Abstract
A multiple comparison approach using whole genome transcriptional arrays was used to identify genes and pathways involved in calorie restriction/dietary restriction (DR) life span extension in Drosophila. Starting with a gene centric analysis comparing the changes in common between DR and two DR related molecular genetic life span extending manipulations, Sir2 and p53, lead to a molecular confirmation of Sir2 and p53’s similarity with DR and the identification of a small set of commonly regulated genes. One of the identified upregulated genes, takeout, known to be involved in feeding and starvation behavior, and to have sequence homology with Juvenile Hormone (JH) binding protein, was shown to directly extend life span when specifically overexpressed. Here we show that a pathway centric approach can be used to identify shared physiological pathways between DR and Sir2, p53 and resveratrol life span extending interventions. The set of physiological pathways in common among these life span extending interventions provides an initial step toward defining molecular genetic and physiological changes important in life span extension. The large overlap in shared pathways between DR, Sir2, p53 and resveratrol provide strong molecular evidence supporting the genetic studies linking these specific life span extending interventions.
Introduction
Calorie restriction/dietary restriction (DR) is one of the most robust and well-known life span and health span extending interventions so far described. Despite DR's clear efficacy in a wide range of organisms including yeast, nematodes, flies, spiders, fish and mammals, the difficulty in maintaining a DR diet in humans has severely limited its practical therapeutic use.Citation1–Citation10 The search for more applicable DR-like interventions has focused on understanding the molecular and physiological changes underlying DR's life span extending effects. One such approach is through the examination of transcriptional changes.Citation11 Whole genome transcriptional arrays provide an unbiased method for investigating the molecular mechanisms of complex biological phenomena.
Results and Discussion
Comparative whole genome transcriptional profiling identifies takeout as a new gene involved in longevity determination.
Whole genome transcriptional profiling of DR flies shows a large number of changes in gene expression (>2,000).Citation11 Among this large group of genes are those associated with life span extension as well as genes that change in response to the reduction in caloric or dietary intake, but are not involved in mediating life span. For example, the nearly universal decrease in fertility that accompanies DR is not directly related to life span extension in flies, but a response to the reduction in caloric or dietary intake.Citation12 Methods that can enrich for the transcriptional changes that are related to life span extension have been sought.
The use of a multiple comparison approach based upon a molecular genetic pathway associated with DR life span extension has shown great promise in identifying a subset of genes that may be important in life span determination.Citation11 A comparative transcriptional approach was performed making use of the knowledge that increased neuronal expression of Sir2 or decreased neuronal activity of p53 extends life span in Drosophila in a DR dependent manner.Citation13–Citation15 These molecular genetic manipulations have been proposed to be part of the downstream DR life span extending pathway (, reviewed in ref. Citation14).
The primary benefit of this approach is that these specific molecular genetic interventions (Sir2, p53) would be expected to share changes in gene expression important in DR life span extension, but not necessarily share the changes in DR induced gene expression that are unrelated to life span extension. For example, DR flies have a reduction in fertility, while Sir2 and p53 long-lived flies do not reduce fertility.Citation15,Citation16 Thus, selecting genes that are shared between all three of these DR related life span extending interventions may help identify genes more specifically involved in life span extension.
Comparison of whole genome transcriptional arrays between DR flies, Sir2 and p53 long-lived flies resulted in significant overlap in the genes that were upregulated and downregulated. According to the molecular genetic model proposed (), Sir2 might be more closely related to DR than p53 is related to DR. As predicted, there was a larger overlap of the total number of genes in DR and Sir2 then with DR and p53. Of the 782 genes that are altered in the Sir2 long-lived flies 72% of the upregulated and 61% of the downregulated genes were shared with DR. In p53 long-lived flies, of the 235 genes that were significantly affected 63% of genes upregulated were shared with DR, but only 4% of the downregulated genes were shared with DR.
