Abstract
The differentiation of myeloid progenitors to mature, terminally differentiated cells is a highly regulated process. Here, we showed that conditional disruption of the c-myb proto-oncogene in adult mice resulted in dramatic reductions in CMP, GMP and MEP myeloid progenitors, leading to a reduction of neutrophils, basophils, monocytes and platelets in peripheral blood. In addition, c-myb plays a critical role at multiple stages of myeloid development, from multipotent CMP and bipotent GMP to unipotent CFU-G and CFU-M progenitor cells. c-myb controls the differentiation of these cells and is required for the proper commitment, maturation and normal differentiation of CMPs and GMPs. Specifically, c-myb regulates the precise commitment to the megakaryocytic and granulo-monocytic pathways and governs the granulocytic-monocytic lineage choice. c-myb is also required for the commitment along the granulocytic pathway for early myeloid progenitor cells and for the maturation of committed precursor cells along this pathway. On the other hand, disruption of the c-myb gene favors the commitment to the monocytic lineage, although monocytic development was abnormal with cells appearing more mature with atypical CD41 surface markers. These results demonstrate that c-myb plays a pivotal role in the regulation of multiple stages in adult myelogenesis.
Introduction
Myeloid progenitor cells are derived from pluripotent hematopoietic stem cells in the bone marrow (BM). Hematopoietic stem cells undergo progressive commitment to generate multipotent common myeloid progenitor (CMP) cells, which, in turn, can differentiate into either megakaryocyte-erythrocyte progenitor (MEP) or granulocyte-monocyte progenitor (GMP) cells.Citation1 GMPs give rise to unipotent precusor cells that terminally differentiate into granulocytes or monocytes. The commitment and differentiation of myeloid progenitors to mature cells are highly regulated processes. Transcription factors play a key role in regulating these processes. Dysregulation or mutation of some of these transcription factors is often associated with myeloid leukemias.Citation2
The c-myb proto-oncogene is the founding member of the myb family of transcription factors. It is expressed highest in hematopoietic tissues, but its expression is noted in non-hematopoietic tissues as well.Citation3 Studies using transformed and leukemic cell lines as well as human bone marrow cells treated with antisense oligonucleotides implicate a role for c-Myb in hematopoiesis.Citation4,Citation5 c-Myb appears to be required for granulopoiesis but not monopoiesis,Citation5 and in vitro studies indicate a role for c-myb in mediating the monocyte/granulocyte lineage decision.Citation5 However, these studies have some shortcomings. For instance, the transcriptional program appears to be altered in some of the transformed and leukemic cell lines.Citation6 In addition, in the antisense studies using normal human bone marrow cells, colony-forming unit (CFU) survival and growth arrest of only bipotent CFU-GM (Granulocyte, Monocyte) and unipotent CFU-G and CFU-M progenitors were determined, but CFU survival of multipotent progenitor CFU-Mix/GEMM (granulocyte, erythrocyte, monocyte, megakaryocyte) was not assayed.Citation7 Furthermore, possible nonspecific effects associated with the antisense technology raise some concerns.Citation8 Lastly, it is not completely certain that these in vitro studies reflect myelopoiesis in the whole organism. For instance, the Egr1-knockout mice do not exhibit any myeloid deficits despite a large body of literature implicating the gene in myeloid development.Citation9
The embryonic lethality of homozygous c-myb-null mice at E15.5 emphasizes the critical requirement of c-myb in fetal hematopoiesis, although the precise defect is unclear.Citation10 Several different c-myb-deficient mouse models, which were generated using ENU (N-ethyl-N-nitrosurea) mutagenesis, showed little perturbation in adult myeloid development.Citation3 Except for the M303V mutant that had a point mutation in the transactivation domain and was devoid of eosinophils, myeloid progenitor cells and the number of neutrophils and monocytes were relatively normal in the other ENU-induced hypomorphic animals.Citation11,Citation12 However, on the other hand, the adult knockdown mice, where c-Myb expression was reduced to approximately 5–10% of control animals, had elevated peripheral blood platelets and monocytes but reduced blood neutrophils.Citation13 In addition, these adult knockdown animals had an abnormal hematopoietic cell population with a self-renewal capability, which could confound any study on myeloid development.Citation13 Furthermore, it is not certain that the phenotype of the adult knockdown mice reflects normal adult myeloid development, as a similar phenotype had been observed during fetal development.Citation11 It is therefore possible that defects in fetal hematopoiesis, seen in the c-myb-knockdown mice, could lead to abnormalities in adult hematopoiesis.
Amplification of c-MYB expression has been observed in myeloid leukemias.Citation14 Recent evidence showing genetic alterations in MYB by duplication or translocation in a subset of childhood T-cell acute leukemias provides a direct link for MYB in human cancer.Citation15,Citation16 In human myeloid leukemias, MYB duplication in two myeloid leukemic cell lines, HL60 and Meg01, has been observed.Citation17 In addition, genomic gain of MYB locus is seen in tissue samples from MYST3/MOZ-linked acute myeloid leukemic patients.Citation18 Thus, in order to delineate the role of c-MYB in myeloid leukemias, a clear and better understanding of c-MYB’s role in normal, adult myeloid development is required.
Here, we used the inducible mybf/f/MxCre system to conditionally delete c-myb gene in various adult myeloid progenitor cells to obtain a definitive and a better understanding of the role of c-myb in these populations of the murine BM cells. We demonstrate that c-myb is an important regulator of adult myelogenesis.
Results
Loss of c-Myb activity reduces all peripheral blood cells, including neutrophils, monocytes and platelets
To determine the role of c-myb in myelogenesis, we first examined the peripheral blood profiles of mybf/f/MxCre mice and their littermate controls after the administration of synthetic double-stranded RNA polyinosinic polycytidylic acid (pIpC) to induce the in vivo deletion of c-myb floxed gene. Total white blood cells (WBC) in the pIpC-induced mybf/f/MxCre mice were decreased as compared with control mice (Fig. S1A). In addition, blood analysis indicated that there was a statistically significant reduction in the number of neutrophils and platelets, while there was a modest and non-significant reduction in the number of monocytes, eosinophils, basophils, red blood cells and lymphocytes (Fig. S1A). This modest decline in some peripheral blood populations may reflect the incomplete deletion efficiency of the c-myb floxed allele in blood cells as shown by DNA analysis (Fig. S1B).
