Abstract
Origin recognition complex (ORC) is highly dynamic, with several ORC subunits getting posttranslationally modified by phosphorylation or ubiquitination in a cell cycle-dependent manner. We have previously demonstrated that a WD repeat containing protein ORC-associated (ORCA/LRWD1) stabilizes the ORC on chromatin and facilitates pre-RC assembly. Further, ORCA levels are cell cycle-regulated, with highest levels during G1, and progressively decreasing during S phase, but the mechanism remains to be elucidated. We now demonstrate that ORCA is polyubiquitinated in vivo, with elevated ubiquitination observed at the G1/S boundary. ORCA utilizes lysine-48 (K48) ubiquitin linkage, suggesting that ORCA ubiquitination mediates its regulated degradation. Ubiquitinated ORCA is re-localized in the form of nuclear aggregates and is predominantly associated with chromatin. We demonstrate that ORCA associates with the E3 ubiquitin ligase Cul4A-Ddb1. ORCA is ubiquitinated at the WD40 repeat domain, a region that is also recognized by Orc2. Furthermore, Orc2 associates only with the non-ubiquitinated form of ORCA, and Orc2 depletion results in the proteasome-mediated destabilization of ORCA. Based on the results, we suggest that Orc2 protects ORCA from ubiquitin-mediated degradation in vivo.
Introduction
Origin recognition complex proteins initiate DNA replication by enabling the establishment of the pre-replicative complex (pre-RC) at the origins of DNA replication. This is then followed by the activation of MCM2-7 complex to initiate origin firing and replication.Citation1-Citation4 DNA replication must occur once and only once in each cell cycle, and the components of pre-RC coordinate this process. This is accomplished by ensuring that the pre-RC components are inactivated during S-phase, and their reassembly on the chromatin is prevented until the end of mitosis.Citation5-Citation7 Thus, the regulation of replication licensing factors during the cell cycle is crucial to prevent re-replication.
Studies in different model organisms have shown that ORC is highly dynamic, with several ORC subunits getting posttranslationally modified by phosphorylation or ubiquitination in a cell cycle-dependent manner.Citation5-Citation7 In S. cerevisiae, ORC is associated with the chromatin throughout the cell cycle, although phosphorylation regulates ORC activity.Citation8-Citation10 In S. pombe, ORC association to chromatin increases during mitosis and peaks at M/G1.Citation11 In Xenopus, ORC is not associated with chromatin during S-phase and is also absent from the mitotic chromatin.Citation12-Citation15 In Drosophila, Orc1 is degraded at the end of M-phase by the Fzr/Cdh1-activated anaphase-promoting complex.Citation16 In human cells, however, Orc1 dissociates from chromatin, is ubiquitinated and then degraded during the G1-S phase transition, and is reloaded at the M-G1 transition when new pre-RCs are formed.Citation17-Citation20 Human Orc2 and Orc3 associate predominantly with centric and pericentromeric heterochromatin during the end of S phase, G2 and mitosis.Citation21 A key molecule regulating replication licensing is Cdt1, which accumulates during M and G1, and is degraded during S phase.Citation22,Citation23 In addition, Geminin, a protein expressed in cells from S phase through early mitosis, controls the levels of Cdt1.Citation24-Citation27 Interestingly, Geminin is degraded during mitosis, thus restricting origin licensing to the end of mitosis and beginning of G1, when CDK/DDK activity is kept low.Citation28 We have previously demonstrated that ORCA modulates ORC loading, shows cell cycle dynamics similar to that of human ORC with highest levels observed during G1 and then sequential release from most sites, except at heterochromatic sites during the post-G1 part of the cell cycle.Citation29
Ubiquitination modulates protein function in multiple ways, including signaling a protein for destruction by proteasomes, organelle biogenesis, chromatin modification, DNA repair function and signal transduction in immune system.Citation30-Citation32 Ubiquitin is transferred to the lysine residues of the target protein by the action of E1-activating, E2-conjugating and E3 ubiquitin-ligase enzymes.Citation33-Citation37 The type of E3 ligase in a particular conjugation complex defines the specificity for substrate recognition. The Cullin-RING-dependent E3 ligases (CRLs), as the major E3 ligase group, play essential roles in directing several pre-RC components for destruction.Citation34-Citation37 During G1/S transition, the SCF E3 ligase complex (Skp1-Cul1-F-box protein) degrades Orc1.Citation20 During S phase, Cdt1 undergoes degradation by SCFSkp2-mediated proteolysis through CDK phosphorylationCitation38-Citation41 as well as replication-coupled proteolysis by Cul4-Ddb1Cdt2 ubiquitin ligase and PCNA.Citation42-Citation46 Upon mitosis, Geminin is degraded by anaphase-promoting complex, another E3 CRL ligase that recognizes its target through the destruction box (D-box) sequence.Citation24 Typically, the polyubiquitinated chain of a protein is recognized by the proteasome, which ultimately degrades the tagged protein.Citation47 Ubiquitin-binding proteins comprise an ubiquitin-binding domain that can interact with an ubiquitin moiety or an ubiquitinated protein. Ubiquitin-binding proteins might also be regulated by ubiquitination.Citation48 Recent evidence has shown that a large subset of WD repeat β-propellers are a novel class of ubiquitin-binding domains, and their binding to ubiquitin is important for their function in vivo.Citation49 We investigated if the significant reduction of ORCA levels at the end of G1 are mediated by posttranslational modifications, primarily through ubiquitination-mediated degradation.
