Abstract
Background
MicroRNA-20a (miR-20a) plays a key role in tumorigenesis and progression. But its function is reverse in different kinds of malignant tumor, and its role and mechanism in cutaneous squamous cell carcinoma (CSCC) remains unclear.
Object
To determine the miR-20a’s roles in CSCC and confirm whether LIMK1 is a direct target gene of miR-20a.
Methods
First miR-20a and LIMK1 expression levels were detected in six pairs of CSCC tissues and corresponding normal skin by qRT-PCR. Then MTT assays and colony formation assays were performed to evaluate the impact of miR-20a on cell proliferation. In addition, scratch migration assays and transwell invasion assays were performed to check miR-20a’s effect on cell metastasis. Since LIMK1 (LIM kinase-1) was predicted as a target gene of miR-20a, the changes of LIMK1 protein and mRNA were measured by western blot and qRT-RCR methods after miR-20a overexpression. Moreover the dual reporter gene assay was performed to confirm whether LIMK1 is a direct target gene of miR-20a. Finally LIMK1 mRNA and miR-20a in other 30 cases of CSCC pathological specimens were determined and a correlation analysis was evaluated.
Results
The miR-20a significantly low-expressed in CSCC tissues compared with that in matched normal tissues while LIMK1 has a relative higher expression. MiR-20a inhibited A431 and SCL-1 proliferation and metastasis. Both of LIMK1 protein and mRNA levels were downregulated after miR-20a overexpression. The dual reporter gene assays revealed that LIMK1 is a direct target gene of miR-20a. Furthermore, qRT-PCR results of LIMK1 mRNA and miR-20a in 30 cases of CSCC pathological specimens showed miR-20a is inversely correlated with LIMK1 expression.
Conclusion
Our study demonstrated that miR-20a is involved in the tumor inhibition of CSCC by directly targeting LIMK1 gene. This finding provides potential novel strategies for therapeutic interventions of CSCC.
Introduction
Cutaneous squamous cell carcinoma (CSCC) is the second most common non-melanoma skin cancer, with an incidence rate of 10%.Citation1-Citation3 The occurrence of this cancer is related to the presence of chronic ulcers, scars, and UV exposure. Its morbidity is greater in males than females, and increases with age.Citation4 Most cases of CSCC have a low metastasis rate of approximately 1.9–5%,Citation1,Citation5,Citation6 however, for these patients mortality is high. Indeed, although CSCC can be cured through surgery when it is detected early, prognosis is poor after metastasis. The risk factors associated with CSCC metastasis include tumor diameter, invasion level, degree of differentiation, and location.Citation5,Citation6 Of note, death caused by CSCC may actually be equal to that caused by melanoma, renal cancer, and oropharyngeal cancer; however, it is underestimated because of a lack of spatial statistics.Citation7 Moreover, patients with CSCC also have an elevated death rate of lung cancer, colorectal cancer and breast cancer.Citation8,Citation9 Therefore, study of the molecular mechanisms underlying metastasis in CSCC could be useful to aid its early detection and thereby improve treatment outcome.
MicroRNAs, which regulate gene expression at a post-transcriptional level, are conserved non-coding RNAs of approximately 22 nucleotides.Citation10 Many microRNAs participate in the regulation of CSCC metastasis. For example, the low expression of miR-361-5p in CSCC is negatively correlated with that of VEGF,Citation11 while miR-125b has been shown to inhibit CSCC proliferation, migration, and invasion through downregulation of MMP13.Citation12
MiR-20a (microRNA 20a) belongs to the miR-17-92 cluster, which includes six microRNAs: miR-17-5p, miR-18a, miR-19a, miR-19b, miR-20a, and miR-92a-16.Citation13 MiR-20a plays different roles in different tumors. It promotes the growth, migration, and invasion of gastric cancer cells by targeting EGR2 inhibition, which enhances GC drug resistance.Citation14 In gallbladder carcinoma patients, miR-20a has been shown to be overexpressed in correlation with a lower survival rate.Citation15 Together with the miR-17-92 cluster, miR-20a has also been reported to function as a tumor promoter in osteosarcoma,Citation16 bladder cancer,Citation17 prostate cancer,Citation18 and cervical cancer.Citation19 On the contrary, in oral squamous cancer cellsCitation20 and hepatic cancer,Citation21 miR-20a acts as an inhibitor of tumor metastasis. As yet, no study has explored the role of miR-20a in squamous cell carcinoma of the skin.
The gene LIMK1 (LIM kinase-1) is located on chromosome 7q11.23. It encodes a serine/threonine kinase which regulates actin polymerization through the phosphorylation and inactivation of cofilin, an actin combination factor. LIMK1 is ubiquitously expressed in higher organisms and participates in many biological processes involving the cytoskeleton, such as neurite growth,Citation22 embryo growth,Citation23 thrombogenesis,Citation24 memory formation,Citation25 and cancer development.Citation26 Indeed, the main function of members in the Lim kinase family is to regulate the actin cytoskeleton which is critical for cell motility and migration. In cancer, LIMK1 has been shown to be a metastasis promoter in breast cancer,Citation26 ovarian cancer,Citation27 osteosarcoma,Citation28 and squamous cell cancer.Citation29
In this study, we demonstrated that miR-20a exhibits a tumor-inhibition impact on CSCC. We found that overexpression of miR-20a inhibited both proliferation and metastasis of A431 and SCL-1 CSCC cells. By using TargetScan, PicTar, PITA, miRanda, and miRDB, we predicted that miR-20a may regulate LIMK1. Indeed, two binding sites for miR-20a were identified in LIMK1 by TargetScan, while the Pictar predicted target score was 6.84 and the mirSVR score was –0.6358, suggesting that LIMK1 is a highly likely target of miR-20a. Indeed, similar to the effect observed by miR-20a overexpression, blockade of LIMK1 with si-RNA also suppressed both proliferation and metastasis of A431 cells. Importantly, overexpression of miR-20a reduced LIMK1 expression at the gene transcription and protein levels. Together, these data strongly suggest that LIMK1 is a downstream target gene of miR-20a. The dual reporter gene assays further confirmed our hypothesis, indicating that miR-20a inhibits the LIMK1 pathway in CSCC, and thus may be a new target for its diagnosis and treatment.
