Abstract
DNA polymerases cannot synthesize DNA without a primer, and DNA primase is the only specialized enzyme capable of de novo synthesis of short RNA primers. In eukaryotes, primase functions within a heterotetrameric complex in concert with a tightly bound DNA polymerase α (Pol α). In humans, the Pol α part is comprised of a catalytic subunit (p180) and an accessory subunit B (p70), and the primase part consists of a small catalytic subunit (p49) and a large essential subunit (p58). The latter subunit participates in primer synthesis, counts the number of nucleotides in a primer, assists the release of the primer-template from primase and transfers it to the Pol α active site. Recently reported crystal structures of the C-terminal domains of the yeast and human enzymes’ large subunits provided critical information related to their structure, possible sites for binding of nucleotides and template DNA, as well as the overall organization of eukaryotic primases. However, the structures also revealed a difference in the folding of their proposed DNA-binding fragments, raising the possibility that yeast and human proteins are functionally different. Here we report new structure of the C-terminal domain of the human primase p58 subunit. This structure exhibits a fold similar to a fold reported for the yeast protein but different than a fold reported for the human protein. Based on a comparative analysis of all three C-terminal domain structures, we propose a mechanism of RNA primer length counting and dissociation of the primer-template from primase by a switch in conformation of the ssDNA-binding region of p58.
Introduction
DNA polymerases cannot synthesize DNA without a primer, posing a problem for the initiation of replication.Citation1 A mainstream mechanism to deal with this obstacle is priming with short RNA made by a specialized enzyme called primase.Citation2–Citation4 In eukaryotes, primase functions within a heterotetrameric complex in concert with a tightly bound DNA polymerase α (Pol α). In humans, the Pol α part is comprised of a catalytic subunit (p180) and an accessory subunit B (p70), and the primase part consists of a small catalytic subunit (p49) and a large essential subunit (p58).Citation5 The two subunits form a tightly bound complex. Biochemical studies revealed that the small p49 subunit by itself is unstable and has only limited ability to synthesize oligoribonucleotides. The large p58 subunit stabilizes the p49™p58 complex and contributes to primase activity.Citation5–Citation7 The discovery and structures of an iron-sulfur cluster with four cysteine residues in the C-terminal region of p58 (p58 4Fe-4S domain) was a break-through in understanding the essential role of the large subunit in archaea, yeastCitation8,Citation9 and humans.Citation10,Citation11 The structure of the C-terminal part of the yeast primase large subunit (PriL-CTD) was similar to a domain of the DNA repair enzyme photolyase, encompassing an active site and binding sites for flavin adenine-dinucleotide (FAD) and ssDNA.Citation9 This observation led to a novel primase mechanism, in which the large subunit of primase assists the catalytic subunit in the simultaneous binding of two initial ribonucleotides.Citation9 However, the structural basis for counting the synthesized RNA primer length and switching the RNA primer-template for the elongation by Pol α is unknown. Moreover, the validity of yeast primase as a model for human primase has been questioned since the crystal structure of the C-terminal part of human primase p58 subunit (p58C/3L9Q) was determined by the Chazin group.Citation10 The structures of the iron-sulfur cluster binding domains were similar in both structures, but the proposed DNA binding domains were different: in the structure of human protein it was represented by an α-helix and a β-sheet, while in the yeast protein structure it formed a three-helix bundle. It has been suggested that the differences between yeast and human structures reflect different enzyme architecture and, likely, mechanisms.Citation10 We offer an alternative explanation.
Here, we present the crystal structure of the 4Fe-4S cluster domain containing the C-terminal part of the human primase p58 subunit (p58C/3Q36). Surprisingly, the fragment that forms a β-sheet in the reported structure p58C/3L9Q of the same human primase domainCitation10 is folded in three α-helices in our p58C/3Q36 structure, similarly to yeast primase.Citation9 We propose that these variants represent two bona fide conformations of p58. The observed ability of the C-terminal domain to acquire different conformations might be linked to a mechanism of RNA primer length counting and dissociation from primase, resulting in a switch for elongation by Pol α.
Results
Comparison with the yeast DNA primase.