To increase the deletion efficiency of the c-myb floxed allele, we performed our studies using myb-/f/MxCre mice, where one of the c-myb allele was null and the other floxed. Inclusion of the two pIpC-induced myb-/f/MxCre mice and their littermate controls resulted in a statistically significant decrease in neutrophils, platelets, basophils, monocytes and lymphocytes (). In contrast, the blood profiles of mybf/f and myb-/f are similar (data not shown). The results from peripheral blood profiles of pIpC-induced mybf/f/MxCre mice are in agreement with a reduction of neutrophils and monocytes in the BMCitation19 and spleen (data not shown) of these mice.
Table 1. Peripheral blood cell counts of pIpC-administered mybf/f/MxCre and myb-/f/MxCre mice
Interestingly, the number of platelets in peripheral blood was significantly reduced when the c-myb gene was disrupted (Fig. S1A and Table S1). In addition, not only was there a reduction in the number of blood platelets, the size of these mutant platelets as shown by the mean platelet volume (MPV) was larger than that of the littermate controls (Fig. S1A and Table S1), suggesting the possibility of other alterations in megakaryocyte development when the c-myb gene is disrupted. Hence, our data from peripheral blood analysis are consistent with our findings in the BM, suggesting that c-myb is required for the development of adult myeloid lineagesCitation19 (Fig. S1; ).
In vivo disruption of c-myb activity results in a dramatic decrease in the myeloid progenitor CMPs, GMPs and MEPs
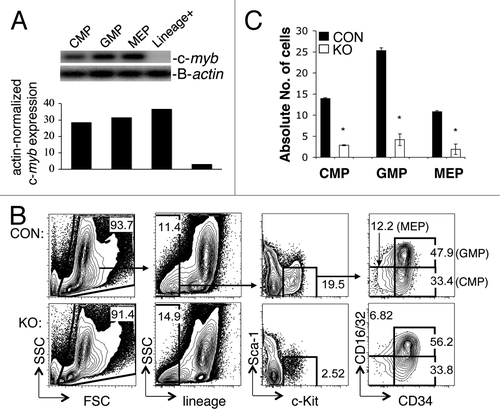
To begin to understand the nature of the myeloid deficiency in the bone marrow and peripheral blood cell counts as a result of loss of c-myb expression, we examined the number of various myeloid progenitor cells in pIpC-treated mybf/f/MxCre mice and their littermate controls. We performed flow cytometric staining to identify the three myeloid progenitors: common myeloid progenitor (CMP), granulocyte-monocyte progenitor (GMP) and megakaryocytic-erythroid progenitor (MEP) cells.Citation1 shows, by RT-PCR, high expression of c-myb in various BM-derived myeloid progenitor cells CMPs, GMPs and MEPs. The percentages of CMPs, GMPs and MEPs in the pIpC-treated mybf/f/MxCre mice were quite similar to control animals (). However, due to the huge cellular loss in the lineage-c-Kit+Sca-1- (LKS-) myeloid compartment (~53%) and in the BM (~59%) of the pIpC-treated mybf/f/MxCre miceCitation19 (), the converted percentages to absolute cellular numbers of CMPs, GMPs and MEPs were actually decreased by more than 80% (). This number (80%) was derived by multiplying the percentage of remaining total bone marrow cells (41%) with the percentage of LKS- cells (47%), which represents the percentage of remaining cells (20%). This represents a loss of 80%. No abnormality was detected in the pIpC-treated mybf/+/MxCre mice (data not shown). These results suggest that c-myb is required for the development of adult myeloid progenitor CMP, GMP and MEP cells.
Figure 1. c-myb is required for the formation of myeloid progenitor CMP, GMP and MEP cells in the bone marrow of pIpC-induced mybf/f/MxCre mice. (A) RT-PCR showing the level of c-myb expression in various myeloid progenitor cells CMP, GMP and MEP from BM of wild-type mouse. (B) Representative two-color flow cytometric analysis of myeloid progenitor cells CMP, GMP and MEP from 35 pIpC-administered mybf/f/MxCre mice (KO) and 35 pIpC-treated control littermates (CON). (C) Bar graph depicts the absolute number of progenitor cells CMP, GMP and MEP (× 104) in pIpC-administered animals. *, p < 0.001. Numbers are presented as mean ± SEM n = 35 mice for each genotype. Lin+, lineage-positive BM cells.
The requirement of c-myb in CMPs, GMPs and CFU-G and CFU-M precursor cells is intrinsic
Though loss of c-myb expression resulted in the decrease of various myeloid progenitors and mature myeloid cells in the BM and peripheral blood of the pIpC-induced mybf/f/MxCre mice as compared with littermate controls, it was not clear whether this was an intrinsic effect on myelogenesis or an indirect effect due to upstream defects in the hematopoietic stem cells, as we had previously described.Citation19 To determine whether c-myb plays a direct role in myeloid progenitor cells, we performed hematopoietic colony-forming assays using FACS-sorted CMPs and GMPs that had been treated with interferon (IFN) to delete the c-myb floxed gene; some deletion could be seen starting at 12 h after IFN addition and peaked around 24 h (data not shown). After treatment for 16–18 h, cells were plated on semi-solid cytokine-containing medium that allows for the growth and differentiation of multipotent, bipotent and unipotent myeloid progenitor cells.
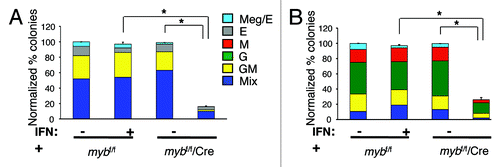
Hematopoietic colonies from IFN-treated mybf/f/MxCre CMPs or GMPs were significantly reduced by 82% and 75%, respectively, with corresponding reductions in all types of colonies as compared with controls (), indicating an important role for c-myb in the growth and/or survival of these cells on semi-solid cytokine-containing medium. Specifically, CFU-Mix and CFU-GM were equally reduced in FACS-purified CMPs, while CFU-GM, CFU-M and CFU-G were proportionally diminished in sorted GMPs (). In addition, disruption of the c-myb gene in purified lineage-negative c-Kit+Sca-1- (LKS-) resulted in a substantial reduction of hematopoietic colonies as seen with CMPs and GMPs (data not shown). Furthermore, addition of G-CSF, M-CSF or GM-CSF to the SCF/IL-3/IL-6 methylcellulose medium did not rescue colony growth (data not shown). In addition, no significant difference in hematopoietic colonies was detected between IFN-treated mybf/+/MxCre and IFN-treated mybf/f CMPs or GMPs (data not shown). Hence, these studies support an intrinsic role for c-myb at multiple stages in myelogenesis, from multipotent CMP and bipotent GMP progenitors to unipotent CFU-G and CFU-M precursor cells.