Results
MiR-20a and LIMK1 expression in CSCC tissue samples
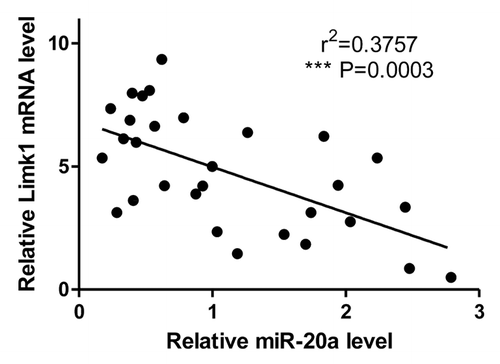
To evaluate the expression of miR-20a in CSCC, we used qRT-PCR to examine its mRNA levels in six CSCC tissues and corresponding normal skin tissues (located >5 cm away from the tumor). The expression of miR-20a was significantly decreased in CSCC tissues when compared with that in normal skin tissue (). Also, we tested the mRNA expression of LIMK1 in CSCC tissue and the corresponding normal tissue. LIMK1 is in lower level in tumor compared with normal skin ().
Figure 1. The expression of miR-20a and LIMK1 in CSCC tissue. (A) MiR-20a expression is lower in CSCC tissue samples than normal matched tissue. The mRNA expression of miR-20a was evaluated by qRT-PCR in six patient-derived CSCC tissue samples (tumor) and matched normal tissues; (B) The mRNA expression of LIMK1 was evaluated by qRT-PCR in six patient-derived CSCC tissue samples (tumor) and matched normal tissues. *P < 0.05, **P < 0.01, ***P < 0.001.
Overexpression of miR-20a inhibits CSCC cell proliferation
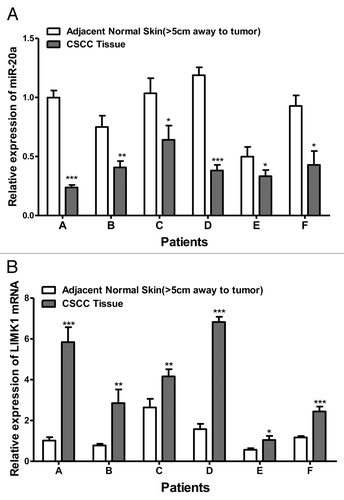
Because the expression of miR-20a was significantly lower in CSCC tissue compared with that in matched normal tissue, we postulated that miR-20a may function as a tumor suppressor. To test this hypothesis we overexpressed miR-20a in the classical skin squamous cancer cell lines, A431Citation30,Citation31 and SCL-1,Citation32,Citation33 through lipofection with the eukaryotic expression plasmid of miR-20a (hsa-mir-20a), and constructed the stable transfected cell line through hygromycin selection. Two control conditions were also included; the no treatment blank and cells transfected with the control plasmid (hsa-mir-20a-NC). The transfection efficiency was confirmed through fluorescence microscopy and also qRT-PCR (). For qRT-PCR, miR-20a expression levels were compared with that of the non-treated blank, set as 1, then the 2-CT was calculated of each group to determine the relative expression level of miR-20a. As shown in , miR-20a was overexpressed in A431 and SCL-1 cells transfected with miR-20a when compared with that in the blank and negative control groups, whereas there was no significant difference in its expression between the negative control and blank groups. Thus, these results show that the transfection efficiency was satisfactory.
Figure 2. The overexpression of miR-20a in CSCC cell lines (A) Validation of hsa-mir-20a overexpression in A431 CSCC cells. (B) Validation of hsa-mir-20a overexpression in SCL-1 CSCC cells. Cells were transfected with the hsa-mir-20a (miR-20a-A431/SCL-1), negative control (NC-A431/SCL-1), or left untreated (A431/SCL-1). Representative bright field microscopy images (100×, top) and fluorescence microscopy images (100×, bottom) are at left. The result of qRT-PCR analysis of miR-20a expression is shown right. MiR-20a expression was determined relative to that in non-treated A431 set as 1.*P < 0.05, **P < 0.01, ***P < 0.001.
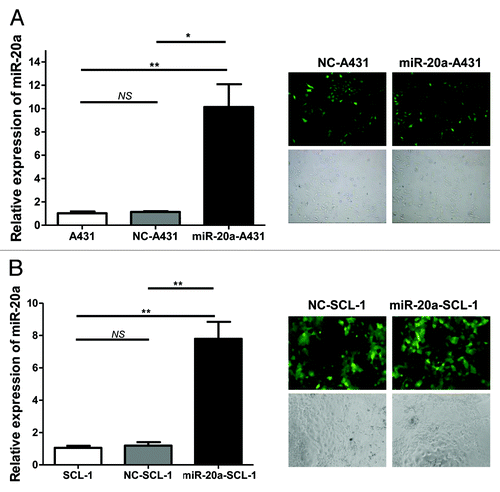
Next, to check whether overexpression of miR-20a affected CSCC cell proliferation, the MTT method was used. Cells transfected with hsa-mir-20a (miR-20a-A431 and miR-20a-SCL-1) grew more slowly than those in either the control (NC-A431 and NC-SCL-1) or non-treated blank (A431 and SCL-1) conditions (), while there was no difference between NC control groups and blank groups. The colony formation assay was also used to evaluate cell proliferation and plating efficiency after overexpression of miR-20a. We found that the number of colonies formed under the miR-20a overexpressed condition was much less than those for non-treatment and NC-treatment (), whereas no obvious difference was detected between non-treatment groups and NC-treatment groups. The results of the MTT and colony formation assays therefore demonstrate that miR-20a overexpression limits CSCC cells proliferation suggesting that it may inhibit CSCC growth.