We demonstrate ORCA is polyubiquitinated in human cells, with an elevated ubiquitination at the G1/S boundary, in agreement with the reduction of ORCA levels at the end of G1. The ubiquitinated ORCA utilizes K48 linkage, consistent with the role of ubiquitin-mediated proteolysis of ORCA. Ubiquitination of ORCA occurs at the WD repeats, a domain required for chromatin association and ORC binding. Ubiquitinated ORCA is chromatin-bound and showed negligible turnover. Further, depletion of Orc2 in human cells causes destabilization of cellular ORCA. However, the presence of a proteasome inhibitor MG132 in Orc2-depleted cells prevents the degradation of ORCA, but not Orc3. Furthermore, Orc2 associates only with non-ubiquitinated form of ORCA, demonstrating that Orc2 association with ORCA prevents its ubiquitin-dependent proteolytic degradation. Finally, we demonstrate that ORCA associates with the E3 ubiquitin ligase Cul4A-Ddb1. We speculate that once ORCA accomplishes the loading of ORC, it dissociates from ORC, gets ubiquitinated, resulting in the reduced levels of ORCA at the G1/S boundary. We suggest that the reduction in ORCA levels in post-G1 cells may be another mechanism cells utilize to ensure that the replication occurs once and only once during each cell division cycle.
Results
ORCA is polyubiquitinated
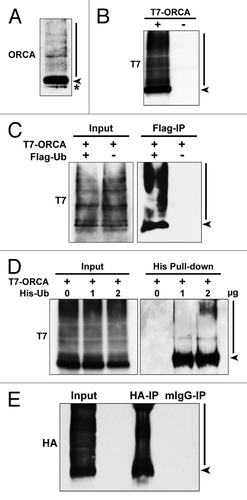
Immunoblot analysis on U2OS whole-cell lysate using ORCA antibody that was exposed for longer duration showed a smear in the high molecular weight region, suggesting the prevalence of posttranslational modification, predominantly ubiquitination, of ORCA (molecular weight of ORCA ~70kDa, ) at the endogenous level. Interestingly, epitope-tagged ORCA (T7 as well as YFP-tagged) also showed this modification, suggesting that this is a posttranslational event ( and data not shown). Further, co-transfection of T7-ORCA and Flag-Ub followed by Flag immunoprecipitation clearly pointed to the fact that a fraction of ORCA is polyubiquitinated (). Similarly, T7-ORCA was co-transfected with varying concentrations of 6 × His-Ub in U2OS cells. His-tag pull-down confirmed the ubiquitination of ORCA (). HA immunoprecipitation of U2OS whole-cell lysates co-expressing HA-ORCA and Flag-Ub under denaturing conditions further confirmed ubiquitination of ORCA ().
