Abstract
In line with their broad-based effects, microRNAs (miRNAs), small non-coding RNA molecules ~22 nucleotides long that silence target mRNAs, are thought to act as oncogenes or tumor suppressor genes based on their inhibition of tumor-suppressive and oncogenic mRNAs, respectively. We and others previously showed that global downregulation of miRNAs, a common feature of human tumors, is functionally relevant to oncogenesis as impairment of miRNA biogenesis enhanced transformation in both cancer cells and a K-Ras-driven model of lung cancer. The dysregulation of miRNA biosynthesis in cancer emerges as a cancer-specific mechanism that enhances its tumorigenic capacity. These observations are further supported by the fact that frameshift mutations of TARBP2 occur in sporadic and hereditary carcinomas with microsatellite instability and that DICER1 mutations are associated with familial pleuropulmonary blastoma. Accordingly, it was reported that reduced expression of miRNA-processing factors is associated with poor prognosis in lung cancer and ovarian cancer. Recently we have also demonstrated the presence of Exportin 5 (XPO5) inactivating mutations in tumors with microsatellite instability. This observed genetic defect is responsible for nuclear retention of pre-miRNAs, thereby reducing miRNA processing. The characterized mutant form of the XPO5 protein lacks a C-terminal region that contributes to the formation of the pre-miRNA/XPO5/Ran-GTP ternary complex and the protein itself, as well as pre-miRNAs accumulating in the nucleus of cancer cells. Most importantly, the restoration of XPO5 function reverses the impaired export of pre-miRNAs and has tumor suppressor features. Our data suggest a cancer-specific mechanism to guide the subcellular distribution of miRNA precursors and prevent them from being processed to the active mature miRNA. The control of the miRNA biosynthesis pathway is emerging as an important mechanism in defining the spatiotemporal pattern of miRNA expression in cancer cells.
Introduction
MicroRNAs (miRNAs) are regulatory RNAs that silence mRNAs in a sequence-specific manner. Originally discovered in Caenorhabditis elegans, almost a thousand human miRNAs are now known to repress target mRNAs. Biogenesis of miRNAs in mammalian systems involves multiple steps, including transcription of primary miRNA (pri-miRNA), cleavage of pri-miRNA to precursor miRNA (pre-miRNA), nucleocytoplasmic export of pre-miRNA, cleavage of pre-miRNA to miRNA duplex and formation of functional RISC.Citation1,Citation2 Pri-miRNAs are transcribed by RNA polymerase II and cleaved by the Microprocessor Complex, containing at least Drosha (RNAase III endonuclease) and DGCR8 in humans (a double-stranded RNA-binding protein). This complex recognizes the double-stranded RNA structure of the pri-miRNA and specifically cleaves at the base of the stem loop, releasing a ∼70-nucleotide precursor pre-miRNA. This pre-miRNA is then exported through the Exportin-5 (XPO5) pathway into the cytoplasm. XPO5 protein in a complex with Ran protein recognizes and binds the pre-miRNA molecule, exporting it to the cytoplasm along a GTP to GDP gradient. Once in the cytoplasm it is further processed into a mature miRNA duplex by DICER1, a second RNase III endonuclease, together with its catalytic partner TAR Binding Protein (TRBP). The miRNA duplex is then loaded into a multi-complex, the RNA-induced silencing complex (RISC), which is comprised of at least TRBP, DICER1 and one Argonaute (Ago2 in humans). Most animal miRNAs exhibit imperfect homology with their targets, which inhibits translation, thereby controlling some of the most important cellular processes such as proliferation, apoptosis and development, amongst others.
Regulation of miRNA Processing
It is clear that miRNA processing is regulated in a complex manner, and we are only beginning to understand its true nature. In principle, miRNA abundance could be controlled at the transcription of the pri-miRNA, during any of the biogenesis steps, or with the turnover of the mature miRNA. Early studies by several laboratories found that mature miRNA expression does not always correlate with expression of the pri-miRNA, especially in the context of oncogenesis.Citation3–Citation8 However, PCR and northern blot have been used to demonstrate that the expression of mature miRNAs is generally correlated with pri-miRNA expression in normal tissues.Citation9–Citation12 On the other hand, several studies have shown that miRNAs are present at the level of the precursor but are not processed to the mature miRNA in cancer cells and primary tumors. For example northern blot analysis demonstrated that the precursors of miR-143 and miR-145 were expressed in colorectal tissues and tumors; however, the mature miRNA was detectable only in normal colorectal tissue.Citation13 Pre-miR-138-2 was shown by northern blot and in situ hybridization to be expressed in HeLa cells, but the mature miR-138 was undetectable.Citation4 Also, it has been reported that a wide discrepancy exists between the levels of precursor and mature miRNA, suggesting that unknown mechanisms that control processing play a critical role in regulating the expression of the active, mature miRNA.Citation7 It was also demonstrated that mutationsCitation14 or SNPsCitation15 within the miRNA precursors can interfere with miRNA processing. Although we believe this to be an unlikely explanation of the lack of miRNA processing, the widespread nature of the phenomenon makes it important to demonstrate that cancer cells avoid being controlled by miRNAs that impair their biosynthesis.