The human p58C/3Q36 contains 12α-helices () and shares a folding topology with the crystal structure of S. cerevisiae PriL-CTDCitation9 ( and B), with a root-mean-square deviation (rmsd) of 1.07 Å and 1.17 Å for the 159 matched Cα atoms of the A chain and B chain, respectively. The largest discrepancy in the folding was observed in the conformation of N-terminal residues 271–282. The helix α4 is one turn longer in p58C/3Q36 due to a two-residue insertion at the C-terminus of α4. In addition, position of the three-helical bundle comprised of helices α9, α10 and α11 has a shift with a maximum value of 3.8 Å. The electron density was not obtained for the part of the loop α6α7 (residues 355–361), indicating a potential disorder of this loop in p58C/3Q36. The corresponding loop is structured in PriL-CTD. On the other hand, the part of the loop α11α12 corresponding to ordered residues 433–438 in the p58C structure is missing from chain A of PriL-CTD and has a different conformation in chain B of PriL-CTD. In spite of the above-described differences, the overall similarities observed for the topology of the yeast and human primase domains point to the same overall mode of action of the two eukaryotic DNA primases.
Comparison with the human DNA primase.
The comparison of the p58C/3Q36 structure presented here with the structure of p58C/3L9Q revealed a surprising difference in the folding of the N-terminal tail up to residues 275 and 319–361, which contain the helices α4, α5 and α6 ( and C). In the p58C/3L9Q structure, the corresponding residues form a three-stranded antiparallel β-sheet. The rest of structure (residues 276–318 and 362–455) superimposes well with the rmsd for Cα atoms of 0.49 Å.
An interconversion of the α-helices and β-sheet within p58C raised questions about the origin of the differences and its potential functional implications. The crystallization conditions are among the factors that might cause the observed structural differences. In order to elucidate the effect of crystallization conditions, we examined the effect of crystallization solution on a secondary structure of the p58C domain using CD spectroscopy. Major differences in crystallization conditions were in pH and in salt. Increasing the pH up to 9.0 and adding 200 mM Li2SO4 did not affect the content of the secondary structure, which remained predominantly helical (). This indicates that other factors such as crystal packing interactions may account for the differences in protein conformation. Indeed, an examination of the crystal structure of p58C revealed only minor involvement of the loop α4α5 within the region spanning helices α4, α5 and α6 in the crystal packing interactions (). In the PriL-CTD structure, in spite of formation of distinct sets of intermolecular interactions by each of the two independent molecules, the overall conformation of the α4, α5 and α6 regions was the same (), indicating to the absence of the crystal packing effect on PriL-CTD folding. However, unlike p58C/3Q36 and PriL-CTD, the corresponding region in p58C/3L9Q is involved in extensive intermolecular interactions, resulting in the formation of a tetramer (). The most notable are the interactions between the β-strands of the opposing molecules, which result in the formation of an incomplete β-barrel. The above-described observations indicate that the specific set of intermolecular interactions in p58C/3L9Q is causing the switch of helices α4, α5 and α6 into β-strands.
Discussion
Within the Pol α-primase complex, the p58 primase subunit interacts with Pol α,Citation5,Citation12 initiates the primer synthesis, counts the number of nucleotides in a newly synthesized primer and then mediates the transfer of the primer-template to the Pol α active site.Citation6,Citation7 Fulfillment of these transactions, especially the dissociation of the RNA primer-template from primase and its transfer to Pol α, requires significant conformational changes in the primase structure. In this respect, the predisposition of p58C to acquire two different conformations is highly intriguing, because the region of p58C exhibiting the conformational switch has been shown to interact with DNA.Citation9,Citation10 Similar to observations of the PriL-CTD structure, the all-helical p58C/3Q36, rather than the β-sheet-containing p58C/3L9Q, has a fold similar to ssDNA-bound DNA photolyase ().Citation13 Based on this observation, we assume that the p58C/3Q36 structure represents a conformation capable of DNA-binding, and the p58C/3L9Q structure represents a conformation that is not able to bind DNA. Existence of two different conformations of p58C provides a basis for a plausible model of primer length counting and primer-template dissociation from DNA primase. According to this model, Pol α-primase complex binds template DNA and initiates the RNA primer synthesis (). Then the extension of RNA primer length up to 7–10 ribonucleotides (unit length) introduces sterical hindrance (). This happens only in presence of interaction with Pol α because without such interaction primase synthesizes the RNA primers with multiplied unit length.Citation14 The sterical hindrance with participation of Pol α induces the switch of p58C helices α4, α5 and α6 to β-strands. As a consequence, the RNA primer-template dissociates from primase and then transfers for extension by Pol α ().