Figure 3. MiR-20a inhibits CSCC cell growth. CSCC cells were transfected with hsa-mir-20a or the corresponding controls, non-treated blank and negative control, then (A–B) MTT assays were performed on A431 and SCL-1 cells at the indicated time points. (C–D) The colony formation assay was also performed using identically treated A431 and SCL-1 cells. The upper three images are representative results for each condition. The pictures below show the colony formation rate (colony number/1000 × 100%) of each group, as indicated. **P < 0.01, ***P < 0.001.
Overexpression of miR-20a inhibits CSCC cell migration and invasion
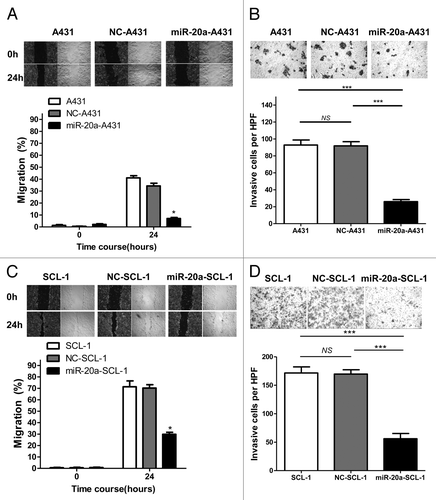
After we found that miR-20a can inhibit CSCC cell proliferation, we explored whether it also affected cell migration and invasion. We used the scratch migration assay to test the migratory ability of CSCC cells with and without overexpression of miR-20a. The results showed that the remaining scratch areas of non-treated blank groups and NC-treated groups were much less than that of the miR-20a-overexpressed groups () which indicated that the migratory ability of CSCC cells was inhibited by miR-20a. We also performed transwell invasion assays to explore the impact of miR-20a on the invasive capacity of CSCC cells. The results showed that the number of invading cells in CSCC cells overexpressing miR-20a was much less than that in either the non-treated blank groups or NC-treated groups (). In short, miR-20a can inhibit CSCC cells migration and invasion, indicating that it suppresses metastasis in CSCC.
Figure 4. MiR-20a inhibits CSCC cell migration and invasion. (A and C) The scratch migration assay was performed in non-treated blank (A431/SCL-1), negative control transfected (NC-A431/NC-SCL-1), and hsa-mir-20a transfected (miR-20a-A431/miR-20a-SCL-1) cells. Migration of cells into the scratch-induced gap was monitored at the indicated time points. Representative microscopic images (40×) are on the left or top. To more clearly show the differences, black and white images were generated according to edge enhancing processing. The crawling areas of cells presented in images were quantified to determine the migration rate of each group at 0, 24h, or at 48 and 72 h. (B and D) The invasive cell number was counted in at least five different high power fields (HPF) of view. Representative microscopy images of invasive cells from the non-treated blank (A431/SCL-1) group, negative control (NC-A431/NC-SCL-1) group, and hsa-mir-20a (miR-20a-A431/miR-20a-SCL-1) group (100×); *P < 0.05, ***P < 0.001.
LIMK1 is a target gene of miR-20a
LIMK1, which exhibits tumor promoting functions in many kinds of cancer, is known as a metastasis stimulator in skin squamous cell carcinoma.Citation29 To confirm its tumor acceleration affect in skin squamous cell carcinoma, we transfected A431 cells with si-RNA against LIMK1 (Fig. S2A) and tested the effects on proliferation and invasion by MTT and transwell invasion assays, respectively. The results showed that inhibition of LIMK1 correlated with suppression of the growth and invasion abilities of A431 cells (Fig. S2B and C).
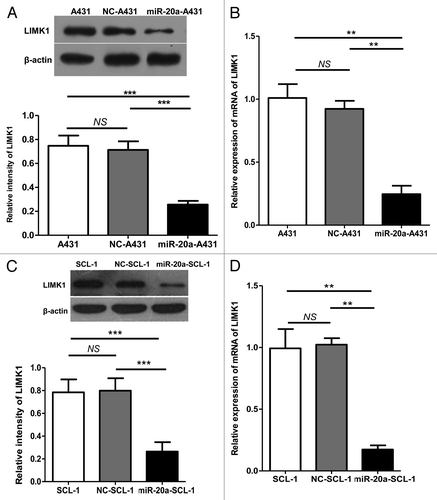
Next, through multiple microRNA target gene prediction software packages, such as TargetScan, we identified LIMK1 as a target of miR-20a. To test whether miR-20a inhibited tumor proliferation and migration through LIMK1 suppression we used qRT-PCR and western blot analyses to examine the expression of LIMK1 in A431 and SCL-1 (non-treated blank); NC-A431 and NC-SCL-1; miR-20a-A431 and miR-20a-SCL-1 cell groups. In correspondence with our hypothesis, the results showed that LIMK1 gene and protein expression levels declined when miR-20a was overexpressed ().
Figure 5. LIMK1 expression was suppressed by miR-20a overexpression. (A and C) Western blot of LIMK1 protein expression in CSCC cells after transfection with negative control (NC-A431/NC-SCL-1) or hsa-mir-20a (miR-20a-A431/miR-20a-SCL-1). Non-treated blank CSCC cells (A431/SCL-1) were also included while determination of β-actin protein expression was used as a control for input and normalization. A representative blot and quantification are shown. (B and D) qRT-PCR was used to determine the relative expression of LIMK1 in identically transfected cells. *P < 0.05, **P < 0.01, ***P < 0.001.
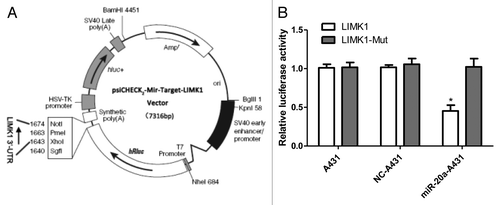
To confirm that LIMK1 is a direct target gene of miR-20a, we amplified the LIMK1 3′ UTR containing the target sequence (Fig. S1A); in which there are two possible biding sites (the underline position). This was either left intact or mutated (Fig. S1B) sequence then inserted into a psiCHECK2 luciferase reporter vector (). As shown in , miR-20a suppressed the luciferase activity of the LIMK1 3′ UTR, while mutation of the miR-20a binding sites blocked this action in A431 cells. When LIMK1 has a mutation or reaction with miR-20a-NC or no treat A431 group, the luciferase activity basically the same and have no significant difference. These results suggest that miR-20a can combine to LIMK1’s 3′ UTR and LIMK1 is a miR-20a target gene that is inhibited by miR-20a at the posttranscriptional level.