The three-way comparison of DR with Sir2 and p53 long-lived flies revealed an overlap of 21 genes, 20 upregulated and one downregulated. The 20 genes upregulated included several with interesting relationships to chromatin structure, circadian rhythm, neural activity, detoxification/chaperone activity, muscle maintenance, immune function, growth factor activity and feeding behavior/response to starvation. One of these genes, takeout, which is involved in feeding behavior and response to starvationCitation17,Citation18 was found to be upregulated in another DR related life span extending genetic intervention, Indy long-lived flies.Citation19,Citation20 takeout was found to be upregulated in all other specific genetically altered long-lived flies tested including rpd3, chico and methuselah.Citation21–Citation23 Finally, confirmation of the power of the whole genome transcriptional comparative approach for identifying genes important in life span extension was directly demonstrated by showing that specific overexpression of takeout alone extends life span.Citation11,Citation24
Gene set analysis reveals that changes in important physiological systems are conserved in DR, Sir2 and p53 life span extending conditions.
A primary goal of the comparative whole genome transcriptional approach is to identify physiological pathways important in extending life span in DR that can be targeted for therapeutic intervention. The identification of changes in specific genes shared by related life span extending interventions is one method for identifying physiological systems important in life span determination. However, the criteria that the same intervention in different genetic backgrounds or related interventions in the same genetic background alter the exact same genes may be too stringent a requirement, resulting in the omission of important genes and pathways.Citation25
Different strains of flies may alter the same physiological pathways to achieve life span extension, but do so by modifying different specific genes within a pathway. To test this we selected two different wild type fly strains, each known to respond to DR with a 40% increase in life span, and examined their whole genome transcriptional profile in response to DR. One strain (yw, w1118) had 2461 changes in gene expression and the other (Canton-S) had 2721 changes. The shared overlap between the strains was 1473 changes.Citation11 Although the shared overlap is greater than 50% (60 and 54% respectively), nearly 50% of the changes in the expression of specific genes in response to DR are different between these similar strains.
We determined the functional groups that are affected in each strain by identifying which gene ontologies were overrepresented in each strain using GOstat with the Drosophila gene ontology pathway database.Citation26 The yw, w1118 strain revealed 754 gene ontology categories and the Canton-S strain 793 gene ontology categories with a shared overlap of 551. The shared overlap of gene ontology categories is approximately 75%.Citation11 If instead of determining the GOstat categories for the two strains on DR and calculating the intersection of the pathways we determine the intersection at the level of genes and then analyze these genes with GOstat we find 10% less GOstat categories. These studies suggest that approaches that can go beyond the comparison of single genes to the level of gene sets or pathways may be advantageous.
Gene set enrichment analysis (GSEA) identifies additional functional groups conserved in DR, Sir2 and p53 life span extending conditions.
An alternative method for functional group analysis which does not require pre-selecting differentially expressed genes according to arbitrarily set thresholds is the Gene Set Enrichment Analysis (GSEA). GSEA is a bioinformatics tool that identifies gene sets (pathways) that are enriched for up or downregulated genes within a given gene expression experiment.Citation27 This algorithm ranks genes from an RNA transcriptional microarray by the fold-change between treatment and controls, looking for functional categories in which genes are preferentially at the top or at the bottom of the ranked gene list. The GSEA system also makes use of a Monte Carlo strategy to test that the observed up or downregulation is statistically significant, not just a consequence of random noise in the data.
We compared the GSEA approach to the GOstat approach on the two strains treated with DR. The GOstat system requires the experimenter to first determine which genes are changed between the experimental and control group (in this case high calorie food versus low calorie food) by defining a cut off for fold change and statistical confidence (e.g., p value). The subset of genes determined to be “changed” by fold change and p value between the two conditions are then processed in GOstat to determine which physiological pathways are associated with that specific group of genes. This approach does not perform any analysis to weigh the relative significance of a gene's enrichment in the initial data set. It determines what the likelihood of association is between a particular pathway and the group of genes that are specifically provided by the experimenter. Thus, the genes for GOstat analysis first have to be identified by a standard fold enrichment and statistical significance measure. The GSEA approach uses statistical methods directly to examine the ranking of genes based upon fold change and other features of the data. GSEA is designed to look for all changes in gene expression within a pathway, small or large, and bypasses the need to determine a cutoff value for the fold change and statistical significance of individual genes.