Figure 1. ORCA is polyubiquitinated in vivo. (A) Immunoblot analysis of U2OS whole-cell lysates using antibodies against ORCA. The arrowhead indicates unmodified ORCA, and the bar indicates the high molecular weight smear. The asterisk denotes the cross-reacting band. (B) Immunoblot analysis of the whole-cell lysates of U2OS cells transfected with T7-tagged ORCA (+) or control vector (-) using antibodies against T7. Note the high molecular weight smear in (+). (C) Flag immunoprecipitation of U2OS cells co-expressing T7-ORCA and Flag-Ub. Note the polyubiquitinated T7-ORCA as denoted by the high molecular weight smear. Cells only transfected with T7-ORCA were used as the immunoprecipitation control. Note that the arrowhead likely denotes the ORCA that has at least one ubiquitin attached, since it was immunoprecipitated-using FLAG in cells expressing FLAG-Ub. (D) Affinity pull-down in cells expressing T7-ORCA and varying amount of 6 × His-Ub (0, 1, 2 µg). Polyubiquitinated T7-ORCA was pulled down by His tag. (E) HA immunoprecipitation of U2OS cells co-expressing HA-ORCA and Flag-Ub under the denaturing condition (see Materials and Methods). Mouse IgG immunoprecipitation serves as the control.
ORCA ubiquitination is elevated at the G1/S boundary
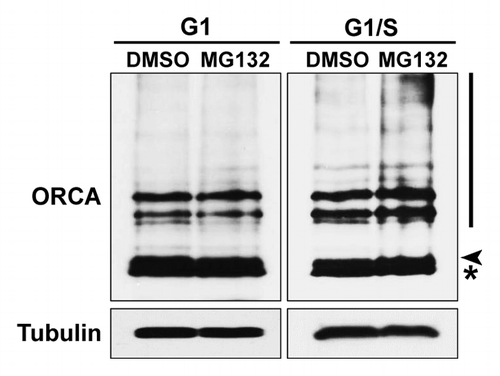
We have previously demonstrated that the total levels, as well as chromatin-associated levels, of ORCA are cell cycle-regulated, with maximum levels observed at G1, decreasing during S-phase and being retained predominantly at the heterochromatic foci during most of S-phase, G2 and mitosis.Citation29 In order to test whether endogenous ORCA levels are regulated by ubiquitin-mediated degradation, preferentially at the G1/S boundary, U2OS cells synchronized at G1 or G1/S were treated with MG132 for 6 h to block the ubiquitin-mediated degradation. Whole-cell lysates from these cells were analyzed by immunoblots using antibodies against ORCA. Ubiquitination of ORCA was low in G1, as there was no dramatic accumulation of the ubiquitinated form of ORCA (high molecular weight smear) upon MG132 treatment compared with the DMSO control (). In contrast, ORCA displayed increased accumulation of the ubiquitinated form upon MG132 treatment at the G1/S boundary, indicating elevated ORCA ubiquitination at the end of G1 ().
Figure 2. Ubiquitination of ORCA is elevated at the G1/S boundary. U2OS cells were synchronized at G1 or G1/S boundary and treated with DMSO or MG132 for 6 h. Whole-cell lysates were analyzed by immunoblots using antibodies against ORCA. Note the accumulation of the high molecular weight smear in lane four (the MG132-treated sample compared with DMSO at the G1/S boundary). Tubulin serves as the loading control. The arrowhead represents unmodified ORCA and the asterisk represents the cross-reacting band.
Ubiquitination of ORCA occurs on the WD repeat-containing domain
The WD domain of ORCA mediates its chromatin association as well as its ORC binding.Citation29 To map the domain that is responsible for ubiquitin-binding as well as to identify the ubiquitination sites on ORCA, we generated several T7-tagged truncation mutants of ORCA-expressing leucine rich repeat (LRR, 1–127aa), LRR + linker (1–270aa), WD alone (270–647aa) and WD + linker (128–647aa) (). Immunoprecipitations were performed using T7 antibody, and the samples were analyzed by immunoblotting. Only the WD-containing ORCA fragments (128–647aa, 270–647aa and full-length) showed prominent high molecular weight smear characteristic of ubiquitination (), which is consistent with recent observations that WD-repeat propellers serve as ubiquitin-binding domains.Citation50 The WD domain of ORCA is not only critical for mediating the association of ORCA to chromatin and pre-RC components, but also for modulating its ubiquitin association.
Figure 3. ORCA ubiquitination occurs on multiple lysine residues in the WD domain. (A) Schematic representation of various truncation mutants of ORCA (adapted and modified from ref. Citation29). T7 epitope was constructed at the N terminus. (B) T7 immunoprecipitation of the ORCA mutants followed by T7 immunoblot analysis. Note that only the WD-containing mutants and the full-length protein display the high molecular weight smear. (C) Schematic representation of all the 14 lysine residues within the WD domain that were mutated into arginine residues. Individual point mutations, several combined mutations and the mutants having all the lysines mutated (T7-ORCA.K-R or T7-ORCA.270–647.K-R) were tested for ubiquitination. (D) Immunoblot analysis of cell extracts expressing T7-vector control, T7-ORCA and T7-ORCA.K-R using T7 antibody. (E) Immunoblot analysis of cell extracts expressing T7-vector control, T7-ORCA.270–647 and T7-ORCA.270–647.K-R using T7 antibody.