Figure 6. LIMK1 is the direct target gent of miR-20a and LIMK1 higher expressed in CSCC which has a reserved relationship of miR-20a. (A) The plasmid structure of psiCHECK2-LIMK1-3′-UTR reporter vector. (B) A431 cells were divided into three groups: non-treated blank (A431), negative control (NC-A431), and hsa-mir-20a (miR-20a-A431), and co-transfected with either the WT LIMK1 reporter vector (LIMK1) or the mutant (LIMK1-Mut) vector, then the luciferase activity was determined and normalized to set A431/LIMK1 as 1. *P < 0.05, **P < 0.01, ***P < 0.001.
Relationship between miR-20a and LIMK1 in CSCC tissue
At last, to further confirm the relationship between LIMK1 and miR-20a in CSCC tissue, we test the mRNA expression of LIMK1 and miR-20a in 30 pathological specimens of CSCC patients and make a correlation analysis (). The results show LIMK1 is higher expressed in CSCC tumor tissue, which means when miR-20a is high expressed, LIMK1 has a lower expression. The correlation analysis of LIMK1 and miR-20a in CSCC tissue confirmed the negative relationship between the miR-20a and LIMK1 which further confirmed miR-20a inhibited LIMK1 in CSCC tissue.
Discussion
MicroRNAs play a key role in tumor development where they act as either suppressors or promoters at the post-transcriptional regulation stage. Also in squamous cell carcinoma of skin, many microRNAs play different role.Citation34 MiR-193b/365a clusterCitation35 and miR-483-3pCitation36 suppress the epidermal squamous cell carcinoma progression. On the contrary, miR-365 is an onco-factor in skin squamous cell carcinoma.Citation37 These microRNAs and their target genes can be treatment targets, early warning labels, or prognosis labels for CSCC’s clinic processing.
The miR-17-92 cluster comprises critical molecules in the central network of tumor control.Citation38 As a member of the miR-17-92 cluster, miR-20a contributes to the regulation of many kinds of tumors. However, its effects in different kinds of cancer can be contradictory. Indeed, miR-20a promotes gastric cancer,Citation14 gallbladder carcinoma,Citation15 and prostate cancer.Citation18 On the contrary, in hepatic cancerCitation21 and oral squamous cancer,Citation20 it acts as a tumor suppressor. To date, there have been no reports of the effects of miR-20a in CSCC. However, in squamous cancer of the cervixCitation39 and squamous cancer of the esophagus,Citation40 miR-20a expression is elevated in the circulation and in tissue in association with a higher risk of tumor metastasis. On the other hand, in oral squamous cancer,Citation20 miR-20a can suppress cell migration. Because the oral squamous cancer tissue is more similar to that of skin squamous cancer than either cervical or esophagus squamous cancer, we speculated that miR-20a likely contributes to tumor suppression in CSCC rather than its promotion.
In this study, we detected the miR-20a level in CSCC tissue and matched normal skin tissue. We found that miR-20a expression was lower in CSCC tissue than in normal tissue, suggesting that miR-20a may play an important role in CSCC. To explore its function in CSCC we overexpressed miR-20a in a typical CSCC cell line, A431 and SCL-1, through transfection of hsa-mir-20a. Compared with either the control group, which was transfected with hsa-mir-20a-NC, or the blank group, which received no treatment; in the miR-20a-A431 group both cell proliferation and migration were significantly suppressed. Indeed through MTT and colony formation assays it was shown that miR-20a inhibited the proliferation of A431 and SCL-1 cells suggesting that it inhibits growth of CSCC. While the results of the transwell invasion and scratch migration assays demonstrated that both migration and invasion were suppressed by miR-20a, suggesting that it may inhibit metastasis in CSCC. In short, for the first time we confirmed that miR-20a is anti-oncogenic in CSCC and could be useful as a therapeutic or diagnostic target in this disease.
To determine how miR-20a elicits its anti-oncogenic function we screened for potential target genes using bioinformatics analysis software packages (Targetscan, PicTar, PITA, miRanda, and miRDB). We found that LIMK1, a tumor metastasis promoter, contained two potential binding sites for miR-20a. LIMK1 is a key cytoskeletal protein and a critical factor in the regulation of metastasis in many kinds of cancer. It is an essential factor for tumor invasion. Indeed, in many kinds of squamous cell cancers, LIMK1 is considered a metastasis promoter.Citation41 For example, in lung squamous cell cancer, knockout of LIMK1 inhibits the migration of sk-mes-1 cells, whereas the activation of Pak1/LIMK1/cofilin pathway has been shown to promote lung squamous cell carcinoma rather than adenocarcinoma.Citation42 Roy et.al found that LIMK1 expression declined in the UV B-induced cell death of A431.Citation29 In our study we transfected A431 cells with si-RNA against LIMK1 and tested the effects by MTT and transwell invasion assays. These experiments found that silencing of LIMK1 suppressed both A431 cell growth and invasion, suggesting that it might function as a tumor promoter, whereas its inhibition could result in impairment of these processes. To test our hypothesis that miR-20a inhibits LIMK1 expression, which in turn prevents the growth and metastasis of CSCC, we used qRT-PCR and western blot to measure LIMK1 mRNA and protein expression levels, respectively, in miR-20a overexpressing A431 and SCL-1 cells. The result show in the two cell lines, the protein and mRNA of LIMK1 rose when miR-20a is overexpressed in A431 and SCL-1, which primarily prove LIMK1 is miR-20a’s target gene in vitro. Also, the dual luciferase activity assay further confirmed that LIMK1 is a direct target gene of miR-20a, suggesting that miR-20a might inhibit CSCC through inhibition of LIMK1. Of note, we compared LIMK1 expression in CSCC tissues with that in normal skin. LIMK1 is higher expressed in CSCC tumor tissue compare with the nearby normal skin, which seems to have a reserved relationship to miR-20a. So we further more test the mRNA of LIMK1 and miR-20a in 30 cases of CSCC tumor tissue. We found that, not only LIMK1 expression was inhibited at both the transcriptional and protein levels in CSCC cells when miR-20a was overexpressed, but also in the CSCC tissue, miR-20a expression was lower than that in tumor tissue whereas LIMK1 expression was higher in tumor tissue. The analysis show there has a reserved relationship between miR-20a and LIMK1 in CSCC tissue. Together these findings support our hypothesis that LIMK1 is a miR-20a target gene and that it may limit CSCC through LIMK1 suppression. Further experiments could be performed to explore about the further mechanism of LIMK1’s pathway.