The outputs of GSEA and GOstat were compared using the Drosophila gene ontology category data set. As noted above using the GOstat approach we determined a fold change cutoff and p valueCitation11 for each of the two different strains on DR, determined the overlap of significant genes from the two different strains on DR and then examined the set of shared genes with GOstat and found 551 categories.Citation11 Using GSEA we independently analyzed each of the two DR raw data sets and found an overlap between the two strains of 713 gene ontology categories. Comparison of the gene ontology categories found between GOstat and GSEA showed an overlap of only 240 gene ontology categories, 44% of the GOstat derived sets and 34% of the GSEA derived sets (Sup. Table 1). 311 gene ontology sets were found only in GOstat and 473 gene ontology sets found only in GSEA. This analysis suggests that the use of both GOstat and GSEA approaches may be advantageous for identifying gene sets important in complex biological phenomena such as DR.
We had previously used GOstat to examine the gene ontology pathways associated with DR, Sir2 and p53.Citation11 We now used GSEA with the KEGG (Kyoto Encylopedia of Genes and Genomes) Drosophlia database to perform a comparative transcriptional pathway analysis between the two different strains on DR.Citation28 The KEGG dataset is particularly well curated for metabolic related pathway categories. Using GSEA on KEGG we found that the Canton-S strain on DR identified 54 categories that were statistically significant, while the yw, w1118 strain identified 72 categories that were statistically significant. All of the 54 KEGG categories in DR Canton-S were included in the 72 categories found in yw, w1118 DR.
We designated the 54 KEGG categories overlap between these two strains as a DR signature and examined the relationship between this DR signature and Sir2 and p53 long-lived flies. As seen in we found a strong overlap between the statistically significant physiological pathways of DR, Sir2 (49 out of 54) and p53 (35 out of 54) (Sup. Table 2). The large overlap in shared pathways between Sir2 and DR signature (91%) and p53 and DR signature (65%) complement previous genetic epistasis experimentsCitation15,Citation16,Citation23 and provide further evidence that Sir2 and p53 life span extending manipulations are closely related to DR.
Tissue specific whole genome transcriptional arrays show a strong association between DR and resveratrol.
Since specific tissues and organs may respond differentially to DR and other life span extending interventions we examined the overlap in KEGG pathways from whole genome transcriptional arrays of DR studies on Canton-S whole body female flies with those from combined head and thorax of Canton-S female flies. Despite the large number of statistically significant KEGG categories in response to DR in whole female bodies (54) and female head/thorax (83) less than 50% (26) of these categories were shared () (Sup. Table 3). Of particular note are the strong opposite effects on pathways of special interest including KEEG defined pathways such as: DNA polymerase, nucleotide excision repair, mismatch repair, base excision repair, proteasome, ubiquitin mediated proteolysis, glycosylphosphatidylinositol GPI anchor biosynthesis and fatty acid elongation in mitochondria. While whole body females showed a statistically significant decrease in these pathways, head/thorax females showed a statistically significant increase in these pathways. Interestingly, oxidative phosphorylation was statistically significantly increased in whole body, but down in head/thorax ().
The complete reversal in the direction of these pathways when comparing whole body and head/thorax on DR highlights the importance of also examining life span extending interventions on a tissue specific level. We therefore examined the relationship between DR and resveratrol treatment using RNA from only head/thorax females. Canton-S female flies from the same cohort were grown on high calorie food, low calorie food and high calorie food with resveratrol. GSEA analysis showed that of the 83 KEGG categories that were statistically significant in head/thorax DR, 81% (67 pathways) were shared with resveratrol (Sup. Table 4). This large overlap is similar to that seen in mice where resveratrol treatment showed an 82% overlap in gene pathways with DR in the liverCitation29 ().