The induction of dramatic conformational changes in the polymerase structure by the newly synthesized RNA was observed before for RNA Pols. WeCitation15 and the Steitz groupCitation16 have shown that the extension by T7 RNA polymerase of the length of RNA in the RNA-DNA hybrid up to 8 bp resulted in polymerase transformation from an initiation complex to an elongation complex, and this transformation was accompanied by an unprecedented rearrangement of the 226 N-terminal amino acid residues (out of total 883). Our current model extends this principle for the first time to primase reaction. Further structural and mutational studies of the Pol α-primase complex is necessary to validate the proposed model and to assess how the postulated conformational changes in p58C depend on primer length.
Materials and Methods
Protein expression and purification.
A PCR-amplified DNA fragment encoding the human p58C domain (residues 266–456) was cloned in pET28 b vector (Novagen). Recombinant protein was expressed in E. coli Bl21 (DE3) cells grown at 37°C. The cells were chilled to 4°C, and after the addition of 0.2 mM ferric chloride hexahydrate, transferred at 30°C and induced with 0.6 mM isopropyl-β-D-thiogalactopyranoside for 16–18 hours. The cells were collected, resuspended in 40 mM HEPES pH 6.5, 500 mM NaCl, 5 mM potassium phosphate, 20% glycerol, 10 mM β-mercaptoethanol, 1 mM phenylmethanesulfonylfluoride and 10 mM MgSO4, and lysed by sonication. The cell debris was removed by centrifugation at 16,000 g for ten minutes. The clarified supernatant was diluted with an equal amount of 20 mM HEPES pH 6.5. The protein was purified on SP-Hi Trap column, dialyzed against 5 mM MES pH 6.2, 40 mM NaCl and 1 mM DTT, and concentrated to ∼15 mg/ml and frozen at −80°C in aliquots.
Crystallization and data collection.
The aggregation and polydispersity of p58C was monitored at 22°C in various buffers using dynamic light scattering technique. The low polydispersity (∼15%) and aggregation state corresponding to a monomeric p58C were obtained in 5 mM MES pH 6.2, 40 mM NaCl and 1 mM DTT. The crystal screens were set with 7 mg/ml−1 protein solution containing 5 mM MES pH 6.2, 40 mM NaCl, 0.5 mM DTT and 0.1 mM potassium hexacyanoferrate(III) at 22°C using sitting-drop vapour diffusion method and 50% diluted solutions of Crystal Screen and Crystal Screen 2 (Hampton Research). Small brownish crystals appeared in a condition #28 of Crystal Screen (100 mM sodium acetate trihydrate, 50 mM sodium cacodylate pH 6.5 and 15% w/v PEG 8000). In further optimization, better quality crystals were obtained in a solution containing 200 mM magnesium acetate tetrahydrate instead of sodium acetate trihydrate. However, in spite of wellshaped parallelepiped geometry, the crystals diffracted as twins.
For cryoprotection, the crystals were soaked in cryoprotectant consisting of 100 mM magnesium acetate tetrahydrate, 50 mM sodium cacodylate pH 6.5, 15% w/v PEG 8000, 11% v/v PEG 400 and 11% v/v glycerol, mounted on a cryoloop and flash cooled in a cryostream. Only one single crystal was found after screening over a hundred twin-diffracting crystals. The 2.5 Å resolution diffraction dataset was collected at the Argonne National Laboratory Advanced Photon Source NE-CAT beamline BL 24ID-E. All intensity data were indexed, integrated and scaled with DENZO and SCALEPACK from the HKL2000 program package.Citation17 The crystal parameters and data processing statistics are summarized in .
Structure determination and refinement.