Figure 2. c-myb is required intrinsically for the growth and differentiation of myeloid progenitor CMP and GMP, and precursor CFU-G and CFU-M cells. Hematopoietic colony assays performed using untreated or 16 h interferon (IFN) treated purified (A) CMPs (n = 9) and (B) GMPs (n = 8) from mybf/f/MxCre mice and littermate controls. The number of colonies from the three groups was normalized to the untreated control, which was set to 100%. Legends for (B) are shown in (A). Granulocytic (G), erythroid (E), monocytic (M), megakaryocytic (Meg) and various (Mix) myeloid CFU colonies are shown. Data are expressed as mean ± SEM *, p < 0.001.
c-myb regulates the functionality of CMPs
To understand the functional role of c-myb in myelogenesis, we sorted CMP and GMP progenitors, treated these cells with type I interferon to delete c-myb floxed alleles in vitro and subjected these cells to FACS analysis after 24 h and 48 h of interferon treatment. To examine the possible roles of c-myb in proliferation, survival and differentiation, we performed BrdU labeling, caspase and surface lineage marker staining on these cells, respectively. Our initial study on purified LKS- showed little or no difference at 24 h following interferon treatment for the above three functional assays, perhaps reflecting the fact that deletion of c-myb floxed allele started at ~12 h after IFN addition and peaked at ~24 h (data not shown). Hence, we performed all our experiments at the 48 h time point.
By surface staining, reduction of c-myb expression by 81%, as assessed by RT-PCR, in CMPs leads to altered expression of surface antigens ( and data not shown). Cell surface expression of CD11b, CD41 (a marker of megakaryocytic differentiation) and CD115 (a mature monocytic differentiation receptor) on IFNα-treated c-myb deleted mybf/f/MxCre CMPs was upregulated, whereas Gr-1 antigen levels was downregulated. The expression of c-Kit remained unchanged as compared with IFN-treated mybf/f CMPs (). We did not detect any differences in the levels of the surface antigens examined in untreated mybf/f/MxCre and untreated mybf/f CMPs (data not shown). To understand these phenotypic changes in further detail, we performed two-color flow cytometric analysis using antibodies directed against the CD11b and CD41 or Gr-1 surface markers (). On the other hand, CD11b is expressed during development in both the monocytic and granulocytic pathways.Citation20 Gr-1 is expressed on granulocytes, with increasing levels of expression correlating with maturity; immature monocytes also express Gr-1, but at intermediate levels and only transiently during development.Citation21,Citation22 The two-color FACS plots of CD11b and CD41 demonstrate that disruption of the c-myb gene led to increases in single positive CD11b and CD41 cells as well as to double positive CD11b+CD41+ cells (), suggesting that loss of c-Myb activity induces differentiation toward the granulo-monocytic and megakaryocytic pathways, and also leads to aberrant differentiation as exhibited by presence of double positive CD11b+CD41+ cells.
Figure 3. c-myb regulates the functionality of CMPs. Following 48 h interferon treatment to induce the disruption of c-myb, the purified CMPs were stained for surface expression of various markers. Shown are the representative (A) overlay histograms and (B and E) two-color flow cytometric analysis of various surface antigens on CMPs, purified from mybf/f/MxCre mice and littermate controls. Representative overlay histograms of antigens on (C) the CD11b+Gr-1+ and (D) the CD11b+Gr-1- cell compartments. Legends for (C and D) are same as (A). Panels are representative of three independent experiments. The percentages for each surface marker in the indicated bar region of the histograms are specified for IFN-treated mybf/f (black) and IFN-treated mybf/f/MxCre (gray) cells.
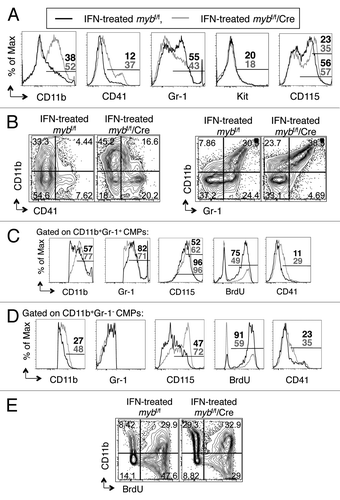
To examine the granulo-monocytic pathway in greater detail, we performed two-color FACS analysis of CD11b and Gr-1. These results indicate that loss of c-Myb activity leads to a shift toward single positive CD11b and a decrease in single positive Gr-1 cells, possibly suggesting that absence of c-myb expression may favor monocytic differentiation (). As monocytes also express Gr-1 during development, we scrutinized the CD11b+Gr-1+ compartment of IFNα-induced mybf/f/MxCre and control CMPs by surface expression of CD115, CD11b and Gr-1 (). Overlay histograms show that loss of c-myb expression led to an increase in surface expression of CD11b and CD115 (). In addition, an increase in cells expressing intermediate levels of Gr-1, with a slight decrease in cells expressing high levels of Gr-1 surface antigen was observed as a function of c-myb deletion (), indicating that the granulocytic developmental pathway is blocked, while the monocytic differentiation pathway is favored. Consistent with the observation that loss of c-myb induced differentiation, the proliferative capacity, which is inversely correlated with maturity,Citation21,Citation23 was decreased in the CD11b+Gr-1+ disrupted c-myb cells as compared with control cells as measured by BrdU incorporation (). Furthermore, compared with control cells, the mutant cells in the CD11b+Gr-1- compartment also expressed elevated surface CD115 and CD11b antigens and exhibited decreased uptake of BrdU (). These studies indicate that loss of c-myb activity led to an increase in the frequency of CD11b+Gr-1- cells that are committed to the monocytic development. A closer examination of the CD11b+Gr-1- and the CD11b+Gr-1+ populations reveals another difference in these mutant cells as compared with control cells (). Specifically, there was an increase in surface expression of the CD41 marker on both of these populations in the c-myb disrupted cells as compared with control cells (), suggesting possibly further aberrant development in both the monocytic and granulocytic pathways. Together, our data indicate that c-myb regulates the commitment to the megakaryocytic and granulo-monocytic pathways. Also, c-myb is required for the differentiation of granulocytes and for the normal development of monocytes.
