Abstract
Cyclin D1 overexpression is a common feature of many human malignancies. Genomic deletion analysis has demonstrated a key role for cyclin D1 in cellular proliferation, angiogenesis, and cellular migration. To investigate the mechanisms contributing to cyclin D1 functions, we purified cyclin D1a-associated complexes by affinity chromatography and identified the PACSIN 2 (protein kinase C and casein kinase substrate in neurons 2) protein by mass spectrometry. The PACSIN 2, but not the related PACSIN 1 and 3, directly bound wild-type cyclin D1 (cyclin D1a) at the carboxyl terminus, and failed to bind cyclin D1b, the alternative splicing variant of cyclin D1. PACSIN 2 knockdown induced cellular migration and reduced cell spreading in LNCaP cells expressing cyclin D1a. In cyclin D1-/- mouse embryonic fibroblasts (MEFs), cyclin D1a, but not cyclin D1b, reduced the cell spreading to a polarized morphology. siPACSIN 2 had no effect on cellular migration of cyclin D1-/- MEFs. Cyclin D1a restored the migratory ability of cyclin D1-/- MEFs, which was further enhanced by knocking down PACSIN 2 with siRNA. The cyclin D1-associated protein, PACSIN 2, regulates cell spreading and migration, which are dependent on cyclin D1 expression.
Introduction
The orderly transition through the cell cycle is orchestrated by the sequential activation and inactivation of cyclin-dependent kinases. The regulatory subunits of the CDK4/6 kinase are encoded by the D-type cyclins. Cyclin D1 is induced early during G1 phase progression by mitogenic stimuli, which activate the transcription of cyclin D1, through specific DNA sequences in the cyclin D1 promoter and through the assembly of protein complexes.Citation1,Citation2 The cyclin D1/CDK complexes phosphorylate and inactivate the retinoblastoma (pRb) protein promoting nuclear DNA synthesis. We have previously shown that the nuclear respiratory factor 1 (NRF-1) is phosphorylated in a cyclin D1-dependent manner. Phosphorylation of NRF-1 represses its transactivation function thereby inhibiting mitochondrial metabolism.Citation3
The heterodimeric partner of cyclin D1, CDK4/6, is associated in high molecular weight cytoplasmic complexes containing Hsp90 and Cdc37.Citation4,Citation5 Hsp90 and Cdc37 facilitate appropriate CDK4 folding, inducing competence for D-type cyclin binding. The sequential association requires assembly factors to bring cyclin D1 and CDK4 together.Citation6,Citation7 Both p21CIP1 and p27KIP1 facilitate assembly of the cyclin D/CDK complexes.Citation8 Additional components that contribute to the molecular weight of holoenzymes have been identified in complexes ranging from 150 to 200 kDaCitation4,Citation9,Citation10 including Hsc70.Citation11
In keeping with a role for cyclin D1 in diverse human cancers, including breast, colon, prostate and hematopoetic malignancies,Citation2 mice deficient in cyclin D1 are resistant to oncogene-induced tumorigenesis. Gastrointestinal tumors induced by mutation of the Apc gene are reduced in number by cyclin D1 deficiency.Citation12 Consistent with findings that the cyclin D1 antisense abrogates mammary epithelial cell growth induced by ErbB2,Citation13 cyclin D1-deficient mice are resistant to mammary gland tumorigenesis induced by Ras or ErbB2.Citation14 Cyclin D1-deficient cells demonstrate reduced cellular survival and DNA synthesis,Citation15 increased mitochondrial size and activity,Citation3,Citation16 and reduced cellular migration of diverse cell types including blood vessels, bone marrow macrophages, fibroblasts and mammary epithelial cells.Citation17 It has been predicted that the diverse functions regulated by cyclin D1 in cellular differentiation, proliferation and migration may be governed by subpartitioning of cyclin D1 into distinct subcellular compartments, or through physical association with distinct binding proteins. In this regard, cyclin D1 associates with the p160 coactivator SRC-1 (AIB-1) to regulate estrogen receptor α activity. In association with transcription factors, cyclin D1a regulates CDK-independent transcriptional activities of the androgen receptor, CEBPα and PPARγ.Citation2,Citation18 Cyclin D1 recruitment to transcription factor binding sites in the contect of local chromatin by ChIP assays is associated with the recruitment of histone modifying enzymes including HDAC1, HDAC3, p300/CBP, SUV39 and HP1α.Citation19–Citation21
A common polymorphism of the human cyclin D1 gene has been associated with an increased rate of cancer development.Citation22–Citation24 The polymorphism (A870G) is located at the splice donor region at the exon 4-intron 4 boundary and modulates the efficiency of alternate splicing. Alternate splicing results in distinct carboxyl terminal amino acid sequences. Characterization of the functional properties of the canonical cyclin D1a and the alternate cyclin D1b isoform has revealed that each encodes subunits with a similar capacity to phosphorylate pRb, but distinguishable abilities to regulate cellular migration. Cyclin D1a promotes migration of fibroblasts and mammary epithelial cells.Citation24,Citation25 However, the cyclin D1b isoform is defective in promoting migration.Citation24,Citation26 The mechanism responsible for these distinct functions of cyclin D1a and cyclin D1b is unknown, although a postulated mechanism includes distinct interaction partners.
In order to determine adapter proteins regulating cyclin D1 function, we immunopurified cyclin D1a-associated proteins. Mass spectrometry and sequence analysis identified PACSIN 2 as a cyclin D1a-associated protein. PACSIN family members (also called syndapins) have been shown to function as cytoplasmic adapter proteins in focal adhesions. Herein, PACSIN 2 co-localized in membrane ruffles with cyclin D1a. The current studies demonstrate the physical association of cyclin D1a, but not cyclin D1b, with NPF motifs of PACSIN 2. We show that endogenous PACSIN 2 represses cellular migration in a cyclin D1a-dependent manner.
