Abstract
Overexpression of DNA methyltransferase 1 (DNMT-1) is observed mostly in pancreatic cancer and it can cause tumor suppressor genes silencing in this disease. Recent studies suggest that abnormal expressions of microRNAs (miRs) are involved in pathogenesis of different types of human cancers including pancreatic cancer. In this study we aimed to investigate the effect of miR-148b and -152 on reverting the tumorigenic phenotype of pancreatic cancer cell lines.
In order to investigate whether miR-148b and -152 are involved in the regulation of DNMT-1, luciferase reporter assay was used and confirmed that the DNMT-1 mRNA could be a target for miR-148b and miR-152. Furthermore, overexpression of miR-148b and -152 in pancreatic cancer cell lines (MIA PaCa-2 and AsPC-1) decreased DNMT-1 expression (53% and 59% respectively), returned DNA methylation to normal patterns and induced re-expression of tumor suppressor genes, like BNIP3 (4.7- and 3.8-fold) and SPARC (5.3- and 2.9-fold) for miR-148b and -152 respectively. Moreover, the introduced miR-148b and -152 could inhibit the proliferation of MIA PaCa-2 (35% and 37% respectively) and AsPC-1 (39% and 40% respectively) cell lines. The apoptosis rates of MIA PaCa-1 after treatment with miR-148b and -152 were 10% and 8% respectively; while these rates in AsPC-1 were 16% and 11% respectively. Conclusively these findings mean that miRs that are targeting DNMT-1 and modifying methylation status of tumor suppressor genes such as BNIP3 and SPARC can be applied in killing the pancreatic cancer cells and decreasing the tumorigenicity of these cells.
Introduction
Pancreatic cancer (PC) is the fourth leading cause of cancer death in the USA. It is one of the most devastating cancers among all types of solid cancers. In 2013, it is estimated that 45 220 Americans will be diagnosed with PC.Citation1 One of the main factors contributing to the poor survival in PC is the lack of early diagnostic and prognostic markers. At the time of diagnosis, about 40% to 50% of patients have metastatic PC in advanced stages. Only 15% to 20% of patients suffer from small, non-metastatic disease that can be considered for resection. The 5-y survival rate is less than 5% after diagnosis. Additionally, this disease shows an intrinsic chemotherapeutic resistance and remains unresponsive to most chemotherapeutic agents.Citation2,Citation3 Therefore, a deeper understanding of the biological mechanisms responsible for the development of this type of cancer and its progression is necessary for implanting effective therapeutic modalities and early detection approaches.
Over the past decades, many studies have been devoted to discover genetic mutations involved in the pathogenesis of PC. These genes include K-RAS, CDKN1A/P16, TP53, SMAD4, DPC4, MADH4, and BRACA2.Citation4 Moreover in correlation with these genetic alterations, many changes in gene expression and specific signaling pathways (such as the aberrant activation of the Hedgehog and Wnt transduction) have been characterized in PC.Citation5,Citation6
Epigenetic changes, including DNA hypermethylation and hypomethylation, leading to transcriptional modifications, in the affected genes, have also been involved in PC development and progression.Citation7 These suggest that PC arise from accumulation of genetic and epigenetic alterations.
Recent works have verified that DNA methylations give rise to transcriptional silencing of tumor suppressor genes (TSG), thus it may play an important role during carcinogenesis.DNA methylation mainly occurs at the C5 position of CpG dinucleotides by three DNA methyltransferases (DNMTs), namely DNMT-1, DNMT3A, and DNMT3B. DNMT3A and 3B are responsible for de novo DNA methylation, while DNMT-1 is the “maintenance” methyltransferase that ensures faithful transmission of the methylation profile from maternal to daughter cells during cell division.Citation8 Overexpression of DNMT-1 has been reported in many types of cancer.Citation9,Citation10 In human cancer cells DNMT-1 can be blamed for both de novo and maintenance methylation of tumor suppressor genes.Citation11 The mechanism of overexpression of DNMT-1 has remained unclear. However, a study has shown that deletion of P53 gene in the HCT116 human colon carcinoma cell line resulted in increase of DNMT-1 transcript and translation.Citation12 Another report showed that dysregulation of p53/Sp1 could lead to DNMT-1 overexpression in lung cancer.Citation13 Analysis of clinical tissue samples revealed that increased in expression of DNMT-1 was significantly correlated with the malignant potential and poor prognosis of some human cancers.Citation14 Previous reports have demonstrated that the level of DNMT-1 expression in PC was considerably higher compared with their corresponding noncancerous mucosa. Also progressive dysregulation of DNMT-1 expression is observed during the development of the disease.Citation15,Citation16 However, the underlying mechanism of DNMT-1 overexpression in human cancer cells, especially in PC cells, remains unclear.
MicroRNAs (miRs) are a class of small non-coding RNAs which participate in posttranscriptional regulation of gene expression in eukaryotic organisms. miRs function via complete or partial matching to target mRNA at their 3′ untranslated regions (UTR). This usually results in gene silencing through translational repression or target degradation. Currently more than 1500 miRs have been identified in human being which are believed to influence and control the expression of approximately 30% of human genes and most genetic pathways.Citation17 It is becoming clear that deviations from normal pattern of miR expression are associated with various human diseases, like lung, colon and PCs.Citation18,Citation19 Hence mutations in miRs, dysfunction of their biogenesis, and dysregulation of their target interactions may play roles in carcinogenesis in pancreas.Citation19,Citation20
In the current study, we investigated the role of miR-148b and -152 in the regulation of DNMT-1 and whether it can return normal pattern methylation of TSGs in PC cells. Additionally, we characterized several factors, including modulation of the expression of TSGs after returning the methylation patterns of their promoters, as well as apoptosis induction and inhibition of PC cells proliferation.