In summary, for the first time our study demonstrates that miR-20a expression is dramatically decreased in CSCC tissues compared with that in matched normal tissues. Overexpression of miR-20a, or silencing of LIMK1, limits the proliferation, migration, and invasiveness of CSCC cells. Moreover, upregulation of miR-20a inhibits the expression of LIMK1, which was confirmed as a target gene of miR-20a by the dual luciferase activity assay. As miR-20a’s target gene, LIMK1 has a lower expression in CSCC tissue and has a reserved relationship to miR-20a level. Our studies for the mechanism of miR-20a and its target LIMK1 might contribute to the early diagnosis, prognosis prediction or the treatment of CSCC.
Materials and Methods
Cell lines and tissue samples
Six paired tissues of CSCC and matched normal skin (located >5 cm away from the tumor) were collected from the Department of Surgery, following informed consent from each of the patients and approval from the Ethics Committee of The Third Xiangya Hospital of Central South University. The 30 cases of CSCC tissue for test of mRNA of LIMK1 and miR-20a were collected from the pathology department of The Third Xiangya Hospital of Central South University. The human cell line A431, derived from CSCC tissue of an 85 y old female, was bought from the ATCC. The human cell line SCL-1 (from Free University of Berlin) was kindly provided by the Dermatology Department of China Medical University. The cells were maintained in Dulbecco’s modified Eagle medium supplemented with 10% fetal bovine serum at 37 °C with 5% CO2.
Plasmids and cell transfection
The eukaryotic expression plasmid of fluorescently labeled miR-20a (hsa-mir-20a, PMUE214000076) and the negative control (hsa-mir-20a-NC) were bought from GeneChem Biotechnology. The sequence of pre-hsa-mir-20a is: 5′-GUAGCACUAA AGUGCUUAUA GUGCAGGUAG UGUUUAGUUA UCUACUGCAU UAUGAGCACU UAAAGUACUG C-3′, and the plasmid was testified by gene sequencing. The siRNA targeting LIMK1 (5′-GACTTGCGTA GCCTTAAGA-3′)Citation22 and negative control siRNA were purchased from GenePharma Biotechnology. The psiCHECK2 vector was purchased from Promega, and constructs expressing either intact LIMK1 or LIMK1 with a mutation in the putative binding site (underlined) for miR-20a were generated as follows. The cDNAs encoding LIMK1 (forward 5′-ccgCTCGAGG GCAGAGGCCA AGTTCCA-3′, reverse 5′-atttGCGGCC GCTCCCACCT CCCTAAGTCA T-3′) and mutated LIMK1 (forward 5′-ccgGCGGCCG CGGCAGAGGC CAAGTTCCA-3′, reverse 5′-atttCTCGAG TCCCACCTCC CTAAGTCAT-3′) were showed in Figure S1B and were amplified by PCR. The amplified sequences were inserted into psiCHECK2-vector within XhoI/NotI sites. Plasmids were introduced to A431 and SCL-1 cells by the lipofectamine method (Lipofectamine RNAiMAX; Invitrogen) when they attained 40–50% confluence in 6 cm culture dishes. Forty-eight hours after transfection, fluorescence microscopy was used to observe transfection efficiency of miR-20a. The hygromycin (Invitrogen) was used for selection of the stable miR-20a overexpression transfected A431 and SCL-1 cells. The qRT-PCR was used to test the expression level of miR-20a and LIMK1.
cDNA synthesis and quantitative real time PCR (qRT-PCR)
The sequences of miR-20a and LIMK1 were acquired from GenBank. The primer sequences of LIMK1 were as follows: forward 5′-TTACGCCCCT TTCCACA-3′ and reverse 5′-CCCAGACTCC CCATTTG-3′. While those for miR-20a were: forward 5′GCTGCCGTAA AGTGCTTATA GTG3′ and reverse 5′AGAGCAGGGT CCGAGGTA3′. Total RNA from cells and tissues were extracted using TRIzol (Dingguo Biotechnology). The purity and integrity of RNA was examined on a Micro-UV Spectrophotometer and RNA electrophoresis after RNA purification. After DNase (RQ1 RNase-free DNase, Promega) treatment to resolve the DNA in total RNA, it was reverse-transcribed into cDNA with a TOYOBO Reverse Transcription System on an ABI-9600 PCR Amplifier (PerkinElmer) detected by ABI PRISM 7700 (PerkinElmer). Samples were compared by using the relative CT method, where the relative expression of miR-20a was normalized to that of U6, while that of LIMK1 was normalized to β-actin expression.
Western blot
Cells were harvested 48 h after transfection, and proteins were extracted and then quantified with a BCA protein assay kit and separated by 10% sodium dodecyl sulfate-PAGE. Gels were transferred to nitrocellulose membranes and probed with 0.5 mg/mL anti-LIMK1 antibody (Bioss Biotechnology Company). Following incubation with a rabbit radish peroxidase-conjugated secondary antibody, immunoreactive bands were visualized by electrochemiluminescence.
MTT assay and plate colony formation assay
For the MTT assay, cells were collected 8 h after transfection and seeded in 96-well plates at 3000/well. MTT (5 mg/mL) was then added at 24, 48, 72, 96, and 120 h later. Following 4 h of incubation, the MTT-media mixture was removed and replaced with 150 µL DMSO to dissolve the formazan crystals. The OD value at 490 nm was then determined and used to construct a growth curve to assess cell proliferation.