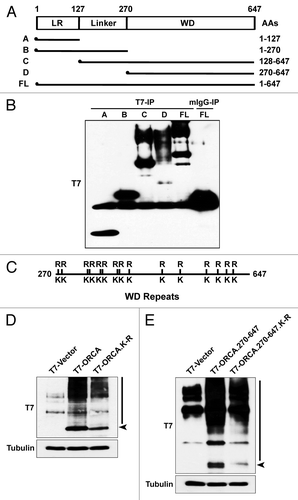
Further, to define the lysine residues in the WD domain of ORCA that are linked to ubiquitin, we generated single point mutations as well as multiple point mutations spanning the entire WD domain of ORCA. Fourteen lysine residues (K) that were present within the WD region (270–647aa) were mutated to arginine residues (R) in various permutations and combinations. Cells were transfected with T7-ORCA, T7-ORCA.270–647 or the mutants T7-ORCA.K-R, T7-ORCA.270–647.K-R, where all of the lysine residues were substituted by the arginine residues within the WD domain (). While T7-ORCA as well as T7-ORCA.270–647 fragments showed robust ubiquitination () none of the single point mutations were able to abolish the ubiquitination (data not shown). The mutants T7-ORCA.K-R, as well as T7-ORCA.270–647.K-R, where all of the lysines were substituted by arginine, showed marginally reduced ubiquitination (), but not complete loss of ubiquitination. It is possible that non-lysine residues are utilized for ubiquitin linkage, as has been observed in case of mammalian substrates for endoplasmic reticulum-associated degradation.Citation51
K48 ubiquitin linkage on ORCA targets it for proteasome-mediated degradation
We have demonstrated that ORCA gets polyubiquitinated. The most abundant polyubiquitin chain has been demonstrated to be the lysine-48 (K48) linkage, which signals the target protein for degradation via the proteasome pathway.Citation52 The other common linkage is lysine-63 (K63), which plays a role in mediating various cellular functions, including DNA repair, signal transduction, transcription and endosomal trafficking.Citation53-Citation56 In order to test the ubiquitin linkage on ORCA, we conducted immunoblot analysis using either K48-specific or K63-specific ubiquitin antibodies. T7-ORCA-expressing cells showed increased K48 linkage compared with the control (, short, non-saturated exposure; and , long, saturated exposure of an independent experiment) and exhibited further accumulation upon MG132 treatment (), consistent with ORCA being targeted for ubiquitin-mediated proteasomal degradation. Immunoprecipitation of ORCA using HA antibodies in cells co-transfected with Flag-Ub and HA-ORCA, followed by immunoblot analysis with K48 antibody () or K63 antibody (), clearly demonstrated K48 linkage in ORCA. These results demonstrate that ORCA poly-ubiquitination is primarily K48, suggesting that ORCA ubiquitination is required for its degradation via the proteasomal machinery. Though the protein levels of ORCA are cell cycle-regulated, its transcript levels did not show any significant variation during different stages of the cell cycle as detected by quantitative real time PCR (data not shown).
Figure 4. ORCA polyubiquitin chain is formed through lysine-48 of the ubiquitin. (A) Immunoblot analysis of the whole-cell lysates of U2OS cells transfected with T7 control vector or T7-tagged ORCA using antibodies against K48 ubiquitin. Short, non-saturated exposure shows the high molecular weight smear in T7-ORCA-expressing cells. (B) Immunoblot analysis of the whole-cell lysates of U2OS cells transfected with T7 control vector or T7-tagged ORCA [with (+) or without (-) MG132 treatment] using antibodies against K48 ubiquitin. Note the accumulation of the high molecular weight smear upon MG132 treatment. Long, saturated exposure shows the presence of K48 ubiquitin in all the samples (including controls). (C and D) HA immunoprecipitations in U2OS cells expressing HA-ORCA with (+) or without (-) Flag-Ub. Immunoblot analysis using K48-specific (C) or K63-specific (D) antibodies. Note the polyubiquitinated HA-ORCA is in the form of K-48 linkage.