Results
DNMT-1 is a direct target of miR-148b and miR-152
We analyzed the 3′-UTR of DNMT-1 as potential target sites for different miRNA binding sites in online tools. For this issue we combined bioinformatic algorithms including miRanda (http://www.microrna.org/microrna/searchMirnas.do),Citation21 PicTar (http://www.pictar.mdc-berlin.de/),Citation22 Microcosm (http://www.ebi.ac.uk/enright-srv/microcosm/htdocs/targets/v5/), and TargetScan 4.0 (http://www.targetscan.org/).Citation23 Interestingly, a group of miRs including miR-148b and -152 showed sequence complementarity to the same region in the 3′-UTR of DNMT-1 gene.
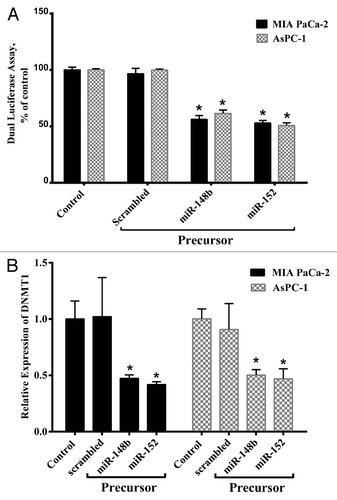
To verify these miRs target interactions, we cloned the DNMT-1 complementary site into the 3′-UTR of the Renilla luciferase gene in psiCHECK™-2. This plasmid was co-transfected with miR-148b and -152 and scrambled in MIA PaCa-2 and AsPC-1 cells. As shown in , miR-148b and -152 markedly reduced the luciferase activity compared with the scrambled oligonucleotide. Transfecting miR-148b and -152 into MIA PaCa-2 cell line decreased the luciferase activity to 44% ± 0.51% and 48% ± 0.41% respectively in comparison with the control (P < 0.05), while in AsPC-1 cell, the luciferase activity decreased to 41% ± 0.81% and 50% ± 0.71% respectively. These results suggested that DNMT-1 can be considered as a candidate target gene for miR-148b and -152 and also its 3′-UTR is a functional target site for these miRs in PC cells.
Figure 1. Mir-148b and -152 directly target DNMT-1. (A) MIA PaCa-2 and AsPC-1 cells were transfected with the Renilla and firefly luciferase expression construct psiCHECK™-2 harboring DNMT-1 3′-UTR and miR-148b, miR-152, or scrambled miR. After 24 h, Dual-Luciferase assays were performed and the decrease in relative Renilla luciferase activity was observed, while no effect was detected with the scrambled miR. (B) MIA PaCa-2 and AsPC-1 were transfected with miR-148b and -152 or scrambled. After 48 h, DNMT-1 expression was evaluated by real-time quantitative PCR. The decrease in relative mRNA expression was evident with miR-148b and miR-152, while no effect was detected with the scrambled miR. Data represents mean ± SE from 3 independent experiments performed in triplicates. *P < 0.05 vs. the control group.
MiR-148b and miR-152 have the ability to downregulate DNMT-1
To confirm the hypothesis that overexpression of miR-148b and -152 downregulate DNMT-1 mRNA expression in PC cell lines, we transfected pre-miR-148b, -152, or a scrambled oligonucleotide into MIA PaCa-2 and AsPC-1 and measured levels of DNMT-1 mRNA by quantitative real-time PCR. As shown in , the overexpression of miR-148b and -152 caused reductions in the level of DNMT-1 mRNA in MIA PaCa-2 to 0.47 ± 0.031 and 0.418 ± 0.025 respectively, and in AsPC-1 to 0.50 ± 0.049 and 0.46 ± 0.090 respectively.
Overexpression of miR-148b and -152 decreases the methylation level of BNIP3, SPARC, PENK, and TFPI-2 promoter regions in MIA PaCa-2 and AsPC-1 cell lines
The observation that miR-148b and -152 target DNMT-1 suggested that expression of these miRs contributes to the DNA epigenetic modifications of promoters of target genes (i.e., SPARC, BNIP3, TFPI-2, and PENK). In order to evaluate the effects of the methylation changes on gene expression, we first analyzed the methylation pattern of SPARC, BNIP3, TFPI-2, and PENK promoters by HRM curve analysis. During each HRM analysis standard methylated controls (0%, 25%, 50%, 75%, and 100%) were analyzed parallel to the samples ().
Figure 2. Normalized HRM standard curves and difference plot of Bnip3 gene. (A) Templates with different ratios (from 100% to 0%) of methylated DNA were amplified by BNIP3-specific primer and subjected to HRM analysis. (B) Difference plot of the same sample. Fluorescence of each control sample was normalized as differential signal against the unmethylated control. (C) Measurement of methylation status of BNIP3 gene in the MIA PaCa-2 cell line transfected with miR-148b. Fluorescence of each sample was normalized as differential signal against unmethylated control. After 48 h the percentage of DNA methylation for BNIP3 decreased from 100% to 50%. The difference in melt profiles are associated with different methylation states.
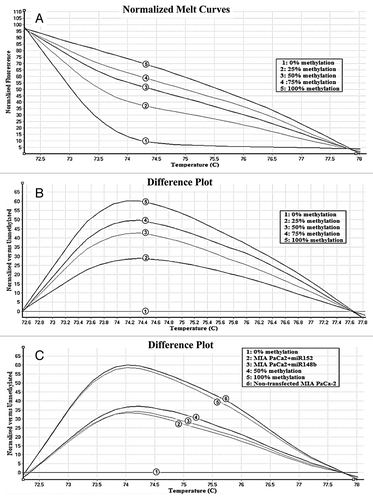
Overexpression of miR-148b and -152 decreases the DNA methylation levels of SPARC, BNIP3, TFPI-2, and PENK promoter regions in AsPC-1 and MIA PaCa-2 cells ().