The crystals belong to monoclinic space group C2 and contain two molecules of p58C per asymmetric unit. The structure was determined by molecular replacement method using the coordinates of PriL-CTD (PDB code 3LGB),Citation9 as a search model. The discrepancy between the folding of p58C/3Q36 and p58C/3L9QCitation10 (the latter structure became available after our structure determination) raised the possibility of bias toward the starting model derived from PriL-CTD. To eliminate this possibility, we rebuilt the structure using the starting model with omitted helices α1, α4, α5 and α6. The electron density for the omitted parts again appeared as for α-helices and not as for β-strands as in p58C/3L9Q. After several rounds of manual model rebuilding with Turbo-Frodo software, the structure has been refined to an R of 22.6% and Rfree of 25% at a resolution of 2.5 Å. Structure determination and refinement was performed using CNS version 1.1.Citation18 The structure has a good geometry with 89.7% of the non-glycine residues located in the most favored regions and the other 10.3% located in the additionally allowed regions of the Ramachandran plot (). The electron density was not observed for the residues 266–270, 355–361 and 456 in the first molecule and 266–270, 354–360 and 456 in the second molecule, and these residues were not included in the final model. The figures containing molecular structures were prepared with PyMOL software. Three-dimensional structure alignments were performed with Swiss-PdbViewer software.Citation19
CD spectroscopy.
For the Circular Dichroism (CD) titration experiments, the p58C protein was diluted to a final concentration of 2 mg/ml. All CD spectroscopy measurements with the Jasco J-815 spectropolarimeter were performed in the far UV region (190–250 nm) at one hour after protein addition and mixing with respective buffers at 22°C. Five measurements from each sample have been obtained, averaged and processed using Jasco's Spectra Manager software, and analyzed with DicroWeb software.Citation20
Accession numbers.
Atomic coordinates and structure factors of human p58C/3Q36 have been deposited in the Protein Data Bank with accession number 3Q36.
Abbreviations
| Pol α | = | DNA polymerase α |
| p180 | = | human DNA polymerase α catalytic subunit |
| p70 | = | human DNA polymerase α accessory subunit B |
| p49 | = | human primase small catalytic subunit |
| p58 | = | human primase large essential subunit |
| PriL-CTD | = | C-terminal part of the yeast primase |
| FAD | = | flavin adenine-dinucleotide |
| p58C/3L9Q | = | C-terminal part of the human primase p58 subunit deposited in PDB with an id code of 3L9Q |
| p58C/3Q36 | = | C-terminal part of the human primase p58 subunit deposited in PDB with an id code of 3Q36 |
| rmsd | = | root-mean-square deviation |
| unit length | = | RNA primer with a length of up to 7–10 ribonucleotides |
| CD | = | circular dichroism |
Figures and Tables
Figure 1 A stereoview of the cartoon representation of human p58C/3Q36. the 4Fe-4S cluster and the side chains of coordinated cysteine residues are shown as sticks.
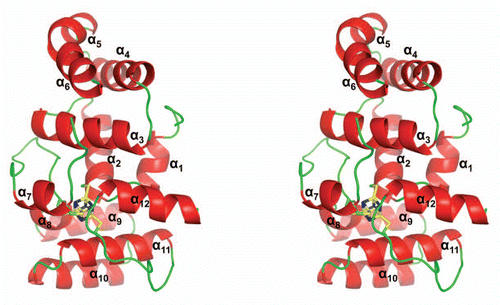
Figure 2 Comparison of the human p58C/3Q36 structure determined in current work with the corresponding domain structures reported for the yeast and human proteins. (A) Structure-based amino acid sequence alignment of the human and yeast primase 4Fe-4S domains. The sequences corresponding to α-helices and β-strands are highlighted by grey and yellow color, respectively. Conserved residues are shown in red and residues not in the final model are highlighted by cyan color. Residues involved in 4Fe-4S cluster coordination are indicated by triangles. (B and C) Comparison of the human p58C/3Q36 with the structures of the (B) yeast (3LGB) and (B) human (3L9Q) 4Fe-4S cluster domains. The labeled regions exhibiting the significant differences are discussed in the text
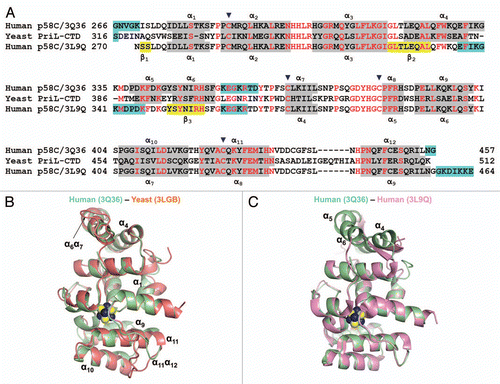
Figure 3 Examples of Far UV spectra of human p58C/3Q36 obtained at lower and high pH. The spectra are shown for the protein dialyzed against MES pH 6.5 and 40 mM NaCl (smooth line), and bicine pH 9.0 and 200 mM Li2SO4 (dotted line).