![Figure 4. ORCA polyubiquitin chain is formed through lysine-48 of the ubiquitin. (A) Immunoblot analysis of the whole-cell lysates of U2OS cells transfected with T7 control vector or T7-tagged ORCA using antibodies against K48 ubiquitin. Short, non-saturated exposure shows the high molecular weight smear in T7-ORCA-expressing cells. (B) Immunoblot analysis of the whole-cell lysates of U2OS cells transfected with T7 control vector or T7-tagged ORCA [with (+) or without (-) MG132 treatment] using antibodies against K48 ubiquitin. Note the accumulation of the high molecular weight smear upon MG132 treatment. Long, saturated exposure shows the presence of K48 ubiquitin in all the samples (including controls). (C and D) HA immunoprecipitations in U2OS cells expressing HA-ORCA with (+) or without (-) Flag-Ub. Immunoblot analysis using K48-specific (C) or K63-specific (D) antibodies. Note the polyubiquitinated HA-ORCA is in the form of K-48 linkage.](/cms/asset/3347c4b8-2560-4f63-b054-48a39a28113c/kccy_a_10921870_f0004.gif)
Orc2 protects ORCA from ubiquitin-mediated proteolysis
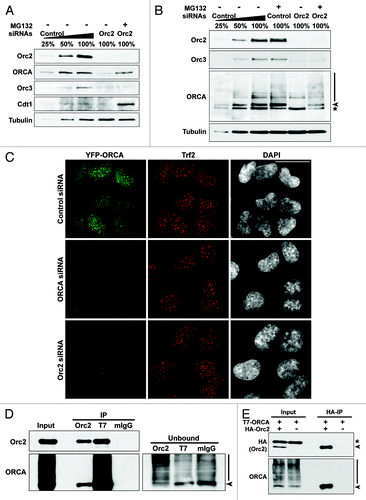
We have recently demonstrated that ORCA is required for ORC binding to chromatin.Citation29 Similarly, Orc2 is required for the cellular stability of ORCACitation57,Citation58 and Orc3 ().Citation21 We have also performed depletion of ORCA as well as Orc2 in YFP-ORCA-expressing stable cell line. YFP-ORCA labeling was significantly decreased in Orc2-depleted cells, corroborating our immunoblot analysis that ORCA levels are destabilized upon Orc2 depletion (). ORCA and ORC associate with heterochromatic structures including at telomeres and centromeres.Citation29 Immunofluorescence analysis using Trf2 antibody in YFP-ORCA-expressing cells showed no significant changes in Trf2 labeling at telomeres in ORCA siRNA and Orc2 siRNA-treated cells (). Next, we addressed if the ORCA destabilization was due to ubiquitin-mediated degradation of ORCA. The presence of a proteasome inhibitor MG132 in Orc2-depleted cells partially rescued ORCA, but not Orc3 levels (, U2OS osteosarcoma cells; and , WI38 primary diploid fibroblasts). MG132-treated control cells also showed further stabilization and increased accumulation of ubiquitinated forms of ORCA (). Polyubiquitinated form of ORCA was apparent in control and MG132-treated control and Orc2-siRNA treated cells. Stabilization of Cdt1 upon MG132 is shown as the positive control. It is interesting to note that Orc2 associates with the WD domain of ORCA, the domain that shows ubiquitination. Based on these data, we hypothesize that Orc2, by interacting with ORCA, prevents the proteasome-mediated degradation of ORCA, possibly by preventing ubiquitin linkage within the WD domain. This would suggest that Orc2 associates with the non-ubiquitinated form of ORCA. To address this, we performed immunoprecipitation of Orc2 in T7-ORCA-expressing cells and examined if Orc2 can associate with ubiquitinated as well as non-ubiquitinated forms of ORCA. It was evident that Orc2 associated only with the non-ubiquitinated ORCA, and the ubiquitinated ORCA was not found to be associated with Orc2 (). Furthermore, in the unbound fraction (following the Orc2 immunoprecipitation), the non-ubiquitinated ORCA (arrowhead) was depleted, whereas the ubiquitinated form of ORCA was enriched, suggesting that Orc2 associated predominantly with the non-ubiquitinated form of ORCA. Immunoprecipitation using T7 antibody in T7-ORCA-expressing cells clearly showed the presence of ubiquitinated ORCA (). Similar results were obtained when HA-Orc2 and T7-ORCA were co-expressed and immunoprecipitation using HA antibody was performed. Orc2 associated only with non-ubiquitinated form of ORCA but not with ubiquitinated ORCA, demonstrating that Orc2 binding to ORCA prevents ubiquitination of ORCA ().