Table 1. The effect of miR-148b and -152 on demethylation of TSG
Overexpression of miR-148b and -152 increases the expression level of SPARC and BNIP3 genes
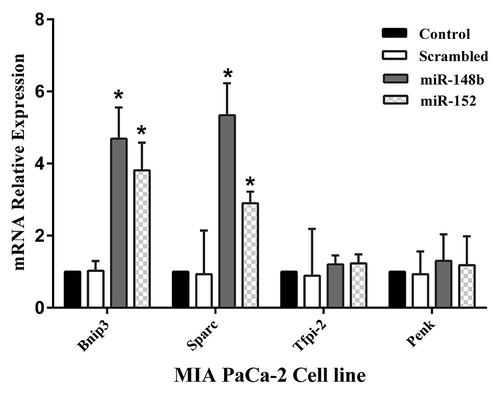
In the next step, in order to determine whether miR-148b and -152 regulate the expression level of SPARC, BNIP3, TFPI-2, and PENK genes by altering promoter methylation status of these genes, we examined the expression levels of these genes. Transfection of MIA PaCa-2 cells with precursors of miR-148b and -152 after 48 h increased the expression of BNIP3 to 4.7 ± 0.86 and 3.8 ± 0.76 respectively (P < 0.05) and SPARC to 5.3 ± 0.9 and 2.9 ± 0.32 respectively (P < 0.05) in comparison to non-transfected cells (). Similar effects of miR-148b and -152 were also seen in AsPC-1 cells. However in the expression level of PENK and TFPI-2 genes no significant change was seen.
Figure 3. MIA PaCa-2 and AsPC-1 were transfected with miR-148b and -152 or scrambled. After 48 h SPARC, BNIP3, TFPI-2, and PENK expression was evaluated by real-time quantitative PCR. MiR-148b and -152 induced increased expression of BNIP3 and SPARC genes, while these miRs showed no effect on TFPI-2 and PENK gene expression. Data represents mean ± SE from 3 independent experiments performed in triplicates. *P < 0.05 vs. the control group.
MiR-148b and -152 inhibit the proliferation of MIA PaCa-2 and AsPC-1 cells and induce apoptosis by targeting DNMT-1
In order to understand the functional role of miR-148b and -152, we evaluated the impact of these miRs on cellular proliferation using MTT proliferation assay in MIA PaCa-2 and AsPC-1 cells. As shown in transfecting miR-148b and -152 into MIA PaCa-2 and AsPC-1 cell line after 72 h decreased proliferation of these cells up to 35% ± 8% and 37% ± 9% (mean ± SE) respectively while these miRs in AsPC-1 cells decreased the proliferation up to 39% ± 13% and 40% ± 11% of control respectively (P < 0.05).We hypothesize that overexpression of miR-148b and -152 can inhibit PC cell proliferation.
Figure 4. MiR-148b and -152 induce cell death and apoptosis in MIA PaCa-2 and AsPC-1. (A) MIA PaCa-2 and AsPC-1 cells were transfected with 100 nM miR-148b and -152 precursors or scrambled. Cell proliferation was assessed by MTT proliferation assay. (B) Percentage of apoptosis is increased after transfection of mir-148b and -152. MIA PaCa-2 and AsPC-1 were transfected with miR-148b and miR-152 or scrambled miR. Two day later, the cells were stained with Annexin-PI. The rate of apoptosis in the cells increased significantly after treatment with miR-148b and miR-152, while no effect was detected with the scrambled. Data represents mean ± SE from 3 independently performed experiments. *P < 0.05 vs. the control group. (C) Representative histograms depicting apoptosis of MIA PaCa-2 cell transfected with miR-148b and -152 or scrambled.
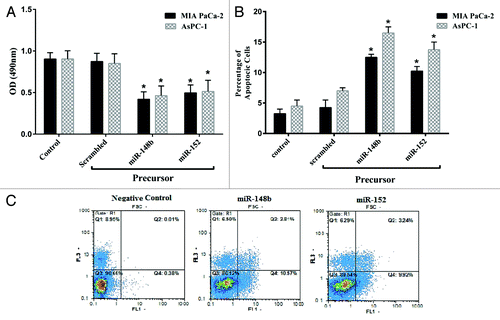
The effect of miR-148b and -152 on the apoptosis of MIA PaCa-2and AsPC-1 cells was evaluated by staining the cells with Annexin-PI. As shown in , the ectopic expression of miR-148b and-152 in comparison to control miR induced apoptosis up to 11% ± 0.7% and 8% ± 1% respectively in MIA PaCa-2 cells while these miRs induced apoptosis in AsPC-1 up to 16% ± 4% and 11% ± 1.5% respectively (P < 0.05). It can be hypothesized that overexpression of miR-148b and -152 can induce apoptosis and reduced cell proliferation in PC cell lines.
Discussion
Hypermethylation at CpG islands of promoters and inactivation of multiple TSGs are common event in pancreatic cancer and it contributes to tumor growth.Citation7,Citation24 In pancreatic cancer, silencing TSGs might be due to abnormal methylation of CpG islands within their promoter region.Citation7,Citation25 The underlying mechanism of hypermethylation in pancreatic cancer remains poorly understood.
One of the suggested mechanisms for the abnormal hypermethylation of these genes is dysregulated expression of DNMT-1 gene. Irregular DNMT-1 expression has been detected in several tumorsCitation26,Citation27 supporting a role for DNMT-1 in tumorigenesis. Previous studies have shown that DNMT-1 expression is upregulated in pancreatic cancerCitation15,Citation16 though the mechanism for DNMT-1 overexpression is not fully elucidated. Possible causes for DNMT-1 overexpression may be alterations in the expression of regulatory miRs.