For the colony formation assay, cells that had been transfected were collected and seeded in 10 cm plates (1000/plates) then maintained in complete culture medium for 21 d. Next, cells were fixed in 4% paraformaldehyde for 15 min and stained with GIMSA dye liquor. They were photographed and the number of clones was calculated.
Scratch migration and transwell invasion assays
For the scratch migration assay, transfected cells were collected and seeded in 6-well plates at 5 × 105/well and cultured for 12 h or until a complete monolayer was achieved. Next, a straight line scratch was made using a 10 µL tip. The migration of cells into the scratch-generated gap was monitored by photographing cultures at 0, 24, or additional 48 and 72 h.
To measure invasive capacity, the transwell invasion assay was performed using transfected cells plated on 8 µm pore size Matrigel-coated membranes, at 105/well in serum-free medium, which were in turn placed in the top chamber of 24-well transwell plates. DMEM containing 10% FBS was added to the bottom chamber as a chemoattractant. After 24 h, cells on the upper surface were removed, while cells attached on the membranes were fixed in 4% paraformaldehyde for 20 min and stained with hematoxylin dye liquor. They were photographed and the number of cells was calculated under a microscope.
Dual luciferase reporter assays
Luciferase reporter assays were performed using the psiCHECK2-LIMK1–3′-UTR vector. Cells were grown to approximately 80% confluence in 6-well plates and co-transfected with either psiCHECK2-LIMK1-3′-UTR or psiCHECK2-LIMK1-Mut-3′-UTR (1 μg) plus either hsa-mir-20a or hsa-mir-20a-NC (1 μg). Cells were incubated with transfection reagent/DNA complexes for 3 h. In a separate set of experiments, cells were incubated for 48 h after transfection. Firefly and Renilla luciferase activities were then evaluated using the Dual-Luciferase Reporter Assay system (Promega), where Renilla luciferase activity was normalized to firefly luciferase activity.
Statistical analysis
All experiments were repeated at least three times, and the results were expressed as the mean ± SD. The results were assessed by one-way ANOVA or the Student t test, where a P value of <0.05 was accepted to indicate statistical significance.
Conclusion
Our work demonstrates that miR-20a is involved in the tumor inhibition of CSCC by targeting LIMK1 gene, which can be a new target for therapy in CSCC clinical treatments.
| Abbreviations: | ||
| CSCC | = | cutaneous squamous cell carcinoma |
| miR-20a | = | microRNA 20a |
| LIMK1 | = | LIM kinase-1 |
| si-LIMK1 | = | siRNA of LIMK1 |
Additional material
Download Zip (392.8 KB)Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
This work was supported by National Natural Science Foundation of China (81372140, 81301688, 81272192, 81171882, and 81071645); Ph.D. Programs Foundation of Ministry of Education of China (20130162110050 and 20130162120093); Natural Science Foundation of Hunan Province (Grant No.13JJ4028); Technology Project of Hunan Province (2012SK3229); Project of the Department of Science and Technology of Hunan Province (2013FJ6003); Program for New Century Excellent Talents in University (NCET-11-0527) ; Fundamental Research Funds for the Central Universities (2011JQ028); Post-doctoral Foundation of Central South University (131425) ; 125 Talent Project of the Third Xiangya Hospital of Central South University and Hunan Provincial Innovation Foundation for Postgraduate. Thanks for the kindly giving of SCL-1 cell line from the Dermatology Department of China Medical University.
References
- Toll A, Masferrer E, Hernández-Ruiz ME, Ferrandiz-Pulido C, Yébenes M, Jaka A, Tuneu A, Jucglà A, Gimeno J, Baró T, et al. Epithelial to mesenchymal transition markers are associated with an increased metastatic risk in primary cutaneous squamous cell carcinomas but are attenuated in lymph node metastases. J Dermatol Sci 2013; 72:93 - 102; http://dx.doi.org/10.1016/j.jdermsci.2013.07.001; PMID: 23928229
- Ya-ning JIAO, Li XIA, Xin-hong GE, Xiu-juan ZHANG, Jian-ping LIU, Li-peng WANG, Ling-di DONG, Nan YU, Xiao-ming Z. The Incidence Analysis of Skin Common Tumor from Five Hospitals in Ningxia from 2002 to 2011. The Chinese Journal of Dermatovenereology 2013; 36 - 8
- Gao TW, Sun DJ, Li CY, He H, Li Q, Liu YS, Diao QC, Huang GS, Hao F, Zhong BY, et al. [Retrospective analysis of 1905 patients with skin cancer from two general hospitals in western China from 1981 to 2000]. Beijing Da Xue Xue Bao 2004; 36:469 - 72; PMID: 15489924
- Rahimi S. Squamous cell carcinoma of skin: a brief review. Clin Ter 2013; 164:143 - 7; PMID: 23698209
- Brougham ND, Dennett ER, Cameron R, Tan ST. The incidence of metastasis from cutaneous squamous cell carcinoma and the impact of its risk factors. J Surg Oncol 2012; 106:811 - 5; http://dx.doi.org/10.1002/jso.23155; PMID: 22592943