Therefore, impairment of these biosynthesis checkpoint control mechanisms of mature miRNAs in cancer cells could lead to an abnormal expression profile of these small non-coding RNAs, thus enhancing the tumorigenic process.
Dysregulation of miRNA in Cancer
The causes of the aberrant miRNA expression pattern in cancer may be due to DNA copy number amplification or deletion,Citation16 inappropriate transactivation,Citation17 genetic mutationCitation18 or epigenetic mechanisms.Citation19,Citation20 Indeed, some miRNA loci often show genomic instability in cancer, and it was also reported that c-Myc broadly repressed the transcription of miRNA genes.Citation21,Citation22
The widespread downregulation of miRNAs is often observed in human malignancies, including breast, prostate and ovarian cancers.Citation23–Citation26 In addition, global repression of miRNA biogenesis by suppression of the key components of miRNA processing machinery, such as Drosha, DGCR8, DICER1, TRBP and XPO5, promotes cellular transformation and tumorigenesis.Citation27–Citation31 While the mechanism(s) remain to be fully elucidated, it suggests that miRNAs might have an intrinsic function in tumor suppression and its downregulation eventually accelerates oncogenesis.
Moreover, it has become evident that some characteristic aspects of cancer-related biological processes, including drug resistance, tumor angiogenesis and metastasis, are associated with miRNA function.Citation32,Citation33 Therefore, understanding the mechanisms behind miRNAs dysregulation in human cancer and their functional consequences might provide new insights for improving the classification, prognosis prediction and treatment of cancer.
Importance of Intracellular Localization
A distinctive stage in evolution was the formation of the cell nucleus, which stores the genetic information encoded by DNA. This compartmentalization resulted in the segregation of key steps in the synthetic pathways from DNA to protein. A similarity exists with the biogenesis of miRNA molecules. The appropriate subcellular localization of the pre-miRNA is essential to their complete processing, their function and regulation. Compartmentalization can control access to binding partners, concentrate cofactors or temporarily segregate components of the pathway away from the rest of the cellular environment. The exquisite spatiotemporal control of miRNA abundance is made possible, in part, by regulation of the miRNA biogenesis pathway. While pri-miRNA processing is a nuclear event, pre-miRNA processing occurs in the cytoplasm. Thus, pre-miRNAs need to transit from the nucleus into the cytoplasm, a process that requires the nuclear export receptor XPO5.Citation34,Citation35
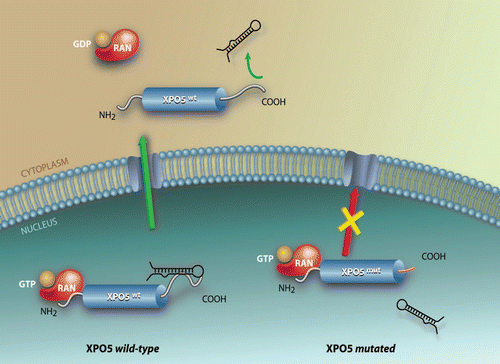
Frameshift mutations occur in protein-coding sequences, which may render affected proteins nonfunctional and thus drive carcinogenesis through the inactivation of tumor suppressor genes. We have reported frameshift mutations in the XPO5 gene in two MSI+ cell lines and primary tumors.Citation31 The mutations found in exon 32 alter and truncate the protein sequence and prevent XPO5 from associating with its pre-miRNA cargo and exiting the nucleus (). In XPO5 heterozygous mutant cells, less pre-miRNA was hence accessible to processing by the cytoplasmic machinery, resulting in decreased mature miRNA levels and enhanced tumorigenicity. Restoration of XPO5 wild-type protein levels in the defective cells rescued pre-miRNA export and processing defects and had tumor suppressor features. These cancer cells exhibited impaired miRNA processing but failed to lose the wild-type XPO5 allele. Recent work has suggested that other components of the miRNA biogenesis pathway, DICER1 and TARBP2, are haploinsufficient tumor suppressors.Citation28–Citation30 Moreover, biallelic deletion was found to impair cell viability, hence preventing the phenomenon of loss-of-heterozygosity (LOH).Citation28–Citation30 This is also the case for XPO5, where mimicking LOH by RNA interference against the XPO5 wild-type transcript rendered cells unviable.Citation31 In addition, the miRISC components AGO2, TNRC6A and TNRC6C can also be mutated in MSI+ cancers,Citation36 although the functional consequences remain to be evaluated. The presence of mutations in the miRNA pathway genes, including TARBP2 and XPO5 genes, in MSI+ cancer samples has also been reported in this separate study of Korean patients.Citation36