There are several facts about the role of miRNAs in aberrant methylation of DNA through regulating DNMTs translation.Citation28 A previous study has proved that miR-29b can indirectly target DNMT-1 and induce global DNA hypomethylation and reexpression of TSGs in acute myeloid leukemia.Citation29
In the present study, we focused on the effect of miR-148b and -152 on pancreatic cancer cells. These miRs have shown to be downregulated in pancreatic cancer cells.Citation19,Citation30 In addition miR-148b and -152 have been predicted to show intriguing complementarity to 3′-UTR of DNMT-1, these predictions have been done by several in silico methods for target gene prediction such as MiRanda,Citation21 PicTar,Citation22 and TargetScan3.1.Citation23
Our study showed that miR-148b and -152 exert its function by specifically targeting DNMT-1 in pancreatic cancer cell lines, and upregulation of these miRs downregulates DNMT-1 (). While overexpression of miR-148b and -152 inhibited the expression of DNMT-1, luciferase activity assay provided evidences that DNMT-1 was a direct target of miR-148b and -152. On the other hand miR-148b and miR-152 are specifically involved in the regulation of DNMT-1 expression. These results are in accordance with Braconi et al. study proving the role of these miRs on controlling DNMT-1 expression in cholangiocarcinoma.Citation31 Another study showed that mir-342 targeted DNMT-1 in colorectal cancer.Citation32 DNMT-1 may be regulated by different miRs in various cell types. This hypothesis is consistent with the proposed combinatorial miRNA target regulation, in which a given miRNA may have multiple different mRNA targets, and a given target might similarly be targeted by multiple miRs.Citation23
Among different DNMTs, DNMT-1 contributes much more to the maintenance of methylation status in the chromosomes hence deregulated expression of DNMT-1 may play a causal role in cancer progression.Citation11,Citation33
In human pancreatic cancer, decreased expression of miRNAs such as miR-148b and -152 can contribute to increased level of DNMT-1. This phenomenon can subsequently lead to inhibition of critical TSGs through altered promoter methylation. Hence we provide insights about whether miR-148b and -152 are involved in the control of DNA methylation by targeting DNMT-1 in pancreatic cancer cells. In our study, we focused on four TSGs, namely, BNIP3, SPARC, PENK, and TFPI-2, in which another studies were observed to be silenced.Citation34-Citation37 Then the methylation level of these genes’ promoters was studied in MIA PaCa-2 and AsPC-1 cells by high resolution melting curve analysis. As shown in , inhibition of DNMT-1 by miR-148b and -152 decreased the methylation level of their promoters, which leads to reexpression of BNIP3 and SPARC genes (). These results indicate that DNMT-1 is necessary and sufficient to maintain aberrant CpG island methylation of BNIP3 and SPARC genes and miR-148b and -152 can restore the hypomethylation status of BNIP3 and SPARC TSGs by targeting DNMT-1 in pancreatic cancer cells. However we did not detect reexpression inTFPI-2 and PENK genes at the mRNA level. One possible cause for this result is the effect of histone modification on the silencing of PENK and TFPI-2 in addition to aberrant promoter methylationCitation38 which could not be restored by miR-148b and miR-152.
MiRNAs have recently been demonstrated to potentially play a significant role in tumorigenesis. Since miRs are negative regulators of gene expression, changes in the amounts of these non-coding RNAs can be tumorigenic if they target mRNAs for either a tumor suppressor or an oncogene.Citation39 To date, hundreds of different miRs regulate many known oncogenic and TSGs involved in the pathogenesis of cancer. It has been shown that miR-148b and -152 are downregulated in various types of cancers including pancreatic cancer.Citation40,Citation41 Reduced expression of these miRs in pancreatic cancer suggests a potential role of these miRs in tumorigenesis. We showed that introducing these miRs suppressed cell growth and induced apoptosis in these cell lines (). These findings suggest that miR-148b and -152 play the tumor-suppressive role. Another possible justification for this phenomenon is partly through decreasing DNMT-1 expression. According to this result, Bitzer et al. reported that inhibition of DNMT-1 through 5-azacytidine in human hepatoma, colon, renal, and lung cancer cells caused apoptosis.Citation42
The demethylation and reexpression of TSGs such as BNIP3 and SPARC after reducing DNMT-1 expression may be responsible for decreased cell proliferation in pancreatic cancer cells.
BNIP3 belongs to the Bcl-2 family and the Bcl-2 homology domain-3-only subfamily.Citation43 Induced expression of BNIP3 has been shown to lead to cell death in Rat-1 fibroblasts and MCF-7 breast carcinoma cells.Citation44 Thus, BNIP3 is considered to be a key regulator of hypoxia-induced cell death. Okamiet al. reported that restoration of Bnip3 in pancreatic cells caused cell death.Citation34 On the other hand SPARC is a TSG that is downregulated in pancreatic cancer cells. Restoration of this gene with miRNA-148b and -152 may also cause growth suppression and apoptosis induction. Yiu et al. have shown that treatment of ovarian cancer cells with exogenous SPARC inhibits cell proliferation and induces apoptosis.Citation45 In addition, expression of SPARC in ovarian cancer cells resulted in reduced tumorigenicity in nude mice, suggesting that SPARC has a tumor-suppressor function.Citation46 In the previous reports, addition of exogenous Sparc protein inhibited the proliferation of pancreatic cancer cells,Citation35,Citation47 suggesting a growth-inhibitory effect of Sparc.
In summary, our results revealed that overexpression of the DNMT-1 in pancreatic cancer cells could be the result of decreased levels of miR-148b and -152. Through downregulating DNMT-1 expression, miR-148b and -152 led to reactivation of TSGs via decreasing methylation of promoters.
Furthermore, miR-148b and -152 can significantly suppress pancreatic cell proliferation and induce apoptosis. Results of this study offer a robust motivation for developing epigenetic therapies that use synthetic miRNAS such as miR-148b and -152, alone or in combination with other treatments, to reactivate TSGs and normalize abnormal patterns of methylation in pancreatic cancer cells. Because decreased expression of miR-148b and -152 is detected in other common human cancers, this approach may be considered for other types of cancers too.
Materials and Methods
Cell culture
MIA PaCa-2 and AsPC-1, human PC cell lines were obtained from National Cell Bank of Iran (Pasteur Institute of Iran). The MIA PaCa-2cells were grown in Dulbecco’s modified Eagle’s medium (DMEM), (Life Technology, 12100046) and AsPC-1 cells were grown in Roswell Park Memorial Institute (RPMI), 1640 medium (Life Technology, 11875119). Media were supplemented with l-glutamine, antibiotics (100 units/mL of penicillin and 100 µg/mL of streptomycin) and 10% fetal bovine serum (Life Technology, 1600044). The cells were maintained at 37 °C in 5% CO2.