To further demonstrate the usefulness of a pathway approach, a gene centric analysis of the same data for resveratrol and DR was performed. Based upon the same fold and p value cutoff as above (1.5-fold, 0.01 p, threshold were validated using qPCR, reviewed in ref. Citation11) there were only 28 genes that changed with resveratrol treatment and 152 genes that changed with DR with a significant overlap of 12 (12/28 or 43% of the genes found with resveratrol are also found in DR, Fisher's exact test p < 10−17). If the stringency in fold change and p value is reduced to 1.2-fold change and 0.05 p value 237 genes are seen to change in the head/thorax with resveratrol and 1,708 with DR. The overlap is 150 genes showing that 63% of the genes changing with resveratrol are now found in DR (Fisher's exact test p < 10−78). While the gene centric approach also supports the close association between resveratrol and DR, these results demonstrate that gene set analysis strategies such as GSEA may be more efficient at assessing small changes in genes that may lead to significant changes in physiological pathways.
Juvenile hormone (JH) regulated genes are altered in DR, Sir2, p53 and resveratrol.
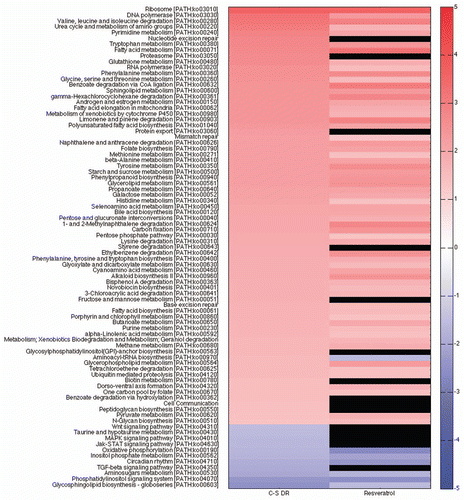
The identification of takeout as a gene involved in lifespan extension combined with the fact that takeout has Juvenile Hormone (JH) binding like domains and JH is thought to be involved in life span determinationCitation18,Citation24,Citation30–Citation32 led us to use the GSEA system to test the hypothesis that Juvenile Hormone (JH) may be important in DR and DR related life span extending interventions (e.g., Sir2, p53, resveratrol). We curated two gene set pathways consisting of genes downregulated by JH or upregulated by JH (reviewed in ref. Citation33–Citation36, Sup. Table 1). We found that in whole body DR both the JH downregulated and JH upregulated categories were statistically significantly upregulated, although the JH downregulated gene set (scores of 2.14 and 2.02) was much more affected then the JH upregulated gene set (scores of 1.24 and 1.27). An increase in expression of JH downregulated genes implies a decrease in JH, while an increase in JH upregulated genes implies an increase in JH. It is known that ecdysone, the other major insect hormone, works in conjunction with JH to regulate expression of some of these genes and a simultaneous change in JH and ecdysone may account for the apparent contradictory results concerning the state of JH signaling in DR of whole female fly bodies. Interestingly, in whole body female Sir2 and p53 long-lived flies and head/thorax from resveratrol treated females only the JH downregulated pathway is statistically significantly increased. An increase in the JH downregulated pathway was also found in the DR head/thorax female flies, but it was just below the level of statistical significance. These data suggest that similar to grasshoppers and butterflies, JH signaling may be decreased in long-lived flies.
Conclusions
These data demonstrate that whole genome transcriptional profiles from molecular genetic components of the DR life span extending pathway (Sir2, p53) can be used to identify individual genes and gene pathways that may be important in DR life span extension. The value of whole genome approaches to directly identify genes important in life span extension was confirmed by using a gene centric approach to identify a small set of candidate genes and then examining one of the identified genes, takeout, known to be important in feeding behaviorCitation17 but not previously known to affect life span. takeout was found to be increased in other molecular genetic life span extending interventions (Indy, Rpd3, chico and mth) and when selectively overexpressed takeout alone extended life span.Citation11
Bioinformatic studies using GOstat and GSEA approaches that emphasize the identification of physiological and metabolic pathways illustrate the value of using both of these pathway centric systems to identify life span extending gene pathways. In addition to identifying specific genes and pathways, comparative studies employing GOstat or GSEA can be used to assess the molecular relatedness between different interventions. The GOstat and GSEA analyses demonstrated the strong molecular relationship between DR, Sir2 and p53 life span extending interventions that had been suggested by genetic epistasis studies and confirmed in flies the molecular association between the life span extending small molecule resveratrol and DR previously shown in mice.Citation29 These studies show that bioinformatic methods utilizing a comparative approach on whole genome transcriptional profiles associated with experimentally well-defined molecular genetic pathways can serve to identify targets for subsequent molecular genetic and pharmacological therapeutic interventions designed to extend healthy life span.
