoding a broad spectrum of arteriogenic cytokines and promote in vitro and in vivo arteriogenesis through paracrine mechanisms. Circ Res 2004; 94:678 - 85; http://dx.doi.org/10.1161/01.RES.0000118601.37875.AC; PMID: 14739163
- Sumi M, Sata M, Toya N, Yanaga K, Ohki T, Nagai R. Transplantation of adipose stromal cells, but not mature adipocytes, augments ischemia-induced angiogenesis. Life Sci 2007; 80:559 - 65; http://dx.doi.org/10.1016/j.lfs.2006.10.020; PMID: 17157325
- Kang Y, Park C, Kim D, Seong CM, Kwon K, Choi C. Unsorted human adipose tissue-derived stem cells promote angiogenesis and myogenesis in murine ischemic hindlimb model. Microvasc Res 2010; 80:310 - 6; http://dx.doi.org/10.1016/j.mvr.2010.05.006; PMID: 20510252
- Cao Y, Sun Z, Liao L, Meng Y, Han Q, Zhao RC. Human adipose tissue-derived stem cells differentiate into endothelial cells in vitro and improve postnatal neovascularization in vivo. Biochem Biophys Res Commun 2005; 332:370 - 9; http://dx.doi.org/10.1016/j.bbrc.2005.04.135; PMID: 15896706
- Liao D, Johnson RS. Hypoxia: a key regulator of angiogenesis in cancer. Cancer Metastasis Rev 2007; 26:281 - 90; http://dx.doi.org/10.1007/s10555-007-9066-y; PMID: 17603752
- de Ferranti S, Mozaffarian D. The perfect storm: obesity, adipocyte dysfunction and metabolic consequences. Clin Chem 2008; 54:945 - 55; http://dx.doi.org/10.1373/clinchem.2007.100156; PMID: 18436717
- Francis MP, Sachs PC, Elmore LW, Holt SE. Isolating adipose-derived mesenchymal stem cells from lipoaspirate blood and saline fraction. Organogenesis 2010; 6:11 - 4; http://dx.doi.org/10.4161/org.6.1.10019; PMID: 20592860
- Forsythe HL, Elmore LW, Jensen KO, Landon MR, Holt SE. Retroviral-mediated expression of telomerase in normal human cells provides a selective growth advantage. Int J Oncol 2002; 20:1137 - 43; PMID: 12011990
- Dumur CI, Ladd AC, Wright HV, Penberthy LT, Wilkinson DS, Powers CN, et al. Genes involved in radiation therapy response in head and neck cancers. Laryngoscope 2009; 119:91 - 101; http://dx.doi.org/10.1002/lary.20005; PMID: 19117295
- Diehl MC, Idowu MO, Kimmelshue KN, York TP, Jackson-Cook CK, Turner KC, et al. Elevated TRF2 in advanced breast cancers with short telomeres. Breast Cancer Res Treat 2011; 127:623 - 30; http://dx.doi.org/10.1007/s10549-010-0988-7; PMID: 20625812
- Brown PD, Levy AT, Margulies IM, Liotta LA, Stetler-Stevenson WG. Independent expression and cellular processing of Mr 72,000 type IV collagenase and interstitial collagenase in human tumorigenic cell lines. Cancer Res 1990; 50:6184 - 91; PMID: 2169338
- Ellerbroek SM, Wu YI, Overall CM, Stack MS. Functional interplay between type I collagen and cell surface matrix metalloproteinase activity. J Biol Chem 2001; 276:24833 - 42; http://dx.doi.org/10.1074/jbc.M005631200; PMID: 11331272
- Debnath J, Muthuswamy SK, Brugge JS. Morphogenesis and oncogenesis of MCF-10A mammary epithelial acini grown in three-dimensional basement membrane cultures. Methods 2003; 30:256 - 68; http://dx.doi.org/10.1016/S1046-2023(03)00032-X; PMID: 12798140
- Wosnitza M, Hemmrich K, Groger A, Gräber S, Pallua N. Plasticity of human adipose stem cells to perform adipogenic and endothelial differentiation. Differentiation 2007; 75:12 - 23; http://dx.doi.org/10.1111/j.1432-0436.2006.00110.x; PMID: 17244018
- Lozito TP, Kuo CK, Taboas JM, Tuan RS. Human mesenchymal stem cells express vascular cell phenotypes upon interaction with endothelial cell matrix. J Cell Biochem 2009; 107:714 - 22; http://dx.doi.org/10.1002/jcb.22167; PMID: 19415687
- Oswald J, Boxberger S, Jørgensen B, Feldmann S, Ehninger G, Bornhäuser M, et al. Mesenchymal stem cells can be differentiated into endothelial cells in vitro. Stem Cells 2004; 22:377 - 84; http://dx.doi.org/10.1634/stemcells.22-3-377; PMID: 15153614