There were no significant changes in polycaspase staining among the four CMP experimental and control groups (data not shown), indicating that apoptosis via caspases at the 48 h time point is not activated when the c-myb gene is disrupted. However, the c-myb deleted CMPs had a modest, but significant, reduction in the proliferative capacity as compared with that of untreated mybf/f/MxCre and IFN-treated and untreated mybf/f CMP controls (Fig. S2A). A detailed analysis to understand the proliferative defect in the c-myb-depleted CMPs showed that the decrease in the proliferative CMPs was accompanied by an increase in cells expressing the monocytic-granulocytic differentiation marker CD11b, further suggesting that c-myb is a regulator of differentiation ().
c-myb is required for the functionality of GMPs
To assess lineage commitment and differentiation potential of GMPs when c-Myb activity is lost, we performed flow cytometric analyses using interferon-treated, purified GMPs to determine the level of surface lineage antigen expression as well as that of c-Kit (). Reduction of c-myb expression by 76%, as assessed by RT-PCR, in GMPs leads to upregulation of CD11b, Gr-1, CD115 and, to a lesser extent, CD41, with little alteration in c-Kit expression ( and data not shown). We did not detect a difference in the levels of any of the markers examined between untreated mybf/f/MxCre and untreated mybf/f GMPs (data not shown). To further assess these phenotypic changes, we performed two-color flow cytometric analysis of CD11b and CD41 or Gr-1 surface markers (). The FACS plot of CD11b and CD41 reveals that approximately 67% of interferon-treated mybf/f GMPs exhibited increased levels of the CD11b surface antigen, while only outliers expressed CD41 (), indicating that differentiation could only occur in the direction of the granulo-monocytic pathway. This observation is consistent with the fact that GMPs are bipotential and more committed than the mutipotent CMPs in certain lineage fate ( and ). On the other hand, disruption of the c-myb gene in GMPs led to additional increases in CD11b expression and a slight increase in CD41 surface antigen (), indicating that loss of c-Myb activity induces differentiation toward the granulo-monocytic pathway.
Figure 4. c-myb is required for the functionality of GMPs. Following 48 h interferon treatment to induce the disruption of c-myb, the purified GMPs were stained for surface expression of various markers. Shown are the representative (A) overlay histograms and (B and E) two-color flow cytometric analysis of various surface antigens on GMPs, purified from mybf/f/MxCre mice and littermate controls. Representative overlay histograms of antigens on (C) the CD11b+Gr-1+ and (D) the CD11b+Gr-1- cell compartments. Legends for (C and D) are same as (A). Panels are representative of three independent experiments. The percentages for each surface marker in the indicated bar region of the histograms are specified for IFN-treated mybf/f (black) and IFN-treated mybf/f/MxCre (gray) cells.
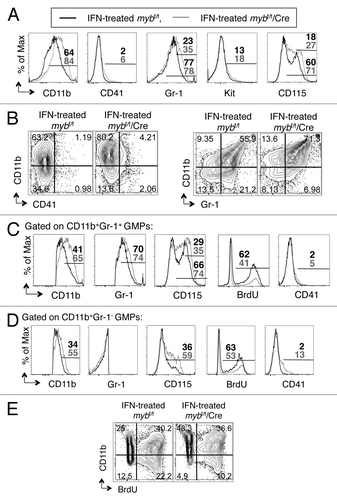
To further assess the altered differentiation potential of c-myb-deleted GMPs, we performed two-color FACS analysis of CD11b and Gr-1 (). Disruption of the c-myb gene resulted in further increase in single positive CD11b and double positive CD11b+Gr-1+ cells as compared with control GMPs (). A closer examination of the CD11b+Gr-1+ compartment indicates that c-myb-deleted cells had increased expression of CD11b, CD115 and Gr-1 compared with control IFNα-treated mybf/f cells (). While the increase in Gr-1 marker indicates further development along the granulocytic lineage, the augmented expression of CD11b and CD115 suggests differentiation toward the monocytic pathway. In the single positive CD11b compartment, IFNα-induced mybf/f/MxCre GMPs had greater surface expression of CD11b, CD115 and CD41 than control cells (), indicating not only increased differentiation toward the monocytic lineage but also some atypical development in the absence of c-Myb activity in GMPs. Furthermore, the decrease in BrdU uptake in both the single positive CD11b and CD11b+Gr-1+ compartments of IFNα-induced mybf/f/MxCre GMPs is further suggested that c-myb-deleted cells experienced increased developmental maturity than control cells (). Together, our data indicate that c-myb regulates commitment to the granulocytic and monocytic pathways and is required for the proper maturation and development of myeloid cells.
Similar to the CMPs, c-Myb does not appear to play a role in apoptosis via caspases at the GMPs, at least at the 48 h time point and under these experimental conditions, as measured by polycaspase staining (data not shown). There is a slight but significant decrease in BrdU incorporation when c-myb gene was deleted in GMPs (Fig. S2B). Further analysis revealed that when c-myb was lost in GMPs, the percentage of proliferating cells was reduced, while the percentage of cells expressing the differentiation marker CD11b was increased, indicating that c-myb is a regulator of differentiation in GMPs ().
Disruption of c-myb in CMP and GMP progenitor cells leads to altered gene expression
To determine the molecular mechanism by which c-myb functions in CMPs and GMPs, we performed DNA microarray analysis on purified lineage-c-Kit+Sca-1- (LKS-) BM cells, as these cells could be obtained in sufficient abundance and in high purity. After the addition of interferon to induce the deletion of the c-myb floxed gene, some deletion could be seen starting at 12 h and peak at 24–48 hr (data not shown). We wanted to assess early changes in gene expression after the loss of c-Myb activity. Thus, LKS- cells were treated with interferon for 18 h to induce the disruption of the c-myb floxed gene. gfi-1, cxcr4, cebpa and bcl2 were shown to be downregulated in interferon-treated LKS- cells when the c-myb gene was disrupted (Table S1). We assessed the expression of these genes in 18 h, interferon-treated mybf/f/MxCre CMPs and GMPs by RT-PCR (). Disruption of c-myb expression in CMPs resulted in significant downregulation of gfi-1, cxcr4, cebpa and bcl2 genes by RT-PCR (). In addition, expression of c-myc was also decreased when c-myb was deleted in CMPs (). On the contrary, in GMP cells, disruption of c-myb gene led to statistically significant downregulation of only cxcr4 and bcl2 genes ().
Figure 5. Disruption of c-myb results in altered gene expression in CMPs and GMPs. RT-PCR analysis of gene expression of (A) CMPs and (B) GMPs after 18 h of interferon treatment is shown. Results reflect at least three independent experiments. In the case of c-myc, results were from two independent sorts. Data on bar graph are β-actin normalized gene expression (y-axis) and are expressed as mean ± SEM *, p < 0.05; **, p < 0.01; ***, p < 0.001.