Abbreviations
| DR | = | dietary restriction |
| JH | = | juvenile hormone |
| GSEA | = | gene set enrichment analysis |
Figures and Tables
Figure 1 DR is a highly pleiotropic process that influences a variety of biological processes, including physiology, fertility, behavior and life span. While the nature of most of these pathways remains unknown, a molecular framework for at least some aspects of DR-dependent life span regulation can be constructed. Under DR conditions (red), rpd3 is down and dSir2 is upregulated. dSir2 activity inhibits Dmp53 (amongst other targets), leading to a portion of the DR related life span extension seen. DR life span extension is larger than life span extension of rpd3, dSir2 or Dmp53 manipulation. Thus each of these interventions define only a portion of the DR life span extension. Reviewed from reference Citation14.
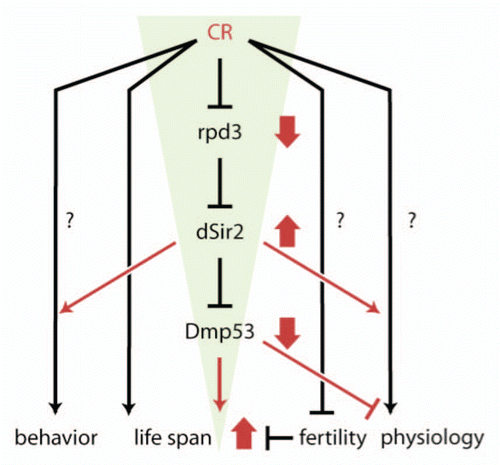
Figure 2 Heat map of KEGG gene sets from Sir2 and p53 long-lived flies show similarity with a DR signature gene set. Red are gene sets that are statistically significantly upregulated, blue are gene sets that are statistically significantly downregulated and black are gene sets that are not statistically significantly changed. RNA is from 10-day old whole body of females.Citation11 Sir2 are flies expressing Sir2 in adult neurons and p53 are flies expressing DN-Dmp53 in adult neurons. yw, w1118 DR flies are genetically identical to Sir2 flies, but without RU486 and genetically very similar to the DN-Dmp53 flies. KEGG categories and scores are in Supplemental Table 2.
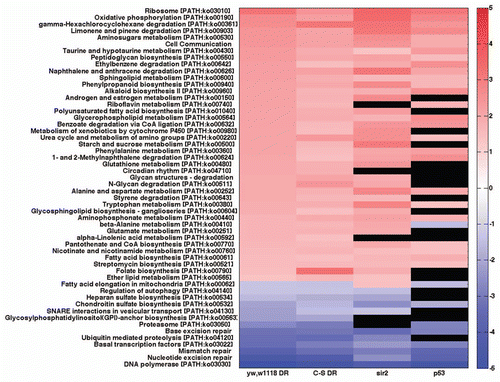
Figure 3 Heat map of KEGG gene sets from DR of female whole body and female head and thorax appear very different. Red are gene sets that are statistically significantly upregulated, blue are gene sets that are statistically significantly downregulated and black are gene sets that are not statistically significantly changed. RNA is from Canton-S (C-S) DR and high calorie fed 10-day old female whole bodiesCitation11 or only heads and thoraces. KEGG categories and scores are in Supplemental Table 3.