Results
Identification of PACSIN as a cyclin D1-binding protein.
In view of the diverse functions of cyclin D1 in DNA synthesis, oncogenesis and migration, we hypothesized that cyclin D1-associated proteins may mediate these functions. Therefore, to identify proteins associated with the cyclin D1a protein, immunopurification of cyclin D1a complexes was conducted using a column pre-loaded with 1 ml of agarose beads conjugated with FLAG antibody (Sigma). 20 mg of cellular extracts were prepared from HEK 293T cells transfected with a vector expressing FLAG-tagged cyclin D1a and loaded on to the column. The complexes co-associating with cyclin D1 were eluted and separated by SDS-PAGE gel followed by silver staining. Bands were excised, eluted and subjected to electro-spray liquid mass spectroscopy. Co-purifying proteins included Cdk4 and heat shock protein, HSC70 that have been previously identified as cyclin D1a associated proteins.Citation11 An additional protein was identified by mass spectrometry with sequences identical to the homolog of chicken FAP52, now identified as PACSIN 2. The PACSIN (Protein Kinase C and Casein Kinase 2 Substrate) family of proteins is structurally conserved and functions as cytoplasmic adaptor proteins.Citation27 The association of PACSIN 2 with cyclin D1a was validated by Immunoprecipitation-western blotting (IP-WB). Protein lysates were prepared from either NIH 3T3 cells or murine liver. IP was conducted using agarose beads pre-conjugated with anti-cyclin D1 (mouse) antibody (Santa Cruz biotechnology, Clone 72-13G), which was followed by WB to detect endogenous Pacsin 2 that bound to cyclin D1. As shown in , Pascin 2 was co-immunoprecipitated with cyclin D1. We further confirmed this observation in cyclin D1-deficient HEK 293T cells. The cells were co-transfected with FLAG-tagged cyclin D1 and Myc-tagged PACSIN 2. IP was conducted for cyclin D1 using a FLAG antibody conjugated to agarose. WB analysis was performed with an anti-Myc antibody. As shown in and C, PACSIN 2 was co-precipitated with cyclin D1.
Cyclin D1 binds to PACSIN 2 through its carboxyl terminus containing an E-rich motif.
To identify the domains of cyclin D1 required for PACSIN 2 binding, cyclin D1 deletion mutant expression vectors were assessed (). Deletion of the carboxyl terminal 28 amino acids of cyclin D1 abolished binding to PACSIN 2. The role of an acidic rich region within the carboxyl terminus of cyclin D1, a stretch of eight amino acids referred to as the “E-rich domain,”Citation28 was examined for PACSIN 2 binding. Deletion of the acidic rich stretch (cyclin D1 ΔE) reduced binding of PACSIN 2 to ∼6% of wild-type (WT) cyclin D1 ( and C). The C-terminal deletion mutant (D1-N1) has an intact E-rich motif and showed ∼45% of WT binding to PACSIN 2, suggesting additional C-terminal sequences (last 8 amino acids of cyclin D1 protein) contribute to complete PACSIN 2 binding. Sequence alignment of D-type cyclins suggested that E278 and E280 within cyclin D1 are conserved in cyclin D2, but not in cyclin D3. Cyclin D1 and cyclin D2, but not cyclin D3 (data not shown), co-precipitated with PACSIN 2 in transfected cells, suggesting a role of these conserved glutamic acids in mediating the protein interaction with PACSIN 2. Point mutations of amino acid residues 278 or 280 reduced binding of PACSIN 2 by 70% ( and D). Similarly, the conserved acidic residues within cyclin D2, when mutated (E272A cyclin D2 or E274A cyclin D2), reduced binding of PACSIN 2 by 70 to 80% (). The cyclin D1b protein failed to bind PACSIN 2 (). Together, these studies demonstrate that PACSIN 2 binds cyclin D1a, but not the alternately spliced form cyclin D1b. Further, these studies demonstrate the importance of the acidic rich region within the carboxyl terminus of cyclin D1 and D2 in physical association with PACSIN 2.
The NPF motifs of PACSIN 2 are required for cyclin D1 binding.
The physical association of cyclin D1 with the PACSIN family members was assessed using IP-WB. HEK293T cells were transduced with expression vectors encoding PACSIN 1, 2 or 3 () and an expression vector encoding cyclin D1 with an amino-terminal FLAG epitope. Cyclin D1 co-precipitated PACSIN 2, but not PACSIN 1 or PACSIN 3 (). In order to identify the binding motifs of PACSIN 2 required for physical interaction with cyclin D1a, a series of point mutants of PACSIN 2 were examined. The amino terminus of PACSIN proteins encodes an F-BAR domain and an SH3 domain is located within the carboxyl terminus. The SH3 motif interacts with proline-rich proteins to mediate protein-protein interactions. The C-terminal SH3 domain of PACSIN 2 regulates interactions with dynamin1, synapsin 1, mSos and N-WASP. The N-terminal F-BAR domain of PACSIN 2 is required for homo-ligomerization and association with curved membranesCitation29–Citation31 and mediates several cytoskeletal rearrangements required for cytokinesis in S. pombe.Citation32 We examined further the domains of PACSIN 2 required for cyclin D1 binding using C-terminal truncation mutants. Deletion of NPF-2 and NPF-3 domains had no significant effect on the interaction between cyclin D1 and PACSIN 2 (). Deletion of all three NPF domains abolished the binding, suggesting that these NPF domains are required for binding. A PACSIN 2 mutant with an internal deletion of a single NPF domain (NPF-1) maintained the ability to bind cyclin D1 (), suggesting that the remaining NPF-2 and NPF-3 may contribute to cyclin D1 binding.