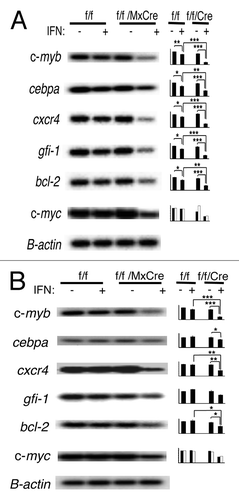
Noteworthy, other genes of interest that were downregulated in the LKS- microarray when c-myb expression was deleted were myeloperoxidase (MPO), neutrophil elastase and myeloblastin (Table S1). All three genes, as well as cxcr4 and bcl2, have been reported as c-myb target genes.Citation24-Citation28 Together, these studies indicate that loss of c-myb expression in LKS-, CMP and GMP cells leads to altered gene expression, affecting many known genes in the myeloid pathways.
Discussion
Our current study, which used the inducible mybf/f/MxCre system to conditionally disrupt c-myb expression in adult BM cells, supports a critical role for c-myb in the regulation of adult myeloid progenitor CMP, GMP, CFU-M and CFU-G cells. Disruption of the c-myb gene resulted in dramatic reductions in CMP, GMP and MEP myeloid progenitor cells in adult mice, leading to a reduction of all peripheral blood cells, significantly in neutrophils, basophils, monocytes and platelets. The requirement of c-myb in myeloid progenitor CMPs and GMPs, as well as precursor CFU-M and CFU-G, is intrinsic, as demonstrated by hematopoietic colony assays and/or in vitro functional assays. In CMPs and GMPs, c-myb acts as a regulator of differentiation. c-myb is required for the proper commitment, normal maturation and differentiation of CMP and GMP progenitor cells. Specifically, c-myb regulates the commitment to the megakaryocytic and granulo-monocytic pathways and governs the granulocytic-monocytic lineage choice. c-myb is required for the commitment to the granulocytic development for early myeloid progenitor cells. The disruption of the c-myb gene favors commitment to the monocytic lineage. However, in this model, monocytic development was abnormal, whereby monocytes appeared more mature, with atypical CD41 megakaryocytic surface markers. Hence, the transcriptional factor c-myb is an important regulator of adult myelogenesis.
Due to the accumulating knowledge in this field, it is perhaps not surprising that conditional disruption of c-myb expression in adult myeloid cells would perturb granulopoiesis. What is surprising about the data presented here was the direct impact of c-myb on monopoiesis. The majority of the published reports suggest that c-myb may not play a direct role in monopoiesis.Citation29-Citation34 Here, we used an inducible, conditional knockout strategy to disrupt c-myb expression in sorted CMPs and GMPs from adult BM cells and show that this gene has an intrinsic role in monopoiesis. First, we showed that the significant decrease in BM and peripheral blood monocytesCitation19 ( and data not shown), in fact, reflect an inherent and essential role of c-myb in monopoieis as shown by the drastic reduction of CFU-GM and CFU-M colonies in the colony-forming assays (). In addition, in vitro functional assays support a role for c-myb in regulating the proper maturation and normal development of monocytes. In these assays, when the c-myb gene was disrupted, CMPs and GMPs expressed increased levels of the CD11b surface marker and the CD115/M-CSF monocytic differentiation receptor as compared with control cells, indicating a biased maturation in the monocytic pathway ( and ). Furthermore, when c-Myb activity was lost, a portion of the CD11b+Gr-1-CD115+ and CD11b+Gr-1+CD115+ cells expressed CD41 surface marker, indicating further aberrant development ( and ). Although the precise defect in fetal hematopoiesis in the systemic c-myb-null embryos is unclear, their fetal livers contained few, but normally appearing, monocytes,Citation10 suggesting that c-myb may not have a direct role in the monocytic developmental pathway. Thus, contrary to most suggestions, c-myb has a direct role in monocytic development: in the absence of c-Myb activity, monocytic precursor cells undergo atypical differentiation.
In the c-myb-knockdown model, it was concluded that c-myb has a role in lineage choice based on the observation that there were an increase in monocytes and a decrease in granulocytes in the fetal liver.Citation29 However, it’s possible that the loss of granulocytes in these mice could be attributed to the critical role of c-myb in other stages along the granulocytic pathway. In support of this notion, in human bone marrow cells, c-myb expression is noted at the myeloblastic, promyelocytic and myelocytic stages.Citation6,Citation29 Our data provide direct evidence for c-myb in governing the granulocytic-monocytic lineage choice in adult myelogenesis and show that there is a direct role for c-myb in granulopoiesis. The significant decrease in BM and blood neutrophils and basophils, as well as the impaired colony growth of CFU-G and CFU-GM, demonstrates a critical and an inherent role for c-myb in granulocytic development. Loss of c-Myb activity skewed the development away from the granulocytic pathway and toward monocytic differentiation. Immature monocytes also express Gr-1, albeit transiently, at an intermediate levels during development, while granulocytes express Gr-1 at an even higher abundance during maturation.Citation21,Citation22 Loss of c-Myb activity by early myeloid progenitors resulted in an increase in cells expressing the CD11b surface marker together with the CD115/M-CSF monocytic receptor, but prevented further upregulation of Gr-1 in the IFN-induced mybf/f/MxCre CMPs (), supporting a direct role for c-myb in lineage choice.