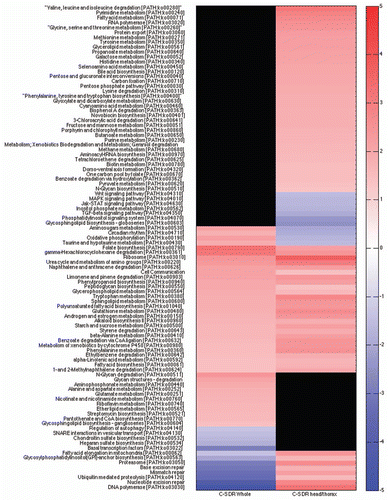
Figure 4 Heat map of KEGG gene sets from DR and resveratrol female head and thorax appear very similar. Red are gene sets that are statistically significantly upregulated, blue are gene sets that are statistically significantly downregulated and black are gene sets that are not statistically significantly changed. RNA is from the same cohort of Canton-S (C-S) 10-day old female heads and thoraces on a DR diet, high calorie food or fed resveratrol. KEGG categories and scores are in Supplemental Table 4.
Additional material
Download Zip (63.9 KB)Acknowledgements
We would like to thank Will Lightfoot for technical assistance. This work was supported by NIA grants AG16667, AG24353 and AG25277 to S.L.H., NIA AG028753 to N.N. and NIA AG029723 to J.B. M.A. was supported by NIA AG030329. S.L.H. is an Ellison Medical Research Foundation Senior Investigator and recipient of a Glenn Award for Research in Biological Mechanisms of Aging.
References
- Wanagat J, Allison DB, Weindruch R. Caloric intake and aging: mechanisms in rodents and a study in nonhuman primates. Toxicol Sci 1999; 52:35 - 40
- Weindruch R. The retardation of aging by caloric restriction: studies in rodents and primates. Toxicol Pathol 1996; 24:742 - 745
- Kayo T, Allison DB, Weindruch R, Prolla TA. Influences of aging and caloric restriction on the transcriptional profile of skeletal muscle from rhesus monkeys. Proc Natl Acad Sci USA 2001; 98:5093 - 5098
- Lane MA, Mattison J, Ingram DK, Roth GS. Caloric restriction and aging in primates: Relevance to humans and possible CR mimetics. Microsc Res Tech 2002; 59:335 - 338
- Chapman T, Partridge L. Female fitness in Drosophila melanogaster: an interaction between the effect of nutrition and of encounter rate with males. Proc R Soc Lond B Biol Sci 1996; 263:755 - 759
- Good TP, Tatar M. Age-specific mortality and reproduction respond to adult dietary restriction in Drosophila melanogaster. J Insect Physiol 2001; 47:1467 - 1473
- Lakowski B, Hekimi S. The genetics of caloric restriction in Caenorhabditis elegans. Proc Natl Acad Sci USA 1998; 95:13091 - 13096
- Lin SJ, Defossez PA, Guarente L. Requirement of NAD and SIR2 for life-span extension by calorie restriction in Saccharomyces cerevisiae. Science 2000; 289:2126 - 2128
- Jiang JC, Wawryn J, Shantha Kumara HM, Jazwinski SM. Distinct roles of processes modulated by histone deacetylases Rpd3p, Hda1p and Sir2p in life extension by caloric restriction in yeast. Exp Gerontol 2002; 37:1023 - 1030
- Guarente L. Sir2 links chromatin silencing, metabolism and aging. Genes Dev 2000; 14:1021 - 1026
- Bauer J, Antosh M, Chang C, Schorl C, Kolli S, Neretti N, et al. Comparative transcriptional profiling identifies takeout as a gene that regulates life span. Aging (Albany NY) 2010; 2:298 - 310
- Mair W, Piper MD, Partridge L. Calories do not explain extension of life span by dietary restriction in Drosophila. PLoS Biol 2005; 3:223
- Bauer JH, Chang C, Morris SN, Hozier S, Andersen S, Waitzman JS, et al. Expression of dominant-negative Dmp53 in the adult fly brain inhibits insulin signaling. Proc Natl Acad Sci USA 2007; 104:13355 - 13360
- Bauer JH, Morris SN, Chang C, Flatt T, Wood JG, Helfand SL. dSir2 and Dmp53 interact to mediate aspects of CR-dependent life span extension in D. melanogaster. Aging 2009; 1:38 - 48
- Rogina B, Helfand SL. Sir2 mediates longevity in the fly through a pathway related to calorie restriction. Proc Natl Acad Sci USA 2004; 101:15998 - 16003