PACSIN 2 represses cellular migration and enhances cell spreading.
In view of the finding that cyclin D1 promotes cellular migration and PACSIN 2 associates with cyclin D1 in membrane ruffles, we examined the role of PACSIN 2 in mammalian cellular migration using prostate cancer LNCaP cells expressing PACSIN 2 and cyclin D1 ( and data not shown). siPACSIN 2 reduced PACSIN 2 abundance and enhanced transwell migration with >2-fold increase (p < 0.001) ( and C). In view of the relationship between cellular migration and cell spreading, we determined the role of endogenous PACSIN 2 on cell spreading. NIH 3T3 cells express abundant PACSIN 2 protein (data not shown) and were more suitable for cell spreading assay considering the consistent and relatively short time for this cell to attach to the culture surface and spread (data not shown). The validated siRNA pool to PACSIN 2 (Santa Cruz Biotechnology) reduced the expression level of PACSIN 2 by 80% ( and H). We compared the spreading efficiency of cells transfected with either siPACSIN 2 or scramble siRNA on fibronectin-coated cell culture dishes. As shown in , a difference in the appearance of cell spreading was observed between control siRNA (Ctrl. siRNA) and siPACSIN 2 expressing cells after 90 min of seeding ( and F). We have previously shown that cyclin D1 promotes cellular migration.Citation25,Citation26 We next determined whether PACSIN 2 inhibition of cellular migration is dependent on cyclin D1 levels. NIH 3T3 and LNCaP cells were transiently transfected with siPACSIN 2 or control. Western blot confirmed a reduction of PACSIN 2 abundance ( and H). Cyclin D1 levels were decreased in cells transfected with siPACSIN 2. Conversely, overexpression of PACSIN 2 in MCF-7 cells increased cyclin D1 protein abundance (). Collectively, these findings suggest that PACSIN 2 may not regulate migration through affecting cyclin D1 abundance because PACSIN 2 inhibits migration; cyclin D1 promotes migration; and PACSIN 2 increases cyclin D1 abundance.
Cyclin D1a is required for PACSIN 2 repression of cellular migration.
Cyclin D1a overexpression enhances cellular migration. A similar phenotype was induced by siPACSIN 2 expression. We examined whether PACSIN 2 repression of cellular migration was cyclin D1 dependent. We first determined whether cyclin D1 re-introduction could restore the migratory ability of cyclin D1−/− MEFs. Cyclin D1−/− MEFs were transduced with a viral expression vector encoding cyclin D1a, cyclin D1b or GFP control. Cyclin D1a, but not cyclin D1b, induced a more polarized morphology (), which is consistent with our prior observation.Citation17 siPACSIN 2 reduced the abundance of PACSIN 2 by >80% compared to scramble control (). siPACSIN 2 increased the transwell migration by 2-fold in cells transduced with cyclin D1a compared to GFP or cyclin D1b (p < 0.001) (). The time-lapse video-microscopy to track the single cell movement was also performed. Cyclin D1+/+ and cyclin D1−/− mouse embryonic fibroblast cells were transfected with vector encoding PACSIN 2. Expression of PACSIN 2 significantly reduced the migration velocity of cyclin D1+/+ cells, but not cyclin D1−/− cells ( and E).
Endogenous PACSIN 2 enhances cell spreading via cyclin D1a.
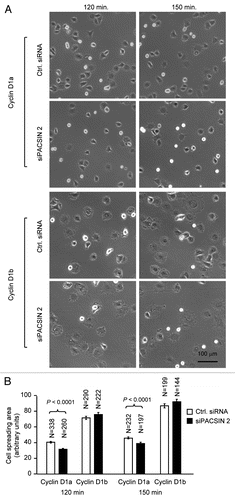
Cyclin D1 promotes cellular migration and MEFs derived from cyclin D1−/− mice showed enhanced cell spreading, we investigated whether the effect of PACSIN 2 on cell spreading is cyclin D1-dependent. Cell spreading assays were conducted on non-coated or fibronectin-coated plastic surface using cyclin D1−/− MEFs transduced with viral expression vector encoding cyclin D1a or cyclin D1b. Re-introduction of physiological levels of cyclin D1a reduced the abnormal spreading of cyclin D1−/− MEFs on both non-coated and fibronectin-coated surface. Data presented here was from fibronectin-coated surface. In contrast, siPACSIN 2 transfected cells spread at a reduced rate with only about 20% of the cells spreading at 60 min and 50% at 90 min. At 60 min after seeding, about half of the cyclin D1−/− +D1a MEFs had spread, similar to that in cyclin D1 wild-type cells. siPACSIN 2 did not further reduce cell spreading in cyclin D1−/− MEFs (data not shown). Knockdown of PACSIN 2 reduced the cell spreading only when cyclin D1 was present (). We had previously shown that cyclin D1b is defective in rescuing the impaired migration phenotype in cyclin D1−/− cells. Due to the slow migratory phenotype of cyclin D1−/− cells, it is not feasible to test the effect of knocking down PACSIN 2 on cellular migration. We compared the spreading efficiency of cyclin D1−/−+D1b to cyclin D1−/−+D1a cells. A well-spreading phenotype was observed at 120 min after cyclin D1−/−+D1b cells were plated. Knocking down PACSIN 2 by siRNA did not affect the cell spreading in cyclin D1b expressing cells.