- Schmults CD, Karia PS, Carter JB, Han J, Qureshi AA. Factors predictive of recurrence and death from cutaneous squamous cell carcinoma: a 10-year, single-institution cohort study. JAMA Dermatol 2013; 149:541 - 7; http://dx.doi.org/10.1001/jamadermatol.2013.2139; PMID: 23677079
- Karia PS, Han J, Schmults CD. Cutaneous squamous cell carcinoma: estimated incidence of disease, nodal metastasis, and deaths from disease in the United States, 2012. J Am Acad Dermatol 2013; 68:957 - 66; http://dx.doi.org/10.1016/j.jaad.2012.11.037; PMID: 23375456
- Song F, Qureshi AA, Giovannucci EL, Fuchs CS, Chen WY, Stampfer MJ, Han J. Risk of a second primary cancer after non-melanoma skin cancer in white men and women: a prospective cohort study. PLoS Med 2013; 10:e1001433; http://dx.doi.org/10.1371/journal.pmed.1001433; PMID: 23630459
- Johannesdottir SA, Lash TL, Jensen AO, Farkas DK, Olesen AB. Mortality in cancer patients with a history of cutaneous squamous cell carcinoma--a nationwide population-based cohort study. BMC Cancer 2012; 12:126; http://dx.doi.org/10.1186/1471-2407-12-126; PMID: 22458954
- Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature 2005; 433:769 - 73; http://dx.doi.org/10.1038/nature03315; PMID: 15685193
- Kanitz A, Imig J, Dziunycz PJ, Primorac A, Galgano A, Hofbauer GF, Gerber AP, Detmar M. The expression levels of microRNA-361-5p and its target VEGFA are inversely correlated in human cutaneous squamous cell carcinoma. PLoS One 2012; 7:e49568; http://dx.doi.org/10.1371/journal.pone.0049568; PMID: 23166713
- Xu N, Zhang L, Meisgen F, Harada M, Heilborn J, Homey B, Grandér D, Ståhle M, Sonkoly E, Pivarcsi A. MicroRNA-125b down-regulates matrix metallopeptidase 13 and inhibits cutaneous squamous cell carcinoma cell proliferation, migration, and invasion. J Biol Chem 2012; 287:29899 - 908; http://dx.doi.org/10.1074/jbc.M112.391243; PMID: 22782903
- Jung YJ, Kim JW, Park SJ, Min BY, Jang ES, Kim NY, Jeong SH, Shin CM, Lee SH, Park YS, et al. c-Myc-mediated overexpression of miR-17-92 suppresses replication of hepatitis B virus in human hepatoma cells. J Med Virol 2013; 85:969 - 78; http://dx.doi.org/10.1002/jmv.23534; PMID: 23532756
- Li X, Zhang Z, Yu M, Li L, Du G, Xiao W, Yang H. Involvement of miR-20a in promoting gastric cancer progression by targeting early growth response 2 (EGR2). Int J Mol Sci 2013; 14:16226 - 39; http://dx.doi.org/10.3390/ijms140816226; PMID: 23924943
- Chang Y, Liu C, Yang J, Liu G, Feng F, Tang J, Hu L, Li L, Jiang F, Chen C, et al. MiR-20a triggers metastasis of gallbladder carcinoma. J Hepatol 2013; 59:518 - 27; http://dx.doi.org/10.1016/j.jhep.2013.04.034; PMID: 23665284
- miR-20a facilitates metastasis of osteosarcoma cells to lung tissue. Bonekey Rep 2012; 1:76; PMID: 23951469
- Yoshino H, Seki N, Itesako T, Chiyomaru T, Nakagawa M, Enokida H. Aberrant expression of microRNAs in bladder cancer. Nat Rev Urol 2013; 10:396 - 404; http://dx.doi.org/10.1038/nrurol.2013.113; PMID: 23712207
- Li X, Pan JH, Song B, Xiong EQ, Chen ZW, Zhou ZS, Su YP. Suppression of CX43 expression by miR-20a in the progression of human prostate cancer. Cancer Biol Ther 2012; 13:890 - 8; http://dx.doi.org/10.4161/cbt.20841; PMID: 22785209
- Zhao S, Yao DS, Chen JY, Ding N. Aberrant expression of miR-20a and miR-203 in cervical cancer. Asian Pac J Cancer Prev 2013; 14:2289 - 93; http://dx.doi.org/10.7314/APJCP.2013.14.4.2289; PMID: 23725129
- Chang CC, Yang YJ, Li YJ, Chen ST, Lin BR, Wu TS, Lin SK, Kuo MY, Tan CT. MicroRNA-17/20a functions to inhibit cell migration and can be used a prognostic marker in oral squamous cell carcinoma. Oral Oncol 2013; 49:923 - 31; http://dx.doi.org/10.1016/j.oraloncology.2013.03.430; PMID: 23602254
- Fan MQ, Huang CB, Gu Y, Xiao Y, Sheng JX, Zhong L. Decrease expression of microRNA-20a promotes cancer cell proliferation and predicts poor survival of hepatocellular carcinoma. Journal of experimental & clinical cancer research. CR (East Lansing, Mich) 2013; 32:21
- Dong Q, Ji YS, Cai C, Chen ZY. LIM kinase 1 (LIMK1) interacts with tropomyosin-related kinase B (TrkB) and Mediates brain-derived neurotrophic factor (BDNF)-induced axonal elongation. J Biol Chem 2012; 287:41720 - 31; http://dx.doi.org/10.1074/jbc.M112.405415; PMID: 23086941
- Lindström NO, Neves C, McIntosh R, Miedzybrodzka Z, Vargesson N, Collinson JM. Tissue specific characterisation of Lim-kinase 1 expression during mouse embryogenesis. Gene Expr Patterns 2011; 11:221 - 32; http://dx.doi.org/10.1016/j.gep.2010.12.003; PMID: 21167960
- Estevez B, Stojanovic-Terpo A, Delaney MK, O’Brien KA, Berndt MC, Ruan C, Du X. LIM kinase-1 selectively promotes glycoprotein Ib-IX-mediated TXA2 synthesis, platelet activation, and thrombosis. Blood 2013; 121:4586 - 94; http://dx.doi.org/10.1182/blood-2012-12-470765; PMID: 23620575