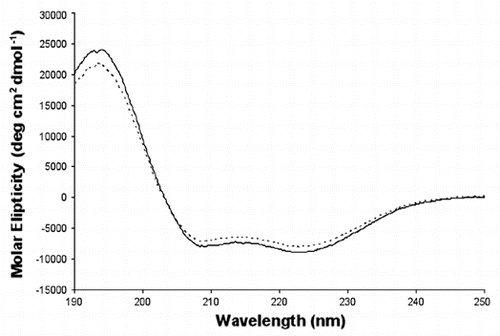
Figure 4 Crystal packing of the primase large subunit 4Fe-4S cluster domains. The figure highlights surrounding of helices α4, α5 and α6 in (A) human p58C/3Q36 and (B) yeast PriL-CTD and (C) β-strands in human p58C/3L9Q.
Figure 5 Comparison of yeast PriL-CTD and human p58C/3Q36 with the ssDNA-binding fragment of DASH cryptochrome 3 from Arabidopsis thaliana (PDB code 2VTB). The yeast PriL-CTD (residues 334–423) and human p58C/3Q36 (285–374) were superimposed with A. thaliana DASH cryptochrome 3 (373–457) with rmsd of 1.36 Å for 72 matched Cα atoms and rmsd of 1.32 Å for 66 matched Cα atoms, respectively. The protein fragments are drawn as cartoons. The FAD and ssDNA bound to a DASH cryptochrome 3 are shown as sticks.
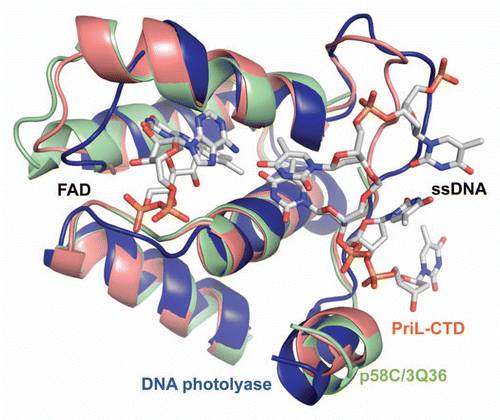
Figure 6 A proposed model of primer length counting and primer-template dissociation from DNA primase. (A) Initiation of primer synthesis. (B) Synthesis of unit length RNA primer causes a sterical hindrance. (C) Sterical hindrance induces the conformational change, resulting in RNA primer-template dissociation from primase and its transfer to Pol α.
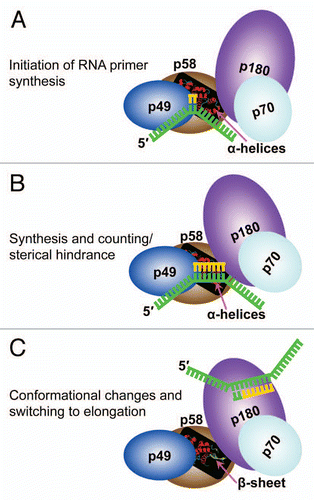
Table 1 Data collection and refinement statistics
Acknowledgements
We thank J. Lovelace and G.E. Borgstahl for the maintenance and management of the Eppley Institute's X-ray Crystallography facility. The primers were synthesized in the Eppley Institute's Molecular Biology Core facility. Both facilities are supported by the Cancer Center Support Grant P30CA036727. This work is supported by Eppley Institute's Pilot Projects and in part by NIGMS grant R01GM082923 to T.H.T. and by NCI grant CA129925 to Y.I.P. This work is also based upon research conducted at the Northeastern Collaborative Access Team beamlines of the Advanced Photon Source, supported by award RR-15301 from the National Center for Research Resources at the National Institutes of Health. Use of the Advanced Photon Source is supported by the US Department of Energy, Office of Basic Energy Sciences, under Contract No. DE-AC02-06CH11357.