Discussion
The abundance of cyclin D1 is a key determinant of both human and murine tumorigenesis.Citation1,Citation33 The identification of cyclin D1 binding partners that contribute to the diverse functions of cyclin D1 governing DNA synthesis, contact-independent growth, angiogenesis and cellular migration, are critical for understanding cyclin D1 function. Proteins involved in cell cycle regulation (CDK4/6, p21CIP1/p27KIP1), mitochondrial biogenesis (NRF-1),Citation3 tumor suppression (pRb, BRCA1),Citation34 DNA-binding transcription factors and co-integrator proteins with histone acetyl transferase (HAT) or histone deacetylase activity (HDAC) associate with cyclin D1 to coordinate diverse functions (reviewed in ref. Citation2). Through identification of polypeptides associated with cyclin D1 by tandem mass spectrometry, the current studies have identified PACSIN 2 as a new cyclin D1-associated protein, which contributes to the induction of cellular migratory directionality.
The current studies demonstrate PACSIN 2 binding to the cyclin D1 and D2, but not cyclin D3 carboxyl terminus. The carboxyl terminal region of cyclin D1a (amino acids 264–295) is structurally divergent from cyclin D3 () and the alternate transcript of cyclin D1, cyclin D1b. PACSIN 2 failed to bind either cyclin D3 or cyclin D1b. Point mutation of amino acids 278 or 280 reduced cyclin D1 binding to PACSIN 2 by greater than 60%. It is likely that the D-type cyclins evolved distinct structures to conduct distinct functions. The alternate transcript of cyclin D1, cyclin D1b, is associated with poor prognosis in head and neck cancer and breast cancer.Citation23,Citation35,Citation36
Cyclin D1-deficient macrophages, fibroblasts and mammary epithelial cells show reduced cellular migration and enhanced adhesion.Citation17,Citation24,Citation37 Reintroduction of cyclin D1 rescued the defect in migration consistent with the role for cyclin D1 in promoting migration of macrophages and blood vessels.Citation17,Citation38 siPACSIN 2 increased cellular migration, indicating a role for PACSIN 2 in repressing cellular migration. Together, these studies are consistent with a model in which endogenous PASCIN 2 serves as a brake to cyclin D1-mediated induction of cellular migration. PACSIN 1 regulates the induction of actin rearrangements mediated by Arp2/3 complexes.Citation39–Citation41 In Xenopus cells, PACSIN 2 co-localizes with ADAM13. PACSIN 2 interacts with the cytoplasmic domain of ADAM13, a cell surface metalloprotease and the two proteins are co-localized in membrane ruffles and cytoplasmic vesicles.Citation42 PACSIN 2 functions to regulate ADAM13 subcellular localization and its catalytic activity.Citation42 The possibility that cyclin D1 association with PACSIN 2 regulates ADAM13 metalloprotease function remains to be determined. All three PACSIN proteins interact with dynamin I, synapsin, synaptojanin and the neural Wiskott-Aldrich syndrome protein (N-WASP). The PACSIN SH3 domain regulates vesicle endocytosis.Citation39,Citation43 The current finding that PACSIN 2 associates with cyclin D1a, suggests a mechanism by which cyclin D1 may contribute to the induction of migration.
Materials and Methods
Plasmids, antibodies and reagents.
Mutants of the D-type cyclin cDNAs (D1, D2 and D3) were generated by PCR and subcloned into p3xFLAG-CMV-10 (Sigma) or MSCV-IRES-GFP. pcDNA3-Myc-PACSIN 1, 2, 3 were described in a previous publication.Citation44 All the mutant constructs for PACSIN 2 were generated by PCR site-directed mutagenesis. The cDNA of PACSIN 2 were amplified by PCR and subcloned into the MSCV-IRES-GFP vector to generate retroviral expression plasmid. Anti-Myc (clone 9E10), anti-cyclin D1 (DCS6), anti-β-actin, siPACSIN 2 (sc-36174(m), sc-36173(h)), control siRNA (sc-27007) were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Anti-PACSIN 2 rabbit serum was previously described in reference Citation44.
Cell culture, transfection and transduction.
The human embryonic kidney 293T (HEK293T), prostate cancer cell line LNCaP and NIH 3T3 were maintained in DMEM containing 1% penicillin/streptomycin and supplemented with 10% fetal bovine serum (FBS). The cyclin D1 wild type (WT) and cyclin D1−/− mouse embryonic fibroblasts (MEFs) were described previously in reference Citation15. For transient transfection, Superfect transfection reagent was used following manufacturer's protocol (Qiagen, Valencia, CA) and with slight modification.Citation45 For siRNA transfection, Oligofectamine was used following the manufacturer's description with minor modification.Citation46 Briefly, cells were placed on 6-well plate at 50% confluence at the time of transfection. 80 pmol siRNA was transfected into cells and analyzed 72 h later, as indicated. The transfection efficiency was detected by control fluor-labeled siRNA.
For cell transduction, retroviruses were prepared by transient co-transfection with helper virus into HEK 293T cells using calcium phosphate precipitation. HEK 293T cells were transfected with plasmid DNA and cultured at 37°C for 12 h, the medium was replaced and after 36 h the supernatant was collected and filtered through a 0.45 µm filter. Cells were infected at approximately 70% confluence in DMEM supplemented with 8 µg/ml of polybrene, then incubated overnight at 37°C, 5% CO2. The following day the media was changed to DMEM medium with 10% FBS and cultured for further assay. A retroviral expression plasmid encoding GFP was included for monitoring infection efficiency.
Protein identification by MS/MS.