- Bauer JH, Poon PC, Glatt-Deeley H, Abrams JM, Helfand SL. Neuronal expression of p53 dominant-negative proteins in adult Drosophila melanogaster extends life span. Curr Biol 2005; 15:2063 - 2068
- Meunier N, Belgacem YH, Martin JR. Regulation of feeding behaviour and locomotor activity by takeout in Drosophila. J Exp Biol 2007; 210:1424 - 1434
- Sarov-Blat L, So WV, Liu L, Rosbash M. The Drosophila takeout gene is a novel molecular link between circadian rhythms and feeding behavior. Cell 2000; 101:647 - 656
- Neretti N, Wang PY, Brodsky AS, Nyguyen HH, White KP, Rogina B, et al. Long-lived Indy induces reduced mitochondrial reactive oxygen species production and oxidative damage. Proc Natl Acad Sci USA 2009; 106:2277 - 2282
- Wang PY, Neretti N, Whitaker R, Hosier S, Chang C, Lu D, et al. Long-lived Indy and calorie restriction interact to extend life span. Proc Natl Acad Sci USA 2009; 106:9262 - 9267
- Lin YJ, Seroude L, Benzer S. Extended life-span and stress resistance in the Drosophila mutantmethuselah. Science 1998; 282:943 - 946
- Clancy DJ, Gems D, Harshman LG, Oldham S, Stocker H, Hafen E, et al. Extension of life-span by loss of CHICO, a Drosophila insulin receptor substrate protein. Science 2001; 292:104 - 106
- Rogina B, Helfand SL, Frankel S. Longevity regulation by Drosophila Rpd3 deacetylase and caloric restriction. Science 2002; 298:1745
- Galikova M, Flatt T. Dietary restriction and other lifespan extending pathways converge at the activation of the downstream effector takeout. Aging (Albany NY) 2010; 2:387 - 389
- Manoli T, Gretz N, Grone HJ, Kenzelmann M, Eils R, Brors B. Group testing for pathway analysis improves comparability of different microarray datasets. Bioinformatics 2006; 22:2500 - 2506
- Beissbarth T, Speed TP. GOstat: find statistically overrepresented Gene Ontologies within a group of genes. Bioinformatics 2004; 20:1464 - 1465
- Subramanian A, Kuehn H, Gould J, Tamayo P, Mesirov JP. GSEA-P: a desktop application for Gene Set Enrichment Analysis. Bioinformatics 2007; 23:3251 - 3253
- Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res 2000; 28:27 - 30
- Pearson KJ, Baur JA, Lewis KN, Peshkin L, Price NL, Labinskyy N, et al. Resveratrol delays age-related deterioration and mimics transcriptional aspects of dietary restriction without extending life span. Cell Metab 2008; 8:157 - 168
- Flatt T, Tu MP, Tatar M. Hormonal pleiotropy and the juvenile hormone regulation of Drosophila development and life history. Bioessays 2005; 27:999 - 1010
- Herman WS, Tatar M. Juvenile hormone regulation of longevity in the migratory monarch butterfly. Proc Biol Sci 2001; 268:2509 - 2514
- Tatar M, Yin C. Slow aging during insect reproductive diapause: why butterflies, grasshoppers and flies are like worms. Exp Gerontol 2001; 36:723 - 738
- Flatt T, Heyland A, Rus F, Porpiglia E, Sherlock C, Yamamoto R, et al. Hormonal regulation of the humoral innate immune response in Drosophila melanogaster. J Exp Biol 2008; 211:2712 - 2724
- Berger EM, Dubrovsky EB. Juvenile hormone molecular actions and interactions during development of Drosophila melanogaster. Vitam Horm 2005; 73:175 - 215
- Dubrovsky EB, Dubrovskaya VA, Berger EM. Juvenile hormone signaling during oogenesis in Drosophila melanogaster. Insect Biochem Mol Biol 2002; 32:1555 - 1565
- Dubrovsky EB, Dubrovskaya VA, Bilderback AL, Berger EM. The isolation of two juvenile hormone-inducible genes in Drosophila melanogaster. Dev Biol 2000; 224:486 - 495