References
- Kornberg A, Baker TA. DNA replication 1992; New York WH Freeman
- Kuchta RD, Stengel G. Mechanism and evolution of DNA primases. Biochim Biophys Acta 2010; 1804:1180 - 1189
- Garg P, Burgers PM. DNA polymerases that propagate the eukaryotic DNA replication fork. Crit Rev Biochem Mol Biol 2005; 40:115 - 128
- Frick DN, Richardson CC. DNA primases. Annu Rev Biochem 2001; 70:39 - 80
- Copeland WC, Wang TS. Enzymatic characterization of the individual mammalian primase subunits reveals a biphasic mechanism for initiation of DNA replication. J Biol Chem 1993; 268:26179 - 26189
- Arezi B, Kirk BW, Copeland WC, Kuchta RD. Interactions of DNA with human DNA primase monitored with photoactivatable cross-linking agents: implications for the role of the p58 subunit. Biochemistry 1999; 38:12899 - 12907
- Zerbe LK, Kuchta RD. The p58 subunit of human DNA primase is important for primer initiation, elongation and counting. Biochemistry 2002; 41:4891 - 4900
- Klinge S, Hirst J, Maman JD, Krude T, Pellegrini L. A iron-sulfur domain of the eukaryotic primase is essential for RNA primer synthesis. Nat Struct Mol Biol 2007; 14:875 - 877
- Sauguet L, Klinge S, Perera RL, Maman JD, Pellegrini L. Shared active site architecture between the large subunit of eukaryotic primase and DNA photolyase. PLoS One 2010; 5:10083
- Vaithiyalingam S, Warren EM, Eichman BF, Chazin WJ. Insights into eukaryotic DNA priming from the structure and functional interactions of the 4Fe-4S cluster domain of human DNA primase. Proc Natl Acad Sci USA 2010; 107:13684 - 13689
- Weiner BE, Huang H, Dattilo BM, Nilges MJ, Fanning E, Chazin WJ. An iron-sulfur cluster in the C-terminal domain of the p58 subunit of human DNA primase. J Biol Chem 2007; 282:33444 - 33451
- Mizuno T, Yamagishi K, Miyazawa H, Hanaoka F. Molecular architecture of the mouse DNA polymerase alpha-primase complex. Mol Cell Biol 1999; 19:7886 - 7896
- Pokorny R, Klar T, Hennecke U, Carell T, Batschauer A, Essen LO. Recognition and repair of UV lesions in loop structures of duplex DNA by DASH-type cryptochrome. Proc Natl Acad Sci USA 2008; 105:21023 - 21027
- Sheaff RJ, Kuchta RD, Ilsley D. Calf thymus DNA polymerase alpha-primase: “communication” and primer-template movement between the two active sites. Biochemistry 1994; 33:2247 - 2254
- Tahirov TH, Temiakov D, Anikin M, Patlan V, McAllister WT, Vassylyev DG, et al. Structure of a T7 RNA polymerase elongation complex at 2.9 A resolution. Nature 2002; 420:43 - 50
- Yin YW, Steitz TA. Structural basis for the transition from initiation to elongation transcription in T7 RNA polymerase. Science 2002; 298:1387 - 1395
- Otwinowski Z, Minor W. Carter CW Jr, Sweet RM. Processing of X-ray Diffraction Data Collected in Oscillation Mode. Macromolecular Crystallography, part A 1997; New York Academic Press 307 - 326
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr 1998; 54:905 - 921
- Guex N, Peitsch MC. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 1997; 18:2714 - 2723
- Lees JG, Miles AJ, Wien F, Wallace BA. A reference database for circular dichroism spectroscopy covering fold and secondary structure space. Bioinformatics 2006; 22:1955 - 1962