MicroRNA transfections
Pre-miR miRNA precursors of pre-miR-148b, pre-miR-152, and control pre-miR precursor (scrambled) were purchased from Ambion (Life Technology, AM17102). Each well of a 24-well plate was seeded by 0.8 × 106 cells 1 d before transfection. When the confluence of seeded cells reached to 80%, they were transfected with 100 nM of synthetic pre-miR-148b, -152, or scrambled oligonucleotide (Ambion) using Lipofectamine 2000 (Life Technology, 11668019), according to the manufacturer protocol. After 6 h, the transfection medium was replaced with fresh medium containing 10% FBS. Transfected cell lines were harvested 24, 48, and 72 h post-transfection.
Luciferase assay
For luciferase reporter experiments, 3′-UTR of DNMT-1 (region 5079–5400, in NM_001130823) was amplified using the following primers: CTCGAGACTA GTTCTGCCCT CCCGTCAC (forward), and GCGGCCGCCA AATCACGAAT ACCCAC (reverse). Bold sequences indicate the endonuclease restriction site. PCR was performed in a total volume of 25 µL with 100 ng of genomic DNA, 10 pmol of each primer, 2 mM MgCl2, 200 mM dNTP, and 1.5 unit Pfu polymerase (Fermentas, EP0501), under the following conditions: 95 °C for 5 min; and 30 cycles of 95 °C for 1 min, 57 °C for 30 s, 72 °C for 1 min, and final extension in 72 °C for 10 min.
After Taq polymerase (Cinnagen) treating, The PCR product was cloned into pTZ57R/T (Fermentas, K1214). Insertion of the PCR product into pTz57R/T was confirmed by colony-PCR with M13 forward and reverse primers.
The product was then digested out from T vector with NotI (Fermentas, ER0571) and Xho1 (Fermentas, ER0691) and inserted into psiCHECK™-2 Vector (Promega, C8021) immediately downstream to the stop codon of renilla luciferase.
Luciferase assays were performed with Dual-Glo® Luciferase Assay System (Promega, E2940). In order to perform this test, 4–5 × 104 of MIA PaCa-2 and AsPC-1 cells were plated into each well of a 48-well plate 1 d before transfection. The cells were transfected with 400 ng psiCHECK™-2 Vector harboring DNMT-1 3′-UTR and 100 nM of each precursor miRs (148b, 152, or scrambled miRs) using Lipofectamine 2000 reagent (Life Technology) according to the manufacturer protocol. Luciferase activities were measured 24 h after transfection using dual-luciferase reporter assay system (Promega). The multiwell plate luminometer Renilla luciferase activity was normalized to that of firefly luciferase.
Genomic DNA bisulfate modification
Genomic DNA was isolated from MIA PaCa-2 and AsPC-1after treatment with each precursor miRs (148b, 152, or scrambled miRs) using the DNeasy® Blood and Tissue Kit (Qiagen, 69581). One and a half micrograms of genomic DNA was modified using EZ DNA methylation Gold kit (Zymo Research, D5006) according to the manufacturer’s instructions. The eluted DNA (10 µL) was used for high resolution melting (HRM) curve analysis. EpiTect Control DNAs (Qiagen, 59655) was used as 100% methylated and unmethylated controls.
Primer design for HRM and real-time PCR analysis
Primers for SPARC, BNIP3, TFPI-2, and PENK were designed with MethylPrimer Express V1.0 software (ABI) and Methprimer (http://www.urogene.org/methprimer/index.html). Then BLAST was performed using methBLAST (http://www.medgen.ugent.be/methBLAST).
Primers for real-time PCR were designed with AlleleID 6. To determine the specificity of each primer BLAST was performed using NCBI Nucleotide software (http://blast.ncbi.nlm.nih.gov/) (). All oligos were purchased from Metabion intentional AG (Lena-Christ-Strasse).
Table 2. Primer sequence for HRM and real-time PCR analysis
HRM analysis
The bisulfite modified DNA was amplified using primer pairs () that specifically target the methylated sequences of the target genes. Standard controls (25%, 50%, and 75%) were made by diluting 100% methylated with different ratios of unmethylated bisulfite-modified control DNA (Qiagen). These standards were included in each experimental run. The reaction mixture was made up of 10 µL Meltdoctor master mix (Life Technology, 4415440), 10 ng of bisulfite-treated DNA, 200 nM of each primer, and PCR grade water in a final volume of 20 μL. PCR cycling and HRM were performed on a Rotor-Gene 6000TM (Corbett Research).
All reactions were performed according to the following conditions: one cycle of 95 °C for 15 min; 40 cycles of 95 °C for 15 s, 58 °C for 30 s, 72 °C for 20 s; and a final denaturation step from 65 °C to 90 °C at intervals (ramps) of 0.1 °C/s.
HRM-curve analysis was performed using the Rotor Gene 6000 software. A differential profile was then evaluated for each sample by comparing fluorescence at the melting point against the value of fluorescence of the unmethylated DNA. All experiments were performed in triplicate. Melting curves were normalized using the HRM software before and after the major fluorescence decrease.
RNA isolation, cDNA synthesis, and quantitative real-time PCR
Total RNA was isolated from MIA PaCa-2 and AsPC-1after transfection with each precursor miRs (148b, 152, or scrambled miRs) using RiboPurTM kit (Life Technology, AM1924). RNA concentration was determined by spectrophotometer (IMPLEN,) and RNA quality was evaluated by determining the A260/A280 ratio. cDNA was synthesized by M-MuLV reverse transcriptase enzyme with 1 µg RNA using the cDNA synthesis kit (Fermentas, EP0351) according to the manufacturer’s recommendations. In each experiment, for evaluation of the DNA contamination, 2 RNA samples selected for negative controls, in which no reverse transcriptase added.