Protein complexes were purified from whole-cell extracts prepared in lysis buffer (50 mM Tris HCl, pH 7.4, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100) supplemented with protease inhibitor on affinity column packed with M2-agarose (Sigma). Following 10–20 column volumes of TBS (50 mM Tris HCl, pH 7.4, 150 mM NaCl) washes, protein complexes were eluted with TBS buffer containing 100 µg/mL 3x FLAG peptide. Purified complexes were concentrated with Microcon YM-10 (Bedford, MA), resolved on 10% SDS-PAGE gel prior to preparation for MALDI mass mapping using instrument PerSeptive DE STR MALDI-TOF instrument.
Immunoprecipitation and immunoblotting analysis.
Cells were lysed in radioimmune precipitation assay buffer (50 mM Tris-HCl pH 7.4, 1% Nonidet P-40, 0.25% sodium deoxycholate, 150 mM NaCl, 1 mM EDTA, 1 mM NaF, 2 mM Na3VO4 and 1 mM phenylmethylsulfonyl fluoride).Citation47 Whole cell lysates (500 µg) were immunoprecipitated with 1 µg of antibodies as indicated. Immunoprecipitates were washed three times with washing buffer (20 mM Hepes pH 7.5, 150 mM NaCl, 1% glycerol, 0.1% Triton X-100 and 1 mM Na3VO4) and resolved by SDS-PAGE gel followed by immunoblotting with the indicated antibodies.
Cellular migration assay.
Cells were transfected with control and siPACSIN 2 and Transwell migration assays were performed as previous described in reference Citation26. Following transfection, the cells were allowed to express siRNAs for 72 h, then trypsinized and seeded on an 8 µm-pore size Transwell filter insert (Costar) coated with ECM (Sigma, St. Louis, MO) at a density of 1 × 104 cells in each well in DMEM containing 10% FBS. After 6 h of incubation at 37°C and 5% CO2, cells adherent to the upper surface of the filter were removed using a cotton applicator. Cells were stained with 0.4% crystal violet dissolved in methanol, and the numbers of cells on the bottom were counted. Data are from at least three experiments done in triplicate (mean ± SEM).
Cell spreading and migration assays.
Cyclin D1−/− MEFs were transduced with retroviral vector expressing GFP, cyclin D1a and cyclin D1b, respectively, followed by transient transfection with siRNA to PACSIN 2. 72 hrs post-transfection, the cells were detached with trypsin and 2 × 105 of cells were re-seeded in the 6-well plate pre-coated with fibronectin (10 µg/ml) and incubated at 37°C. The cells were washed with medium after 30 min and images were collected at the time points indicated. The experiments were carried out in triplicate and the number of attached cells at 30 min was counted. The images were taken with the 20x objective (Axiovert 200, Zeiss) and further analyzed by NIH Image J and the average cell spreading was calculated as previously described in reference Citation48. For time-lapse observation of cell movement, cells on 12-well plates were maintained in DMEM with 10% fetal calf serum (FCS) and HEPES. Cells were placed in an incubator to maintain the temperature at 37°C and CO2 at 5%. The cell movement videos were taken at 5-min intervals by using a Nikon Eclipse TE-300 inverted microscope system. The cell movement velocity was determined by tracing the single cells at different time points using MetaMorph software.
Abbreviations
| MEFs | = | mouse embryonic fibroblasts |
| PACSIN 2 | = | protein kinase C and casein kinase substrate in neurons 2 |
| pRb | = | retinoblastoma |
| CDK | = | cyclin-dependent kinase |
Figures and Tables
Figure 1 Cyclin D1 binds to PACSIN 2 through its C-terminal E-rich motif. (A) Immunoprecipitation (IP)-western blot (WB) was performed to determine the binding of endogenous cyclin D1 and Pacsin 2. Protein lysates were prepared from either NIH 3T3 cells or murine liver tissue. IP was conducted with a cyclin D1 antibody pre-conjugated to agarose. The immunoprecipitates was subjected to WB for detection of Pacsin 2. (B) IP-WB was conducted of cells transfected with either an expression vector for Myc epitope-tagged PACSIN 2 and FLAG-tagged cyclin D1 wild type or mutant expression vectors. The schematic representation shows the acidic-rich region. The C-terminal E-rich motif is required for PACSIN 2 binding. (C) Amino acid sequence of the carboxyl terminal region of cyclin D1. IP-WB of PACSIN 2 association with D-type cyclins, wild type or mutant is as shown. IP was conducted with the FLAG antibody directed to the N-terminus of cyclin Ds with western blot to detect the Myc epitope of PACSIN 2. The amino acid residues required for the binding were mapped to E278 and E280 for cyclin D1 and E272 and E274 for cyclin D2, respectively. (D) Schematic representation of constructs of D-type cyclins that were used for mapping the domain requirement for PACSIN 2 binding.
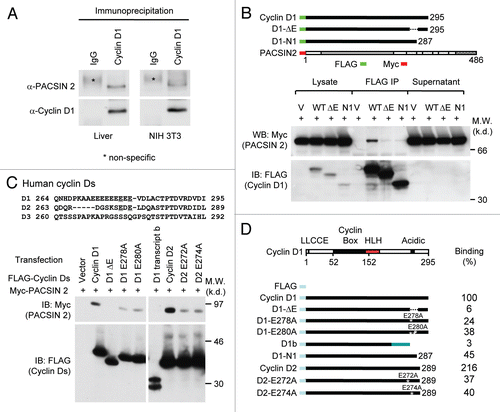
Figure 2 The NPF motifs of PACSIN 2 are required for cyclin D1 binding. (A) Schematic representation of PACSIN 1, 2 and 3. (B) The mammalian expression vectors encoding PACSIN 1, 2 or 3 were co-expressed with FLAG-tagged cyclin D1 and subsequent IP-WB was conducted. The Myc epitope of PACSINs are shown by western blot of cellular lysates. Co-precipitation of cyclin D1 with PACSIN 2 is shown by immunoprecipitation of cyclin D1 with FLAG antibody. (C and D) HEK293T cells were co-transfected with mutant constructs of PACSIN 2 (as indicated) together with vector encoding FLAG-cyclin D1. Immunoprecipitation was performed with an anti-Myc antibody following by western blot using FLAG antibody to detect cyclin D1.