- Nikitina EA, Medvedeva AV, Dolgaia IuF, Korochkin LI, Pavlova GV, Savvateeva-Popova EV. [Participation of GDNF, LIMK1 signal pathways and heat shock proteins in processes of Drosophila learning and memory formation]. Zh Evol Biokhim Fiziol 2012; 48:588 - 96; PMID: 23401971
- Li R, Doherty J, Antonipillai J, Chen S, Devlin M, Visser K, Baell J, Street I, Anderson RL, Bernard O. LIM kinase inhibition reduces breast cancer growth and invasiveness but systemic inhibition does not reduce metastasis in mice. Clin Exp Metastasis 2013; 30:483 - 95; http://dx.doi.org/10.1007/s10585-012-9553-6; PMID: 23239465
- Zhang W, Gan N, Zhou J. Immunohistochemical investigation of the correlation between LIM kinase 1 expression and development and progression of human ovarian carcinoma. J Int Med Res 2012; 40:1067 - 73; http://dx.doi.org/10.1177/147323001204000325; PMID: 22906279
- Zhang H, Wang Y, Xing F, Wang J, Wang Y, Wang H, Yang Y, Gao Z. Overexpression of LIMK1 promotes migration ability of multidrug-resistant osteosarcoma cells. Oncol Res 2011; 19:501 - 9; http://dx.doi.org/10.3727/096504012X13286534482511; PMID: 22715593
- Roy P, Madan E, Kalra N, Nigam N, George J, Ray RS, Hans RK, Prasad S, Shukla Y. Resveratrol enhances ultraviolet B-induced cell death through nuclear factor-kappaB pathway in human epidermoid carcinoma A431 cells. Biochem Biophys Res Commun 2009; 384:215 - 20; http://dx.doi.org/10.1016/j.bbrc.2009.04.100; PMID: 19397895
- Prasad S, Madan E, Nigam N, Roy P, George J, Shukla Y. Induction of apoptosis by lupeol in human epidermoid carcinoma A431 cells through regulation of mitochondrial, Akt/PKB and NFkappaB signaling pathways. Cancer Biol Ther 2009; 8:1632 - 9; http://dx.doi.org/10.4161/cbt.8.17.9204; PMID: 19625778
- Ung N, Putoczki TL, Stylli SS, Ng I, Mariadason JM, Chan TA, Zhu HJ, Luwor RB. Anti-EGFR therapeutic efficacy correlates directly with inhibition of STAT3 activity. Cancer Biol Ther 2014; 15:623 - 32; http://dx.doi.org/10.4161/cbt.28179; PMID: 24556630
- Wu D, Guo Z, Min W, Zhou B, Li M, Li W, Luo D. Upregulation of TCTP expression in human skin squamous cell carcinoma increases tumor cell viability through anti-apoptotic action of the protein. Exp Ther Med 2012; 3:437 - 42; PMID: 22969908
- Xia YH, Li M, Fu DD, Xu SL, Li ZG, Liu D, Tian ZW. Effects of PTTG down-regulation on proliferation and metastasis of the SCL-1 cutaneous squamous cell carcinoma cell line. Asian Pac J Cancer Prev 2013; 14:6245 - 8; http://dx.doi.org/10.7314/APJCP.2013.14.11.6245; PMID: 24377512
- Sand M, Skrygan M, Georgas D, Sand D, Hahn SA, Gambichler T, Altmeyer P, Bechara FG. Microarray analysis of microRNA expression in cutaneous squamous cell carcinoma. J Dermatol Sci 2012; 68:119 - 26; http://dx.doi.org/10.1016/j.jdermsci.2012.09.004; PMID: 23026055
- Gastaldi C, Bertero T, Xu N, Bourget-Ponzio I, Lebrigand K, Fourre S, Popa A, Cardot-Leccia N, Meneguzzi G, Sonkoly E, et al. miR-193b/365a cluster controls progression of epidermal squamous cell carcinoma. Carcinogenesis 2014; 35:1110 - 20; http://dx.doi.org/10.1093/carcin/bgt490; PMID: 24374827
- Bertero T, Bourget-Ponzio I, Puissant A, Loubat A, Mari B, Meneguzzi G, Auberger P, Barbry P, Ponzio G, Rezzonico R. Tumor suppressor function of miR-483-3p on squamous cell carcinomas due to its pro-apoptotic properties. Cell Cycle 2013; 12:2183 - 93; http://dx.doi.org/10.4161/cc.25330; PMID: 24067364
- Zhou M, Liu W, Ma S, Cao H, Peng X, Guo L, Zhou X, Zheng L, Guo L, Wan M, et al. A novel onco-miR-365 induces cutaneous squamous cell carcinoma. Carcinogenesis 2013; 34:1653 - 9; http://dx.doi.org/10.1093/carcin/bgt097; PMID: 23514750
- Olive V, Jiang I, He L. mir-17-92, a cluster of miRNAs in the midst of the cancer network. Int J Biochem Cell Biol 2010; 42:1348 - 54; http://dx.doi.org/10.1016/j.biocel.2010.03.004; PMID: 20227518
- Chen J, Yao D, Li Y, Chen H, He C, Ding N, Lu Y, Ou T, Zhao S, Li L, et al. Serum microRNA expression levels can predict lymph node metastasis in patients with early-stage cervical squamous cell carcinoma. Int J Mol Med 2013; 32:557 - 67; PMID: 23799609
- Liu R, Liao J, Yang M, Shi Y, Peng Y, Wang Y, Pan E, Guo W, Pu Y, Yin L. Circulating miR-155 expression in plasma: a potential biomarker for early diagnosis of esophageal cancer in humans. J Toxicol Environ Health A 2012; 75:1154 - 62; http://dx.doi.org/10.1080/15287394.2012.699856; PMID: 22891887
- Scott RW, Hooper S, Crighton D, Li A, König I, Munro J, Trivier E, Wickman G, Morin P, Croft DR, et al. LIM kinases are required for invasive path generation by tumor and tumor-associated stromal cells. J Cell Biol 2010; 191:169 - 85; http://dx.doi.org/10.1083/jcb.201002041; PMID: 20876278
- Jang I, Jeon BT, Jeong EA, Kim EJ, Kang D, Lee JS, Jeong BG, Kim JH, Choi BH, Lee JE, et al. Pak1/LIMK1/Cofilin Pathway Contributes to Tumor Migration and Invasion in Human Non-Small Cell Lung Carcinomas and Cell Lines. Korean J Physiol Pharmacol 2012; 16:159 - 65; http://dx.doi.org/10.4196/kjpp.2012.16.3.159; PMID: 22802696