Twenty microliters of reverse transcription products were diluted (1:5) and 1 to 2 µL of them were used for real-time PCR. Real-time PCR reactions were performed in a volume of 25 µL containing 140 ng of specific primers () and 12.5 µL SYBR green master mix (Life Technology, 4309155) in the following condition:
Initial denaturing: 95 °C for 10 min, followed by 40 cycles of denaturing at 95 °C for 30 s, annealing at 59 °C for 40 s, and extension at 72 °C for 30 s. Beta actin was used as an internal reference gene. Statistical analysis for relative mRNA expression was performed by REST proposed by Pfaffl.Citation48 Fold change and P value (< 0.05) were estimated by REST, where all requirements of this software was considered.
For each primer, the efficiency of amplification was examined by measuring the degree of amplification from a serially 4-fold diluted cDNA template. A standard curve containing 5 dilution points was measured in triplicate. All PCR assays displayed efficiencies between 1.8 and 2.0.
Cell proliferation analysis
Cell proliferation assay was performed using 3-(4, 5-dimethylthiazole-2-yl)-2, 5-biphenyl tetrazolium bromide (MTT) (Sigma, M2128). Briefly, MIA PaCa-2 and AsPC-1 cells (3 × 104 cells/well in 96-well microplates) were transfected with pre miR-148b, -152, or negative control. After 72 h cells were incubated by MTT (500 µg/mL), the supernatants were discarded and the insoluble formazan was dissolved by dimethylsulphoxide (DMSO). The optical density was measured at 540 nm (reference wavelength: 630 nm) and then cell viability was defined relative to untreated control cells; viability = 100 × (absorbance of treated sample) / (absorbance of control). All experiments were repeated 3 times, and the average results were calculated.
Apoptosis assay
MIA PaCa-2 and AsPC-1 cells (1 × 104 for each well) were seeded into 24-well plate one day before transfection. Thereafter scrambled oligonucleotides, pre-miR-148b or -152 were transfected at a final concentration of 100 nM, by Lipofectamine 2000 (Life Technology), according to the manufacturer’s protocol. After 48 h, cells were harvested and resuspended in binding buffer containing annexin V–FITC and propidium iodide according to the instructions of the supplier (Roche Applied Sciences, 11858777001) and were analyzed by flow cytometery. Each sample was run in triplicate.
Statistical methods
All experiments were repeated at least three times. Results are expressed as the mean ± standard error (SE). Differences between groups were analyzed by analysis of variance (ANOVA) with subsequent post hoc Tukey test for sub-two group comparison. P value < 0.05 was considered significant.
Funding
Funding for this research was provided by the Pasteur Institute of Iran.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin 2013; 63:11 - 30; http://dx.doi.org/10.3322/caac.21166; PMID: 23335087
- Butturini G, Stocken DD, Wente MN, Jeekel H, Klinkenbijl JH, Bakkevold KE, Takada T, Amano H, Dervenis C, Bassi C, et al, Pancreatic Cancer Meta-Analysis Group. Influence of resection margins and treatment on survival in patients with pancreatic cancer: meta-analysis of randomized controlled trials. [discussion] Arch Surg 2008; 143:75 - 83, discussion 83; http://dx.doi.org/10.1001/archsurg.2007.17; PMID: 18209156
- Li J, Wientjes MG, Au JL. Pancreatic cancer: pathobiology, treatment options, and drug delivery. AAPS J 2010; 12:223 - 32; http://dx.doi.org/10.1208/s12248-010-9181-5; PMID: 20198462
- Jimeno A, Hidalgo M. Molecular biomarkers: their increasing role in the diagnosis, characterization, and therapy guidance in pancreatic cancer. Mol Cancer Ther 2006; 5:787 - 96; http://dx.doi.org/10.1158/1535-7163.MCT-06-0005; PMID: 16648548
- Pasca di Magliano M, Biankin AV, Heiser PW, Cano DA, Gutierrez PJ, Deramaudt T, Segara D, Dawson AC, Kench JG, Henshall SM, et al. Common activation of canonical Wnt signaling in pancreatic adenocarcinoma. PLoS One 2007; 2:e1155; http://dx.doi.org/10.1371/journal.pone.0001155; PMID: 17982507
- Feldmann G, Fendrich V, McGovern K, Bedja D, Bisht S, Alvarez H, Koorstra JB, Habbe N, Karikari C, Mullendore M, et al. An orally bioavailable small-molecule inhibitor of Hedgehog signaling inhibits tumor initiation and metastasis in pancreatic cancer. Mol Cancer Ther 2008; 7:2725 - 35; http://dx.doi.org/10.1158/1535-7163.MCT-08-0573; PMID: 18790753
- Omura N, Goggins M. Epigenetics and epigenetic alterations in pancreatic cancer. Int J Clin Exp Pathol 2009; 2:310 - 26; PMID: 19158989
- Denis H, Ndlovu MN, Fuks F. Regulation of mammalian DNA methyltransferases: a route to new mechanisms. EMBO Rep 2011; 12:647 - 56; http://dx.doi.org/10.1038/embor.2011.110; PMID: 21660058
- Dhe-Paganon S, Syeda F, Park L. DNA methyl transferase 1: regulatory mechanisms and implications in health and disease. Int J Biochem Mol Biol 2011; 2:58 - 66; PMID: 21969122