The mutated status of this central component turned out to be crucial in malignant transformation since it clogged pre-miRNA flow between the nucleus and cytoplasm. The disruption of this process resulted in a dramatic deregulation of cellular functions that triggered tumorigenesis. Additionally, we also characterized a minimal region in XPO5 required for pre-miRNA binding and/or recognition and therefore for nuclear envelope transversion. Others have reported that XPO5 is expressed at low levels in many tumor types.Citation37 Therefore, the XPO5 heterozygous mutational event found in human cancer clogs the miRNA-nuclear export complex in the nucleus and prevents the export of pre-miRNAs to the cytoplasm and further processing. Nonetheless, analysis of miRNA array data shows that there are pre-miRNAs that the processing does not seem to alter by this impairment of the export machinery. Recent work in C. elegans has suggested additional nuclear export pathways for pre-miRNAs that could explain why some miRNA levels remained unaffected by this mutation.Citation38 Many pre-miRNAs are also targeted by ADARs at various stages of their processing, and the modification can also prevent export of pre-miRNAs.
Similar to what we demonstrated, the expression of over 200 precursor and mature miRNAs demonstrated a wide discrepancy between in a high percentage of human primary tumors.Citation10 Our results are also in line with the finding that, after profiling 225 precursor and mature miRNAs in 22 human primary tumors and 16 pancreatic and liver tissues/tumors, many of the miRNAs analyzed are processed to the precursor but these precursors are retained in the nucleus.Citation7 However, this study did not provide any possible explanation that could shed light on the mechanism underlying this pre-miRNA processing blockage and nuclear retention. In light of our results we can now speculate about possible impairment at the level of the nucleocytoplasmic pre-miRNA export machinery in some of the analyzed samples. Two other groups have also reported the accumulation of let7 miRNA precursors at various stages during fruit fly and sea urchin development.Citation39,Citation40 Although this could represent a defect in RNA processing, it is also possible that the nuclear export of the let7 precursor, and hence access to cytoplasmic DICER1 and TRBP, are developmentally regulated.
Suppression of miRNA Biogenesis in Human Cancer as a Way to Escape miRNA Regulation
Recent reports show that states of increased proliferation or cellular transformation are associated with widespread occurrence of the production of mRNAs with shortened 3′UTR and fewer miRNA target sites, indicating that a global switch of the use of miRNA-mediated gene regulation could occur as part of a general program for cellular proliferation and transformation.Citation41,Citation42 These findings are consistent with the enhancement of miRNA maturation under DNA damage and the previous findings on widespread decrease of miRNAs in cancer. Avoidance of regulation of gene expression by miRNAs might be a general feature of cancer cells, and a regulatory layer by miRNAs may have an essential function in tumor suppression. Accordingly, it was reported that reduced expression of miRNA processing factors is associated with poor prognosis in lung and ovarian cancer.Citation42,Citation43
Overall, there is increasing evidence from the geneticCitation28,Citation30,Citation31 and functionalCitation27,Citation29 standpoints to suggest a role for miRNA processing machinery genes, such as DICER1, TRBP, DROSHA and XPO5, as tumor suppressor genes in human cancer.