In spite of the lineage bias toward monocytic development, one reason for the decrease in monocytic colonies and peripheral blood monocytes is that increased differentiation coupled with impaired proliferation caused a loss of the myeloid progenitor and monocytic precursor cells. Alternatively, the aberrant development of these committed monocytic cells could lead to their demise after the 48 h time point. Consistent with this notion of cell death, analysis of DNA isolated from the cells remaining on the IFN-induced mybf/f/MxCre LKS-, CMP and GMP methylcellulose plates after 10–12 d demonstrated only the presence of the c-myb floxed allele by PCR (data not shown). While the significant decrease in BM and blood neutrophils as well as peripheral basophils when c-Myb activity is lost could in part be due to the unfavorable lineage bias, aberrant induced differentiation may also account for the decrease in granulocytes. In support of this notion, while the expression of Gr-1 is already elevated in the control GMPs, the c-myb-deleted GMPs expressed even higher Gr-1 on the cell surface (). As indicated in , most of these cells were further downstream from the bipotent stage, further upregulation of Gr-1 marker in c-myb-deleted cells may suggest further progression in granulocytic maturation (), possibly indicating that cells which have progressed further along the granulocytic pathway may have proceeded further along that maturation pathway in the absence of c-Myb activity. Another reason for the decrease in granulocytes is that other points along the granulocytic developmental pathway, aside from what was shown here (CFU-Mix, CFU-GM and CFU-G), may depend on c-myb as is evident by high levels of c-myb expression at the myeloblastic, promyelocytic and myelocytic stages.Citation6
It has been assumed that megakaryocytic development may be c-myb-independent, because this population in the fetal livers of homozygous c-myb-null mice was not affected.Citation10 Since then, four hypomorphic mouse models seem to suggest a role for c-myb in the negative regulation of megakaryocytic development.Citation11-Citation13,Citation29 All four murine models presented with elevated blood platelets, and in the knockdown and M303V mutants, BM megakaryocytes were increased.Citation11-Citation13,Citation29 On the contrary, our c-myb-disrupted mybf/f/MxCre mice had a decreasing megakaryocytic trend in the BM (data not shown), and their blood platelets were dramatically decreased (Fig. S1 and Table S1); thus, our mouse model seems to be at odds with all the other mouse models to date. Our hematopoietic colony assay, performed on CMPs, suggests a role for c-myb in megakarypoietic development. In support of this latter notion, no hematopoietic colonies were derived from CMPs with disrupted c-myb alleles, and analysis of DNA isolated from the cells remaining on the IFN-induced mybf/f/MxCre LKS- or CMP methylcellulose plates after 10–12 d showed only the presence of the c-myb floxed allele by PCR (, data not shown). Furthermore, the size of the platelets was increased in the pIpC-treated mybf/f/MxCre mice (Fig. S1 and Table S1). Interestingly, the M303V mice were also presented with increased platelet size.Citation12 Thus, our data suggest that c-myb may have a previously unappreciated role in megakaryopoiesis.
To better understand the role of c-myb in myelogenesis, we examined the gene expression of sorted CMPs and GMPs. At the molecular level, the statistically significant downregulation of gfi-1, cebpa and c-myc transcription factors in c-myb-disrupted myeloid cells could explain the critical impairment in granulocytes.Citation35-Citation37 In addition, statistically significant inhibition of cxcr4 chemokine receptor expression could critically affect myelogenesis.Citation38 Furthermore, statistically significant downregulation of Bcl-2 could explain the absence of colony formation and could partially account for the decreases in BM and blood myeloid cells.Citation39 Our data in totality support a role for c-myb at multiple stages in the formation of mature myeloid cells: from multipotent CMP and bipotent GMP to unipotent myeloid precursor, CFU-M and CFU-G. Hence, the transcription factor c-myb has a pivotal role as a regulator of differentiation in adult myelogenesis. A better understanding of the role of c-myb in normal myelogenesis will hopefully improve further research into the role of c-myb in myeloid leukemias for therapeutic intervention.
Materials and Methods
Mice
The mybf/f/MxCre mice, which were maintained on a N6 to N10 C57BL/6 genetic background, had been described.Citation19 The mutant mice were genotyped using a three-primer PCR amplification method: mybG2e 5′- att cca gtg gtt ctt gat agc att atc -3′; mybG11e: 5′- gcc gct aag cca caa tgg aag ggc -3′; mybG19e: 5′- cct tga ctc tga gta aga aag taa ac -3′.
Flow cytometry, sorting and antibodies
Freshly isolated single cell suspensions of BM cells were prepared as previously described.Citation19 Stained cells were analyzed using the FacsAria (BD Biosciences) following staining with fluorochrome-conjugated antibodies purchased from BD Biosciences or eBiosciences. For cell sorting, lineage-positive cells were partially depleted via StemSep murine progenitor enrichment cocktail (StemCell Technologies). Then enriched progenitor cells were then stained with lineage cocktail antibodies consisting of CD3, B220, TER119, Gr-1 and CD11b as well as stem/progenitor cell markers: c-Kit, Sca-1, CD34 and CD16/32. Labeled cells were sorted and analyzed on a high-speed cell sorter (FacsAria; BD Biosciences). CMPs were defined as Lin-c-Kit+Sca-1-CD34+CD16/32- and GMPs as Lin-c-Kit+Sca-1-CD34+CD16/32+. Data were analyzed using FlowJo (TreeStar).
Colony assay on methylcellulose
At 16–18 h following ex vivo interferon [IFNα (R&D Systems);2 × 104 units per ml] treatment, ~500 sorted cells were plated onto dishes containing Methocult M3434 (StemCell Technologies), supplemented with rmGM-CSF (2 ng/ml), rhTPO (10 ng/ml) and rmIL-11 (20 ng/ml). Colonies were scored 10–12 d post-plating. In each experiment, the number of colonies for each of the four groups (untreated mybf/f, IFN-treated mybf/f, untreated mybf/f/MxCre and IFN-treated mybf/f/MxCre CMPs/GMPs) was divided by the number of colonies for the untreated mybf/f control; these numbers were then multiplied by 100%. Thus the untreated mybf/f control was set to 100%, enabling statistical comparison among the different independent experiments. The sort purities of the CMPs and GMPs were in the range of 91.1–97.5% and 85.2–93.4%, respectively.
In vivo deletion of the c-myb floxed allele and in vitro functional assay
For in vivo disruption of the c-myb floxed allele, the mybf/f /MxCre and control mice were given a 250 µl of 2 mg/ml pIpC (polyinosinic-polycytidylic acid, Sigma, P-1530) i.p. injection every other day for a total of seven to nine injections and analyzed 1 or 2 d after the last injection. pIpC was dissolved in sterile PBS by heating at 56°C for 30 min and then stored in frozen aliquot at -20°C. For injections, defrosted pIpC solution was heated at 56°C for 8 min and allowed to cool at room temperature. The sort purities of CMPs and GMPs for the three independent experiments for the functional assays were > 90% and > 95%, respectively.
For in vitro deletion, 2 × 104 units of IFNα (R&D Systems) per ml of SCF/IL3/IL6 cytokine-containing DMEM medium (DMEM, 15% heat-inactivated FBS, 10% Wehi supernatant, 50 units/mL penicillin/streptomycin, 2 mM L-glutamine, 100 mg/mL mSCF, 10 ng/mL mIL-3, 10 ng/mL hIL-6) was used. For cell culture, regardless of the purified cell number, a minimum of 200 µl medium was used with a maximum concentration of 1 × 106 cells/ml.
At the indicated time following IFNα treatment, purified cells were pulsed with BrdU (BD Biosciences) in fresh cytokine medium for 2 h at 37°C in a tissue culture incubator under humidified conditions with 5% CO2. Then, the indicated fluorochrome inhibitor of caspases for detecting poly-caspase activity (Invitrogen, V35117 FLICA kit) was added, and cells were returned to the incubator for an additional 1 h. After the incubation, purified cells were washed, stained with surface antibodies and then fixed and permeabilized for anti-BrdU staining as specified by the manufacturer (BD Biosciences).