- De Marzo AM, Marchi VL, Yang ES, Veeraswamy R, Lin X, Nelson WG. Abnormal regulation of DNA methyltransferase expression during colorectal carcinogenesis. Cancer Res 1999; 59:3855 - 60; PMID: 10463569
- Ting AH, Jair KW, Schuebel KE, Baylin SB. Differential requirement for DNA methyltransferase 1 in maintaining human cancer cell gene promoter hypermethylation. Cancer Res 2006; 66:729 - 35; http://dx.doi.org/10.1158/0008-5472.CAN-05-1537; PMID: 16424002
- Peterson EJ, Bögler O, Taylor SM. p53-mediated repression of DNA methyltransferase 1 expression by specific DNA binding. Cancer Res 2003; 63:6579 - 82; PMID: 14583449
- Lin RK, Wu CY, Chang JW, Juan LJ, Hsu HS, Chen CY, Lu YY, Tang YA, Yang YC, Yang PC, et al. Dysregulation of p53/Sp1 control leads to DNA methyltransferase-1 overexpression in lung cancer. Cancer Res 2010; 70:5807 - 17; http://dx.doi.org/10.1158/0008-5472.CAN-09-4161; PMID: 20570896
- Etoh T, Kanai Y, Ushijima S, Nakagawa T, Nakanishi Y, Sasako M, Kitano S, Hirohashi S. Increased DNA methyltransferase 1 (DNMT1) protein expression correlates significantly with poorer tumor differentiation and frequent DNA hypermethylation of multiple CpG islands in gastric cancers. Am J Pathol 2004; 164:689 - 99; http://dx.doi.org/10.1016/S0002-9440(10)63156-2; PMID: 14742272
- Peng DF, Kanai Y, Sawada M, Ushijima S, Hiraoka N, Kosuge T, Hirohashi S. Increased DNA methyltransferase 1 (DNMT1) protein expression in precancerous conditions and ductal carcinomas of the pancreas. Cancer Sci 2005; 96:403 - 8; http://dx.doi.org/10.1111/j.1349-7006.2005.00071.x; PMID: 16053511
- Li A, Omura N, Hong SM, Goggins M. Pancreatic cancer DNMT1 expression and sensitivity to DNMT1 inhibitors. Cancer Biol Ther 2010; 9; Forthcoming http://dx.doi.org/10.4161/cbt.9.4.10750; PMID: 20234167
- Freedman JE, Tanriverdi K. Defining miRNA targets: balancing simplicity with complexity. Circulation 2013; 127:2075 - 7; http://dx.doi.org/10.1161/CIRCULATIONAHA.113.003058; PMID: 23625958
- Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, et al. MicroRNA expression profiles classify human cancers. Nature 2005; 435:834 - 8; http://dx.doi.org/10.1038/nature03702; PMID: 15944708
- Bloomston M, Frankel WL, Petrocca F, Volinia S, Alder H, Hagan JP, Liu CG, Bhatt D, Taccioli C, Croce CM. MicroRNA expression patterns to differentiate pancreatic adenocarcinoma from normal pancreas and chronic pancreatitis. JAMA 2007; 297:1901 - 8; http://dx.doi.org/10.1001/jama.297.17.1901; PMID: 17473300
- Hasanali Z, Stuart A, Yee N, Sharma K, Epner E. The Epigenetics of Gastrointestinal Malignancies. Curr Colorectal Cancer Rep 2012; 8:254 - 62; http://dx.doi.org/10.1007/s11888-012-0147-0
- John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS. Human MicroRNA targets. PLoS Biol 2004; 2:e363; http://dx.doi.org/10.1371/journal.pbio.0020363; PMID: 15502875
- Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell 2003; 115:787 - 98; http://dx.doi.org/10.1016/S0092-8674(03)01018-3; PMID: 14697198
- Krek A, Grün D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M, et al. Combinatorial microRNA target predictions. Nat Genet 2005; 37:495 - 500; http://dx.doi.org/10.1038/ng1536; PMID: 15806104
- Brune K, Hong SM, Li A, Yachida S, Abe T, Griffith M, Yang D, Omura N, Eshleman J, Canto M, et al. Genetic and epigenetic alterations of familial pancreatic cancers. Cancer Epidemiol Biomarkers Prev 2008; 17:3536 - 42; http://dx.doi.org/10.1158/1055-9965.EPI-08-0630; PMID: 19064568
- Shimizu H, Horii A, Sunamura M, Motoi F, Egawa S, Unno M, Fukushige S. Identification of epigenetically silenced genes in human pancreatic cancer by a novel method “microarray coupled with methyl-CpG targeted transcriptional activation” (MeTA-array). Biochem Biophys Res Commun 2011; 411:162 - 7; http://dx.doi.org/10.1016/j.bbrc.2011.06.121; PMID: 21723258
- Mutze K, Langer R, Schumacher F, Becker K, Ott K, Novotny A, Hapfelmeier A, Höfler H, Keller G. DNA methyltransferase 1 as a predictive biomarker and potential therapeutic target for chemotherapy in gastric cancer. Eur J Cancer 2011; 47:1817 - 25; http://dx.doi.org/10.1016/j.ejca.2011.02.024; PMID: 21458988
- Ellis L, Atadja PW, Johnstone RW. Epigenetics in cancer: targeting chromatin modifications. Mol Cancer Ther 2009; 8:1409 - 20; http://dx.doi.org/10.1158/1535-7163.MCT-08-0860; PMID: 19509247
- Fabbri M, Calin GA. Epigenetics and miRNAs in human cancer. Adv Genet 2010; 70:87 - 99; http://dx.doi.org/10.1016/B978-0-12-380866-0.60004-6; PMID: 20920746
- Garzon R, Liu S, Fabbri M, Liu Z, Heaphy CE, Callegari E, Schwind S, Pang J, Yu J, Muthusamy N, et al. MicroRNA-29b induces global DNA hypomethylation and tumor suppressor gene reexpression in acute myeloid leukemia by targeting directly DNMT3A and 3B and indirectly DNMT1. Blood 2009; 113:6411 - 8; http://dx.doi.org/10.1182/blood-2008-07-170589; PMID: 19211935