Figure 5. Orc2 prevents ORCA from proteasome-mediated degradation. (A) Immunoblot analysis of whole-cell extracts from cells treated with Orc2 siRNAs in the absence (-) or presence (+) of proteasomal inhibitor MG132. Varying concentrations (25%, 50% and 100%) of whole-cell extracts treated with luciferase siRNAs (control) are shown to provide information on the percentage of knockdown and reduction of protein levels. Note the destabilization of ORCA and Orc3 in Orc2-depleted U2OS cells and the inhibition of ORCA degradation in the presence of MG132. Orc3 is destabilized upon Orc2 depletion but cannot be stabilized upon MG132 treatment, whereas Cdt1 protein levels are elevated upon MG132 treatment. (B) Depletion of Orc2 in human WI38 cells. Note the degradation of ORCA and Orc3 in Orc2-depleted cells, but stabilization of ORCA, as well as evidence of polyubiquitinated ORCA in the presence of MG132. (C) Depletion of ORCA and Orc2 in YFP-ORCA-expressing cells. Immunofluorescence analysis was performed using Trf2 antibody. The scale bar represents 30 µm. (D) Immunoprecipitation using Orc2, T7 or mouse IgG antibody in T7-ORCA-expressing cells. Note Orc2 associates with non-ubiquitinated ORCA. The arrowhead in unbound fractions denotes the absence of non-ubiquitinated form of ORCA in the Orc2 IP unbound sample. Note the enrichment of ubiquitinated forms of ORCA in the Orc2 IP unbound sample. (E) Immunoprecipitation using HA antibody in cells co-expressing HA-Orc2 and T7-ORCA.
Ubiquitinated ORCA is associated with nuclear sub-structures and bound to nuclear insoluble chromatin fraction
We have thus far demonstrated that ORCA gets polyubiquitinated in the WD domain; this polyubiquitination is elevated at the G1/S boundary and targets it for degradation, and Orc2 binding to ORCA protects it from ubiquitin-mediated proteolysis. We then examined how the localization of ORCA is affected when cells are treated with MG132, so that ORCA can be ubiquitinated but not degraded. In control cells (DMSO-treated), upon paraformaldehyde (PF) fixation, ORCA (as visualized by YFP-ORCA in a stable cell line expressing this fusion protein) showed diffuse nuclear staining (). In MG132-treated cells, ORCA showed large nuclear aggregates that were primarily located in DAPI-less regions (). In order to remove the soluble proteins, cells were detergent pre-extracted (PE) before PF fixation. In DMSO-treated interphase cells, the punctate patterns of ORCA binding to telomeres () were discernible;Citation29 however, in MG132-treated cells, ORCA continued to show nuclear aggregates in DAPI-less regions (). Immunofluorescence using antibodies detecting K48 polyubiquitin chains showed colocalization of ORCA with K48Ub in these foci (), suggesting that upon MG132 treatment, polyubiquitinated ORCA binds to nuclear sub-structures that are resistant to detergent extraction. Recent studies have demonstrated that ubiquitin and ubiquitin-like proteins are present in the nucleoli, some of which play crucial roles in the ribosome biogenesis.Citation59 The functional relevance of the nucleolar accumulation of ubiquitinated proteins remains to be understood. To address if the nuclear sub-structures decorated by ORCA in MG132-treated cells represent nucleoli, we performed immunostaining using Fibrillarin antibody. The ubiquitinated-ORCA-containing nuclear foci did not co-localize with nucleoli, but were generally localized adjacent to nucleoli ().