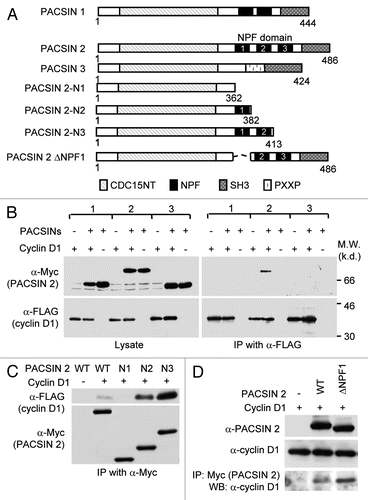
Figure 3 Knockdown of PACSIN 2 reduced cell migration. (A) Prostate cancer cells, LNCaP, were transiently transfected with PACSIN 2 siRNA (siPACSIN 2). 72 hr after transfection, western blot was conducted for PACSIN 2 protein. Actin was included as protein loading control. (B) Transwell migration assays were performed with cells transfected with either control siRNA (Ctrl. siRNA) or siPACSIN 2. (C) Quantitative analysis of trans-migrated cells. (D) 3T3 cells were assessed for cell spreading. Cells transfected with siPACSIN 2 have reduced cell spreading phenotype. (E) Quantitative analysis of cell spreading (arbitrary unit). (G and H) NIH 3T3 and LNCaP cells were transiently transfected with siPACSIN 2 or control. Western blot was performed to determine the expression of cyclin D1 and PACSIN 2. (I) MCF-7 cells were transfected vector encoding PACSIN 2. GDI and β-actin served as protein loading control.
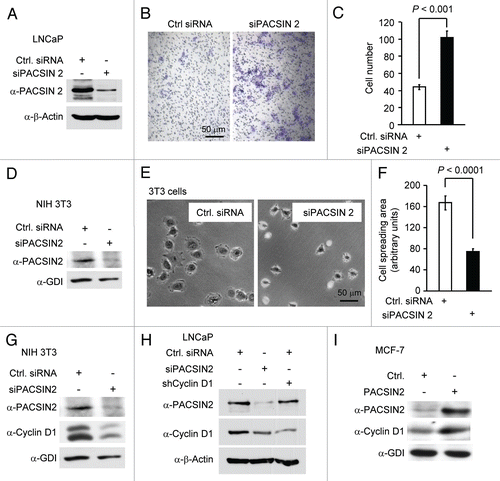
Figure 4 PACSIN 2 repression of cellular migration is dependent on cyclin D1a. (A) Cyclin D1−/− MEFs were transduced with viral expression vector (MSCV-IRES-GFP) encoding cyclin D1a, cyclin D1b or GFP control. (B) Cells were transiently transfected with siPACSIN 2. Western blot was performed with an anti-PACSIN 2 antibody to show a reduction of endogenous PACSIN 2. (C) Transwell migration assays were conducted in triplicate. Cells that migrated were stained and counted. Five fields in each of triplicate wells were randomly recorded. (D and E) The time-lapse video-microscopy was conducted in cyclin D1+/+ and cyclin D1−/− MEFs transfected with a PACSIN 2 expression or control vector. Vector expressing GFP was co-transfected. The GFP-positive cells were further analyzed for cell migration velocity. Student t-test was used for quantitative analysis of cell migration.
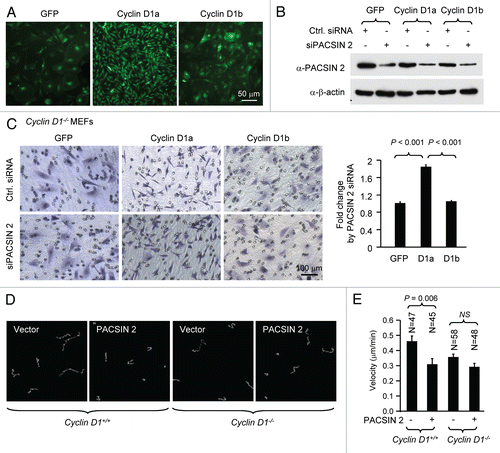
Figure 5 PACSIN 2 regulation of cell spreading depends on cyclin D1 expression. (A) Cyclin D1−/− cells transduced with either cyclin D1a or cyclin D1b retrovirus and transfected with either control or siPACSIN 2 were assessed for cell spreading at different time points after seeding. (B) Quantitative analysis of cell spreading (arbitrary unit). Student t-test was used for quantitative analysis of cell spreading. p-value and number of cells analyzed were indicated in the figure. N is for the number of cells counted. Data are representative of three separate experiments.
Acknowledgements
This work was supported in part by awards from the Susan Komen Breast Cancer Foundation [BCTR0504227 to C.W.], National Institutes of Health [R01CA70896, R01CA75503 and R01CA86072 to R.G.P.]. Work conducted at the Kimmel Cancer Center was supported by the NIH Cancer Center Core grant [P30CA56036 to R.G.P.]. This project was partially supported by the China Scholarship Council. This project is funded in part by the Pennsylvania Department of Health grant [C.W., R.G.P.]. The Department specifically disclaims responsibility for any analyses, interpretations or conclusions.