Peripheral blood count analysis
Blood samples collected from the heart were drawn into EDTA tubes and analyzed within 2 h using a Hemavet 950 FS hematology counter, equipped with a mouse-specific software (Drew Scientific).
Microarray and RT-PCR analysis
Agilent gene array analysis of purified lineage-c-Kit+Sca-1- (LKS-) cells was performed at the DNA Microarray Core Facility at the Fox Chase Cancer Research Center. RNA isolation, RT-PCR conditions, gene amplification primers and oligonucleotide probes used were previously reported.Citation19
Statistical analysis
Data are expressed as means ± SEM. Comparisons were analyzed by using Student's two-pair or unpaired t-test (equal variance). Differences were considered significant when p < 0.05.
Additional material
Download Zip (209.9 KB)Acknowledgments
We thank Dr. Jodene K. Moore for cell sorting and FACS acquisition. We also thank Lisa V. Outterbridge for her help in maintaining the mouse colony. This work was supported by the National Institute of Health Grant 5RO1HL085279 to E.P.R.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplementary Material
Supplementary material may be found here: www.landesbioscience.com/journals/cc/article/21802/
References
- Akashi K, Traver D, Miyamoto T, Weissman IL. A clonogenic common myeloid progenitor that gives rise to all myeloid lineages. Nature 2000; 404:193 - 7; http://dx.doi.org/10.1038/35004599; PMID: 10724173
- Rosenbauer F, Tenen DG. Transcription factors in myeloid development: balancing differentiation with transformation. Nat Rev Immunol 2007; 7:105 - 17; http://dx.doi.org/10.1038/nri2024; PMID: 17259967
- Greig KT, Carotta S, Nutt SL. Critical roles for c-Myb in hematopoietic progenitor cells. Semin Immunol 2008; 20:247 - 56; http://dx.doi.org/10.1016/j.smim.2008.05.003; PMID: 18585056
- Oh IH, Reddy EP. The myb gene family in cell growth, differentiation and apoptosis. Oncogene 1999; 18:3017 - 33; http://dx.doi.org/10.1038/sj.onc.1202839; PMID: 10378697
- Sarrazin S, Sieweke MH. c-Myb as a key player in the control of myeloid cell differentiation. In Frampton J, editor Myb Transcription Factors: Their Role in Growth, Differentiation and Disease The Netherlands: Kluwer Academic Publishers 2004:133-44.
- Bjerregaard MD, Jurlander J, Klausen P, Borregaard N, Cowland JB. The in vivo profile of transcription factors during neutrophil differentiation in human bone marrow. Blood 2003; 101:4322 - 32; http://dx.doi.org/10.1182/blood-2002-03-0835; PMID: 12560239
- Gewirtz AM, Calabretta B. A c-myb antisense oligodeoxynucleotide inhibits normal human hematopoiesis in vitro. Science 1988; 242:1303 - 6; http://dx.doi.org/10.1126/science.2461588; PMID: 2461588
- Burgess TL, Fisher EF, Ross SL, Bready JV, Qian YX, Bayewitch LA, et al. The antiproliferative activity of c-myb and c-myc antisense oligonucleotides in smooth muscle cells is caused by a nonantisense mechanism. Proc Natl Acad Sci USA 1995; 92:4051 - 5; http://dx.doi.org/10.1073/pnas.92.9.4051; PMID: 7732029
- Lee SL, Wang Y, Milbrandt J. Unimpaired macrophage differentiation and activation in mice lacking the zinc finger transplantation factor NGFI-A (EGR1). Mol Cell Biol 1996; 16:4566 - 72; PMID: 8754857
- Mucenski ML, McLain K, Kier AB, Swerdlow SH, Schreiner CM, Miller TA, et al. A functional c-myb gene is required for normal murine fetal hepatic hematopoiesis. Cell 1991; 65:677 - 89; http://dx.doi.org/10.1016/0092-8674(91)90099-K; PMID: 1709592
- Carpinelli MR, Hilton DJ, Metcalf D, Antonchuk JL, Hyland CD, Mifsud SL, et al. Suppressor screen in Mpl-/- mice: c-Myb mutation causes supraphysiological production of platelets in the absence of thrombopoietin signaling. Proc Natl Acad Sci USA 2004; 101:6553 - 8; http://dx.doi.org/10.1073/pnas.0401496101; PMID: 15071178
- Sandberg ML, Sutton SE, Pletcher MT, Wiltshire T, Tarantino LM, Hogenesch JB, et al. c-Myb and p300 regulate hematopoietic stem cell proliferation and differentiation. Dev Cell 2005; 8:153 - 66; http://dx.doi.org/10.1016/j.devcel.2004.12.015; PMID: 15691758
- García P, Clarke M, Vegiopoulos A, Berlanga O, Camelo A, Lorvellec M, et al. Reduced c-Myb activity compromises HSCs and leads to a myeloproliferation with a novel stem cell basis. EMBO J 2009; 28:1492 - 504; http://dx.doi.org/10.1038/emboj.2009.97; PMID: 19360001
- Slamon DJ, Boone TC, Murdock DC, Keith DE, Press MF, Larson RA, et al. Studies of the human c-myb gene and its product in human acute leukemias. Science 1986; 233:347 - 51; http://dx.doi.org/10.1126/science.3014652; PMID: 3014652
- Clappier E, Cuccuini W, Kalota A, Crinquette A, Cayuela JM, Dik WA, et al. The C-MYB locus is involved in chromosomal translocation and genomic duplications in human T-cell acute leukemia (T-ALL), the translocation defining a new T-ALL subtype in very young children. Blood 2007; 110:1251 - 61; http://dx.doi.org/10.1182/blood-2006-12-064683; PMID: 17452517
- Lahortiga I, De Keersmaecker K, Van Vlierberghe P, Graux C, Cauwelier B, Lambert F, et al. Duplication of the MYB oncogene in T cell acute lymphoblastic leukemia. Nat Genet 2007; 39:593 - 5; http://dx.doi.org/10.1038/ng2025; PMID: 17435759
- O’Neil J, Tchinda J, Gutierrez A, Moreau L, Maser RS, Wong KK, et al. Alu elements mediate MYB gene tandem duplication in human T-ALL. J Exp Med 2007; 204:3059 - 66; http://dx.doi.org/10.1084/jem.20071637; PMID: 18070937