- Olson P, Lu J, Zhang H, Shai A, Chun MG, Wang Y, Libutti SK, Nakakura EK, Golub TR, Hanahan D. MicroRNA dynamics in the stages of tumorigenesis correlate with hallmark capabilities of cancer. Genes Dev 2009; 23:2152 - 65; http://dx.doi.org/10.1101/gad.1820109; PMID: 19759263
- Braconi C, Huang N, Patel T. MicroRNA-dependent regulation of DNA methyltransferase-1 and tumor suppressor gene expression by interleukin-6 in human malignant cholangiocytes. Hepatology 2010; 51:881 - 90; PMID: 20146264
- Wang H, Wu J, Meng X, Ying X, Zuo Y, Liu R, Pan Z, Kang T, Huang W. MicroRNA-342 inhibits colorectal cancer cell proliferation and invasion by directly targeting DNA methyltransferase 1. Carcinogenesis 2011; 32:1033 - 42; http://dx.doi.org/10.1093/carcin/bgr081; PMID: 21565830
- Jair KW, Bachman KE, Suzuki H, Ting AH, Rhee I, Yen RW, Baylin SB, Schuebel KE. De novo CpG island methylation in human cancer cells. Cancer Res 2006; 66:682 - 92; http://dx.doi.org/10.1158/0008-5472.CAN-05-1980; PMID: 16423997
- Okami J, Simeone DM, Logsdon CD. Silencing of the hypoxia-inducible cell death protein BNIP3 in pancreatic cancer. Cancer Res 2004; 64:5338 - 46; http://dx.doi.org/10.1158/0008-5472.CAN-04-0089; PMID: 15289340
- Sato N, Fukushima N, Maehara N, Matsubayashi H, Koopmann J, Su GH, Hruban RH, Goggins M. SPARC/osteonectin is a frequent target for aberrant methylation in pancreatic adenocarcinoma and a mediator of tumor-stromal interactions. Oncogene 2003; 22:5021 - 30; http://dx.doi.org/10.1038/sj.onc.1206807; PMID: 12902985
- Ueki T, Toyota M, Skinner H, Walter KM, Yeo CJ, Issa JP, Hruban RH, Goggins M. Identification and characterization of differentially methylated CpG islands in pancreatic carcinoma. Cancer Res 2001; 61:8540 - 6; PMID: 11731440
- Sato N, Parker AR, Fukushima N, Miyagi Y, Iacobuzio-Donahue CA, Eshleman JR, Goggins M. Epigenetic inactivation of TFPI-2 as a common mechanism associated with growth and invasion of pancreatic ductal adenocarcinoma. Oncogene 2005; 24:850 - 8; http://dx.doi.org/10.1038/sj.onc.1208050; PMID: 15592528
- Manuyakorn A, Paulus R, Farrell J, Dawson NA, Tze S, Cheung-Lau G, Hines OJ, Reber H, Seligson DB, Horvath S, et al. Cellular histone modification patterns predict prognosis and treatment response in resectable pancreatic adenocarcinoma: results from RTOG 9704. J Clin Oncol 2010; 28:1358 - 65; http://dx.doi.org/10.1200/JCO.2009.24.5639; PMID: 20142597
- Zhang W, Dahlberg JE, Tam W. MicroRNAs in tumorigenesis: a primer. Am J Pathol 2007; 171:728 - 38; http://dx.doi.org/10.2353/ajpath.2007.070070; PMID: 17724137
- Tsuruta T, Kozaki K, Uesugi A, Furuta M, Hirasawa A, Imoto I, Susumu N, Aoki D, Inazawa J. miR-152 is a tumor suppressor microRNA that is silenced by DNA hypermethylation in endometrial cancer. Cancer Res 2011; 71:6450 - 62; http://dx.doi.org/10.1158/0008-5472.CAN-11-0364; PMID: 21868754
- Zhao G, Zhang JG, Liu Y, Qin Q, Wang B, Tian K, Liu L, Li X, Niu Y, Deng SC, et al. miR-148b functions as a tumor suppressor in pancreatic cancer by targeting AMPKα1. Mol Cancer Ther 2013; 12:83 - 93; http://dx.doi.org/10.1158/1535-7163.MCT-12-0534-T; PMID: 23171948
- Venturelli S, Berger A, Weiland T, Essmann F, Waibel M, Nuebling T, Häcker S, Schenk M, Schulze-Osthoff K, Salih HR, et al. Differential induction of apoptosis and senescence by the DNA methyltransferase inhibitors 5-azacytidine and 5-aza-2′-deoxycytidine in solid tumor cells. Mol Cancer Ther 2013; 12:2226 - 36; http://dx.doi.org/10.1158/1535-7163.MCT-13-0137; PMID: 23924947
- Yasuda M, Theodorakis P, Subramanian T, Chinnadurai G. Adenovirus E1B-19K/BCL-2 interacting protein BNIP3 contains a BH3 domain and a mitochondrial targeting sequence. J Biol Chem 1998; 273:12415 - 21; http://dx.doi.org/10.1074/jbc.273.20.12415; PMID: 9575197
- Chen G, Ray R, Dubik D, Shi L, Cizeau J, Bleackley RC, Saxena S, Gietz RD, Greenberg AH. The E1B 19K/Bcl-2-binding protein Nip3 is a dimeric mitochondrial protein that activates apoptosis. J Exp Med 1997; 186:1975 - 83; http://dx.doi.org/10.1084/jem.186.12.1975; PMID: 9396766
- Yiu GK, Chan WY, Ng SW, Chan PS, Cheung KK, Berkowitz RS, Mok SC. SPARC (secreted protein acidic and rich in cysteine) induces apoptosis in ovarian cancer cells. Am J Pathol 2001; 159:609 - 22; http://dx.doi.org/10.1016/S0002-9440(10)61732-4; PMID: 11485919
- Mok SC, Chan WY, Wong KK, Muto MG, Berkowitz RS. SPARC, an extracellular matrix protein with tumor-suppressing activity in human ovarian epithelial cells. Oncogene 1996; 12:1895 - 901; PMID: 8649850
- Mahadevan D, Von Hoff DD. Tumor-stroma interactions in pancreatic ductal adenocarcinoma. Mol Cancer Ther 2007; 6:1186 - 97; http://dx.doi.org/10.1158/1535-7163.MCT-06-0686; PMID: 17406031
- Pfaffl MW, Horgan GW, Dempfle L. Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res 2002; 30:e36; http://dx.doi.org/10.1093/nar/30.9.e36; PMID: 11972351