References
- Sherr CJ, Roberts JM. CDK inhibitors: Positive and negative regulators of G1-phase progression. Genes and Dev 1999; 13:1501 - 1512
- Fu M, Wang C, Li Z, Sakamaki T, Pestell RG. Minireview: Cyclin D1: Normal and Abnormal Functions. Endocrinology 2004; 145:5439 - 5447
- Wang C, Li Z, Lu Y, Du R, Katiyar S, Yang J, et al. Cyclin D1 repression of nuclear respiratory factor 1 integrates nuclear DNA synthesis and mitochondrial function. Proc Natl Acad Sci USA 2006; 103:11567 - 11572
- Mahony D, Parry DA, Lees E. Active cdk6 complexes are predominantly nuclear and represent only a minority of the cdk6 in T cells. Oncogene 1998; 16:603 - 611
- Stepanova L, Leng X, Parker SB, Harper JW. Mammalian p50Cdc37 is a protein kinase-targeting subunit of Hsp90 that binds and stabilizes Cdk4. Genes Dev 1996; 10:1491 - 1502
- Cheng M, Sexl V, Sherr CJ, Roussel MF. Assembly of cyclin D-dependent kinase and titration of p27Kip1 regulated by mitogen-activated protein kinase kinase (MEK1). Proc Natl Acad Sci USA 1998; 95:1091 - 1096
- Kato Jy, Matsuoka M, Stromm DK, Sherr CJ. Regulation of cyclin D-dependent kinase 4 (cdk4) by cdk4-activating kinase. Mol Cell Biol 1994; 14:2713 - 2721
- LaBaer J, Garrett MD, Stevenson LF, Slingerland JM, Sandhu C, Chou HS, et al. New functional activities for the p21 family of CDK inhibitors. Genes Dev 1997; 11:847 - 862
- McConnell BB, Gregory FJ, Stott FJ, Hara E, Peters G. Induced expression of p16(INK4a) inhibits both CDK4- and CDK2-associated kinase activity by reassortment of cyclin-CDK-inhibitor complexes. Mol Cell Biol 1999; 19:1981 - 1989
- Parry D, Mahony D, Wills K, Lees E. Cyclin D-CDK subunit arrangement is dependent on the availability of competing INK4 and p21 class inhibitors. Mol Cell Biol 1999; 19:1775 - 1783
- Diehl JA, Yang W, Rimerman RA, Xiao H, Emili A. Hsc70 regulates accumulation of cyclin D1 and cyclin D1-dependent protein kinase. Mol Cell Biol 2003; 23:1764 - 1774
- Hulit J, Wang C, Li Z, Albanese C, Rao M, Di Vizio D, et al. Cyclin D1 genetic heterozygosity regulates colonic epithelial cell differentiation and tumor number in ApcMin mice. Mol Cell Biol 2004; 24:7598 - 7611
- Lee RJ, Albanese C, Fu M, D'Amico M, Lin B, Watanabe G, et al. Cyclin D1 is required for transformation by activated Neu and is induced through an E2F-dependent signaling pathway. Mol Cell Biol 2000; 20:672 - 683
- Yu Q, Geng Y, Sicinski P. Specific protection against breast cancers by cyclin D1 ablation. Nature 2001; 411:1017 - 1021
- Albanese C, Wu K, D'Amico M, Jarrett C, Joyce D, Hughes J, et al. IKKalpha regulates mitogenic signaling through transcriptional induction of cyclin D1 via Tcf. Mol Biol Cell 2003; 14:585 - 599
- Sakamaki T, Liu M, Ju X, Zhang X, Li A, Joyce D, Albanese C, Pestell RG. NFκB induction by ErbB2 mediates DNA synthesis and breast turmor growth. Mol Cell Biol 2006;
- Neumeister P, Pixley FJ, Xiong Y, Xie H, Wu K, Ashton A, et al. Cyclin D1 governs adhesion and motility of macrophages. Mol Biol Cell 2003; 14:2005 - 2015
- Wang C, Pattabiraman N, Zhou JN, Fu M, Sakamaki T, Albanese C, et al. Cyclin D1 repression of peroxisome proliferator-activated receptor gamma expression and transactivation. Mol Cell Biol 2003; 23:6159 - 6173
- Fu M, Wang C, Rao M, Wu X, Bouras T, Zhang X, et al. Cyclin D1 represses p300 transactivation through a cyclin-dependent kinase-independent mechanism. J Biol Chem 2005; 280:29728 - 29742
- Fu M, Rao M, Bouras T, Wang C, Wu K, Zhang X, et al. Cyclin D1 inhibits PPARgamma-mediated adipogenesis through HDAC recruitment. J Biol Chem 2005; 280:16934 - 16941
- Bienvenu F, Jirawatnotai S, Elias JE, Meyer CA, Mizeracka K, Marson A, et al. Transcriptional role of cyclin D1 in development revealed by a genetic-proteomic screen. Nature 2010; 463:374 - 378
- Betticher DC, Thatcher N, Altermatt HJ, Hoban P, Ryder WDJ, Heighway J. Alternate splicing produces a novel cyclin D1 transcript. Oncogene 1995; 11:1005 - 1011
- Hosokawa Y, Arnold A. Mechanism of cyclin D1 (CCND1, PRAD1) overexpression in human cancer cells: analysis of allele-specific expression. Genes Chromosomes Cancer 1998; 22:66 - 71
- Li Z, Wang C, Jiao X, Katiyar S, Casimiro MC, Prendergast GC, et al. Alternate Cyclin D1 mRNA Splicing Modulates p27KIP1 Binding and Cell Migration. J Biol Chem 2008; 283:7007 - 7015