- Murati A, Gervais C, Carbuccia N, Finetti P, Cervera N, Adélaïde J, et al. Groupe Francophone de Cytogénétique Hématologique. Genome profiling of acute myelomonocytic leukemia: alteration of the MYB locus in MYST3-linked cases. Leukemia 2009; 23:85 - 94; http://dx.doi.org/10.1038/leu.2008.257; PMID: 18818702
- Lieu YK, Reddy EP. Conditional c-myb knockout in adult hematopoietic stem cells leads to loss of self-renewal due to impaired proliferation and accelerated differentiation. Proc Natl Acad Sci USA 2009; 106:21689 - 94; http://dx.doi.org/10.1073/pnas.0907623106; PMID: 19955420
- Lübbert M, Herrmann F, Koeffler HP. Expression and regulation of myeloid-specific genes in normal and leukemic myeloid cells. Blood 1991; 77:909 - 24; PMID: 1847312
- Hestdal K, Ruscetti FW, Ihle JN, Jacobsen SE, Dubois CM, Kopp WC, et al. Characterization and regulation of RB6-8C5 antigen expression on murine bone marrow cells. J Immunol 1991; 147:22 - 8; PMID: 1711076
- Lagasse E, Weissman IL. Flow cytometric identification of murine neutrophils and monocytes. J Immunol Methods 1996; 197:139 - 50; http://dx.doi.org/10.1016/0022-1759(96)00138-X; PMID: 8890901
- Liu Q, VanHoy RW, Zhou JH, Dantzer R, Freund GG, Kelley KW. Elevated cyclin E levels, inactive retinoblastoma protein, and suppression of the p27(KIP1) inhibitor characterize early development of promyeloid cells into macrophages. Mol Cell Biol 1999; 19:6229 - 39; PMID: 10454569
- Britos-Bray M, Friedman AD. Core binding factor cannot synergistically activate the myeloperoxidase proximal enhancer in immature myeloid cells without c-Myb. Mol Cell Biol 1997; 17:5127 - 35; PMID: 9271390
- Oelgeschläger M, Nuchprayoon I, Lüscher B, Friedman AD. C/EBP, c-Myb, and PU.1 cooperate to regulate the neutrophil elastase promoter. Mol Cell Biol 1996; 16:4717 - 25; PMID: 8756629
- Lutz PG, Houzel-Charavel A, Moog-Lutz C, Cayre YE. Myeloblastin is an Myb target gene: mechanisms of regulation in myeloid leukemia cells growth-arrested by retinoic acid. Blood 2001; 97:2449 - 56; http://dx.doi.org/10.1182/blood.V97.8.2449; PMID: 11290610
- Rushton JJ, Davis LM, Lei W, Mo X, Leutz A, Ness SA. Distinct changes in gene expression induced by A-Myb, B-Myb and c-Myb proteins. Oncogene 2003; 22:308 - 13; http://dx.doi.org/10.1038/sj.onc.1206131; PMID: 12527900
- Wolff L, Schmidt M, Koller R, Haviernik P, Watson R, Bies J, et al. Three genes with different functions in transformation are regulated by c-Myb in myeloid cells. Blood Cells Mol Dis 2001; 27:483 - 8; http://dx.doi.org/10.1006/bcmd.2001.0409; PMID: 11259171
- Emambokus N, Vegiopoulos A, Harman B, Jenkinson E, Anderson G, Frampton J. Progression through key stages of haemopoiesis is dependent on distinct threshold levels of c-Myb. EMBO J 2003; 22:4478 - 88; http://dx.doi.org/10.1093/emboj/cdg434; PMID: 12941699
- Ferrari S, Donelli A, Manfredini R, Sarti M, Roncaglia R, Tagliafico E, et al. Differential effects of c-myb and c-fes antisense oligodeoxynucleotides on granulocytic differentiation of human myeloid leukemia HL60 cells. Cell Growth Differ 1990; 1:543 - 8; PMID: 2088479
- Ness SA, Marknell A, Graf T. The v-myb oncogene product binds to and activates the promyelocyte-specific mim-1 gene. Cell 1989; 59:1115 - 25; http://dx.doi.org/10.1016/0092-8674(89)90767-8; PMID: 2688896
- Beug H, Leutz A, Kahn P, Graf T. Ts mutants of E26 leukemia virus allow transformed myeloblasts, but not erythroblasts or fibroblasts, to differentiate at the nonpermissive temperature. Cell 1984; 39:579 - 88; http://dx.doi.org/10.1016/0092-8674(84)90465-3; PMID: 6096011
- Beug H, Blundell PA, Graf T. Reversibility of differentiation and proliferative capacity in avian myelomonocytic cells transformed by tsE26 leukemia virus. Genes Dev 1987; 1:277 - 86; http://dx.doi.org/10.1101/gad.1.3.277; PMID: 2824281
- Burk O, Klempnauer KH. Estrogen-dependent alterations in differentiation state of myeloid cells caused by a v-myb/estrogen receptor fusion protein. EMBO J 1991; 10:3713 - 9; PMID: 1718743
- Hock H, Hamblen MJ, Rooke HM, Traver D, Bronson RT, Cameron S, et al. Intrinsic requirement for zinc finger transcription factor Gfi-1 in neutrophil differentiation. Immunity 2003; 18:109 - 20; http://dx.doi.org/10.1016/S1074-7613(02)00501-0; PMID: 12530980
- Zhang DE, Zhang P, Wang ND, Hetherington CJ, Darlington GJ, Tenen DG. Absence of granulocyte colony-stimulating factor signaling and neutrophil development in CCAAT enhancer binding protein alpha-deficient mice. Proc Natl Acad Sci USA 1997; 94:569 - 74; http://dx.doi.org/10.1073/pnas.94.2.569; PMID: 9012825
- Hoffman B, Amanullah A, Shafarenko M, Liebermann DA. The proto-oncogene c-myc in hematopoietic development and leukemogenesis. Oncogene 2002; 21:3414 - 21; http://dx.doi.org/10.1038/sj.onc.1205400; PMID: 12032779
- Ma Q, Jones D, Borghesani PR, Segal RA, Nagasawa T, Kishimoto T, et al. Impaired B-lymphopoiesis, myelopoiesis, and derailed cerebellar neuron migration in CXCR4- and SDF-1-deficient mice. Proc Natl Acad Sci USA 1998; 95:9448 - 53; http://dx.doi.org/10.1073/pnas.95.16.9448; PMID: 9689100
- Frampton J, Ramqvist T, Graf T. v-Myb of E26 leukemia virus up-regulates bcl-2 and suppresses apoptosis in myeloid cells. Genes Dev 1996; 10:2720 - 31; http://dx.doi.org/10.1101/gad.10.21.2720; PMID: 8946913