References
- Kim VN. MicroRNA biogenesis: coordinated cropping and dicing. Nat Rev Mol Cell Biol 2005; 6:376 - 385
- Winter J, Jung S, Keller S, Gregory RI, Diederichs S. Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat Cell Biol 2009; 11:228 - 234
- Thomson JM, Newman M, Parker JS, Morin-Kensicki EM, Wright T, Hammond SM. Extensive post-transcriptional regulation of microRNAs and its implications for cancer. Genes Dev 2006; 20:2202 - 2207
- Obernosterer G, Leuschner PJ, Alenius M, Martinez J. Post-transcriptional regulation of microRNA expression. RNA 2006; 12:1161 - 1167
- Blenkiron C, Goldstein LD, Thorne NP, Spiteri I, Chin SF, Dunning MJ, et al. MicroRNA expression profiling of human breast cancer identifies new markers of tumor subtype. Genome Biol 2007; 8:214
- Wulczyn FG, Smirnova L, Rybak A, Brandt C, Kwidzinski E, Ninnemann O, et al. Post-transcriptional regulation of the let-7 microRNA during neural cell specification. FASEB J 2007; 21:415 - 426
- Lee EJ, Baek M, Gusev Y, Brackett DJ, Nuovo GJ, Schmittgen TD. Systematic evaluation of microRNA processing patterns in tissues, cell lines and tumors. RNA 2008; 14:35 - 42
- Pawlicki JM, Steitz JA. Subnuclear compartmentalization of transiently expressed polyadenylated primicroRNAs: processing at transcription sites or accumulation in SC35 foci. Cell Cycle 2009; 8:345 - 356
- Schmittgen TD, Jiang J, Liu Q, Yang L. A highthroughput method to monitor the expression of microRNA precursors. Nucleic Acids Res 2004; 32:43
- Jiang J, Lee EJ, Gusev Y, Schmittgen TD. Real-time expression profiling of microRNA precursors in human cancer cell lines. Nucleic Acids Res 2005; 33:5394 - 5403
- Meng F, Henson R, Lang M, Wehbe H, Maheshwari S, Mendell JT, et al. Involvement of human micro-RNA in growth and response to chemotherapy in human cholangiocarcinoma cell lines. Gastroenterology 2006; 130:2113 - 2129
- Lee J, Li Z, Brower-Sinning R, John B. Regulatory circuit of human microRNA biogenesis. PLoS Comput Biol 2007; 3:67
- Michael MZ, O'Connor SM, van Holst Pellekaan NG, Young GP, James RJ. Reduced accumulation of specific microRNAs in colorectal neoplasia. Mol Cancer Res 2003; 1:882 - 891
- Calin GA, Ferracin M, Cimmino A, Di Leva G, Shimizu M, Wojcik SE, et al. A MicroRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. New Engl J Med 2005; 353:1793 - 1801
- Duan R, Pak C, Jin P. Single nucleotide polymorphism associated with mature miR-125a alters the processing of pri-miRNA. Hum Mol Genet 2007; 16:1124 - 1131
- Calin GA, Croce CM. MicroRNA signatures in human cancers. Nature Rev Cancer 2006; 6:857 - 866
- Dews M, Homayouni A, Yu D, Murphy D, Sevignani C, Wentzel E, et al. Augmentation of tumor angiogenesis by a Myc-activated microRNA cluster. Nat Genet 2006; 38:1060 - 1065
- Jazdzewski K, Murray EL, Franssila K, Jarzab B, Schoenberg DR, de la Chapelle A. Common SNP in pre-miR-146a decreases mature miR expression and predisposes to papillary thyroid carcinoma. Proc Natl Acad Sci USA 2008; 105:7269 - 7274
- Lujambio A, Ropero S, Ballestar E, Fraga MF, Cerrato C, Setien F, et al. Genetic unmasking of an epigenetically silenced microRNA in human cancer cells. Cancer Res 2007; 67:1424 - 1429
- Lujambio A, Calin GA, Villanueva A, Ropero S, Sánchez-Céspedes M, Blanco D, et al. A microRNA DNA methylation signature for human cancer metastasis. Proc Natl Acad Sci USA 2008; 105:13556 - 13561
- Calin GA, Sevignani C, Dumitru CD, Hyslop T, Noch E, Yendamuri S, et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci USA 2004; 101:2999 - 3004
- Chang TC, Yu D, Lee YS, Wentzel EA, Arking DE, West KM, et al. Widespread microRNA repression by Myc contributes to tumorigenesis. Nat Genet 2008; 40:43 - 50
- Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, et al. MicroRNA expression profiles classify human cancers. Nature 2005; 435:834 - 838
- Marton S, Garcia MR, Robello C, Persson H, Trajtenberg F, Pritsch O, et al. Small RNAs analysis in CLL reveals a deregulation of miRNA expression and novel miRNA candidates of putative relevance in CLL pathogenesis. Leukemia 2008; 22:330 - 338
- Ozen M, Creighton CJ, Ozdemir M, Ittmann M. Widespread deregulation of microRNA expression in human prostate cancer. Oncogene 2008; 27:1788 - 1793
- Zhang L, Volinia S, Bonome T, Calin GA, Greshock J, Yang N, et al. Genomic and epigenetic alterations deregulate microRNA expression in human epithelial ovarian cancer. Proc Natl Acad Sci USA 2008; 105:7004 - 7009
- Kumar MS, Lu J, Mercer KL, Golub TR, Jacks T. Impaired microRNA processing enhances cellular transformation and tumorigenesis. Nat Genet 2007; 39:673 - 677
- Melo SA, Ropero S, Moutinho C, Aaltonen LA, Yamamoto H, Calin GA, et al. A TARBP2 mutation in human cancer impairs microRNA processing and DICER1 function. Nat Genet 2009; 41:365 - 370
- Kumar MS, Pester RE, Chen CY, Lane K, Chin C, Lu J, Kirsch DG, et al. Dicer1 functions as a haploinsufficient tumor suppressor. Genes Dev 2009; 23:2700 - 2704
- Hill DA, Ivanovich J, Priest JR, Gurnett CA, Dehner LP, Desruisseau D, et al. DICER1 mutations in familial pleuropulmonary blastoma. Science 2009; 325:965
- Melo SA, Moutinho C, Ropero S, Calin GA, Rossi S, Spizzo R, et al. A genetic defect in exportin-5 traps precursor microRNAs in the nucleus of cancer cells. Cancer Cell 2010; 18:303 - 315
- Kovalchuk O, Filkowski J, Meservy J, Ilnytskyy Y, Tryndyak VP, Chekhun VF, et al. Involvement of microRNA-451 in resistance of the MCF-7 breast cancer cells to chemotherapeutic drug doxorubicin. Mol Cancer Ther 2008; 7:2152 - 2159
- Tavazoie SF, Alarcon C, Oskarsson T, Padua D, Wang Q, Bos PD, et al. Endogenous human microRNAs that suppress breast cancer metastasis. Nature 2008; 451:147 - 152
- Yi R, Qin Y, Macara IG, Cullen BR. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev 2003; 17:3011 - 3016
- Lund E, Guttinger S, Calado A, Dahlberg JE, Kutay U. Nuclear export of microRNA precursors. Science 2004; 303:95 - 98
- Kim MS, Oh JE, Kim YR, Park SW, Kang MR, Kim SS. Somatic mutations and losses of expression of microRNA regulation-related genes AGO2 and TNRC6A in gastric and colorectal cancers. J Pathol 2010; 221:139 - 146
- Yi R, Doehle BP, Qin Y, Macara IG, Cullen BR. Overexpression of exportin 5 enhances RNA interference mediated by short hairpin RNAs and microRNAs. RNA 2005; 11:220 - 226
- Bussing I, Yang JS, Lai EC, Grosshans H. The nuclear export receptor XPO-1 supports primary miRNA processing in C. elegans and Drosophila. EMBO J 2010; 29:1830 - 1839
- Hutvagner G, McLachlan J, Pasquinelli AE, Balint E, Tuschl T, Zamore PD. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science 2001; 293:834 - 838
- Pasquinelli AE, Reinhart BJ, Slack F, Martindale MQ, Kuroda MI, Maller B, et al. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature 2000; 408:86 - 89
- Mayr C, Bartel DP. Widespread shortening of 3′UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell 2009; 138:673 - 684
- Sandberg R, Neilson JR, Sarma A, Sharp PA, Burge CB. Proliferating cells express mRNAs with shortened 3′ untranslated regions and fewer microRNA target sites. Science 2008; 320:1643 - 1647
- Karube Y, Tanaka H, Osada H, Tomida S, Tatematsu Y, Yanagisawa K, et al. Reduced expression of Dicer associated with poor prognosis in lung cancer patients. Cancer Sci 2005; 96:111 - 115
- Merritt WM, Lin YG, Han LY, Kamat AA, Spannuth WA, Schmandt R, et al. Dicer, Drosha and outcomes in patients with ovarian cancer. New Engl J Med 2008; 359:2641 - 2650