- Li Z, Jiao X, Wang C, Ju X, Lu Y, Yuan L, et al. Cyclin D1 induction of cellular migration requires p27(KIP1). Cancer Res 2006; 66:9986 - 9994
- Li Z, Wang C, Jiao X, Lu Y, Fu M, Quong AA, et al. Cyclin D1 regulates cellular migration through the inhibition of thrombospondin 1 and ROCK signaling. Mol Cell Biol 2006; 26:4240 - 4256
- Plomann MMM, Schael S. The PACSIN proteins and their role in membrane trafficking. Landes Bioscience 2009;
- Reutens AT, Fu M, Wang C, Albanese C, McPhaul MJ, Sun Z, et al. Cyclin D1 binds the androgen receptor and regulates hormone-dependent signaling in a p300/CBP-associated factor (P/CAF)-dependent manner. Mol Endocrinol 2001; 15:797 - 811
- Kessels MM, Qualmann B. Syndapin oligomers interconnect the machineries for endocytic vesicle formation and actin polymerization. J Biol Chem 2006; 281:13285 - 13299
- Halbach A, Morgelin M, Baumgarten M, Milbrandt M, Paulsson M, Plomann M. PACSIN 1 forms tetramers via its N-terminal F-BAR domain. FEBS J 2007; 274:773 - 782
- Wang Q, Navarro MV, Peng G, Molinelli E, Lin Goh S, Judson BL, et al. Molecular mechanism of membrane constriction and tubulation mediated by the F-BAR protein Pacsin/Syndapin. Proc Natl Acad Sci USA 2009; 106:12700 - 12705
- Fankhauser C, Reymond A, Cerutti L, Utzig S, Hofmann K, Simanis V. The S. pombe cdc15 gene is a key element in the reorganization of F-actin at mitosis. Cell 1995; 82:435 - 444
- Sherr CJ. Cancer cell cycles. Science 1996; 274:1672 - 1677
- Wang C, Fan S, Li Z, Fu M, Rao M, Ma Y, et al. Cyclin D1 antagonizes BRCA1 repression of estrogen receptor alpha activity. Cancer Res 2005; 65:6557 - 6567
- Holley SL, Parkes G, Matthias C, Bockmuhl U, Jahnke V, Leder K, et al. Cyclin D1 polymorphism and expression in patients with squamous cell carcinoma of the head and neck. Am J Pathol 2001; 159:1917 - 1924
- Millar EK, Dean JL, McNeil CM, O'Toole SA, Henshall SM, Tran T, et al. Cyclin D1b protein expression in breast cancer is independent of cyclin D1a and associated with poor disease outcome. Oncogene 2009; 28:1812 - 1820
- Li Z, Jiang J, Wang Z, Zhang J, Xiao M, Wang C, et al. Endogenous interleukin-4 promotes tumor development by increasing tumor cell resistance to apoptosis. Cancer Res 2008; 68:8687 - 8694
- Holnthoner W, Pillinger M, Groger M, Wolff K, Ashton AW, Albanese C, et al. Fibroblast growth factor-2 induces Lef/Tcf-dependent transcription in human endothelial cells. J Biol Chem 2002; 277:45847 - 45853
- Qualmann B, Roos J, DiGregorio PJ, Kelly RB. Syndapin I, a synaptic dynamin-binding protein that associates with the neural Wiskott-Aldrich syndrome protein. Mol Biol Cell 1999; 10:501 - 513
- Modregger J, Ritter B, Witter B, Paulsson M, Plomann M. All three PACSIN isoforms bind to endocytic proteins and inhibit endocytosis. J Cell Sci 2000; 113:4511 - 4521
- Qualmann B, Kelly RB. Syndapin isoforms participate in receptor-mediated endocytosis and actin organization. J Cell Biol 2000; 148:1047 - 1062
- Cousin H, Gaultier A, Bleux C, Darribere T, Alfandari D. PACSIN2 is a regulator of the metalloprotease/disintegrin ADAM13. Dev Biol 2000; 227:197 - 210
- Simpson F, Hussain NK, Qualmann B, Kelly RB, Kay BK, McPherson PS, et al. SH3-domain-containing proteins function at distinct steps in clathrin-coated vesicle formation. Nat Cell Biol 1999; 1:119 - 124
- Ritter B, Modregger J, Paulsson M, Plomann M. PACSIN 2, a novel member of the PACSIN family of cytoplasmic adapter proteins. FEBS Lett 1999; 454:356 - 362
- Balicki D, Reisfeld RA, Pertl U, Beutler E, Lode HN. Histone H2A-mediated transient cytokine gene delivery induces efficient antitumor responses in murine neuroblastoma. Proc Natl Acad Sci USA 2000; 97:11500 - 11504
- Li Y, Dowbenko D, Lasky LA. AKT/PKB phosphorylation of p21Cip/WAF1 enhances protein stability of p21Cip/WAF1 and promotes cell survival. J Biol Chem 2002; 277:11352 - 11361
- Bouras T, Fu M, Sauve AA, Wang F, Quong AA, Perkins ND, et al. SIRT1 deacetylation and repression of P300 involves lysine residues 1,020/1,024 within the cell cycle regulatory domain 1. J Biol Chem 2005; 280:10264 - 10276
- Katiyar S, Jiao X, Wagner E, Lisanti MP, Pestell RG. Somatic excision demonstrates c-Jun induces cellular migration and invasion through induction of stem cell factor. Mol Cell Biol 2007; 27:1356 - 1369