Figure 6. The polyubiquitinated ORCA associates with detergent-resistant chromatin fraction. (A) YFP-ORCA-expressing U2OS cells were incubated with DMSO and MG132, and were then fixed directly by paraformaldehyde (PF) or pre-extracted with detergent before fixation (PE) to remove soluble proteins. Note that YFP-ORCA localized in the form of aggregated foci upon MG132 treatment, which are resistant to detergent extraction. (B) Immunostaining of DMSO and MG132-treated YFP-ORCA-expressing cells with K48 ubiquitin antibody. (C) Immunostaining of DMSO and MG132-treated YFP-ORCA-expressing cells with Fibrillarin shows that the ORCA-containing foci do not localize to the nucleolar structures in the cells. (D) Biochemical fractionation of YFP-ORCA expressing cells treated with DMSO or MG132 followed by immunoblot analysis using GFP antibody. (S) represents cytosolic fraction, and (P) denotes chromatin-bound fraction. Note the high molecular weight smear of YFP-ORCA predominantly on the chromatin fraction (P) upon MG132 treatment. (E) Snapshots of the FRAP analyses on DMSO or MG132-treated YFP-ORCA-expressing cells. The insets denote where the photobleach was conducted. (F) Quantitations of FRAP data show a highly dynamic rate of ORCA on chromatin, with the recovery time T(1/2) = 0.30 ± 0.07s to a 0.93 ± 0.04 recovery fraction, whereas, in MG132-treated cells, no more than 20% recovery after photobleach was observed. Scale bars in (A, B, C and E) represent 10 µm.
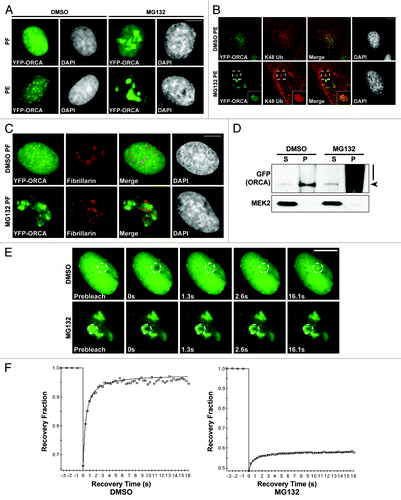
Biochemical fractionations of DMSO-treated and MG132-treated YFP-ORCA-expressing cells were performed to ascertain the association of ORCA to chromatin. ORCA was present in the pellet fraction (P), suggesting that ORCA is associated with nuclear insoluble/chromatin fraction. In MG132-treated cells, the majority of the ubiquitinated ORCA was in the pellet fraction (P), suggesting that ubiquitinated ORCA is bound to nuclear insoluble matrix/chromatin ().
We next conducted fluorescence recovery after photobleaching (FRAP) analysis to study the dynamics of ORCA and ubiquitinated ORCA in YFP-ORCA-expressing stable cells. The YFP signal of a defined region in the nucleus was irreversibly bleached, and the recovery kinetics of fluorescence intensity into the bleached region was assessed (). ORCA showed extremely fast and almost complete recovery kinetics with a T(1/2) = 0.3 ± 0.07s (mobile fraction 0.93 ± 0.04) (). In contrast, ubiquitinated ORCA (MG132-treated) failed to recover (less than 20% recovery, ), suggesting that ubiquitinated ORCA did not exchange and was largely immobile.
Cul4A-Ddb1 E3 ligase associates with ORCA
A recent study has demonstrated that Cul4-Ddb1 ligase interacts with multiple WD-containing proteins including WDR5, Cdt2 and EED, and that Cul4-Ddb1 ligase regulates histone methylation.Citation50 Several groups have also reported that WD-repeat proteins directly bind Ddb1 and serve as the substrate-recruiting module of E3.Citation60,Citation61 It has also been shown recently that WD propellers define an ubiquitin-binding domain that regulated turnover of F-box proteins.Citation49 Multiple pre-RC components are degraded by the ubiquitin-mediated degradation pathway during the G1/S boundary in order to prevent re-replication.Citation3 We tested if Cul4/Ddb1 associates with ORCA and if this, in turn, modulates ORCA ubiquitination. Immunoprecipitation using ORCA antibody in cells transiently expressing T7-ORCA and Myc-Cul4A demonstrated that ORCA interacts with Cul4A (). Similarly, co-transfection of T7-ORCA and Myc-Cul4A, followed by T7 IP, confirmed the interaction between ORCA and Cul4A (). Moreover, co-transfection of T7-ORCA with Myc-Ddb1, followed by ORCA IP, demonstrated that ORCA associates with Ddb1 as well (). Reverse IPs using Myc antibody in cells transiently expressing Myc-Cul4A or Myc-Ddb1 further confirmed the association of ORCA with this E3 ligase complex (). Finally, we mapped the domains of ORCA to which Cul4A and Ddb1 bind. Different truncation mutants of T7-ORCA () were co-transfected with Myc-Cul4A, and immunoprecipitations using T7 antibody were performed. Only the WD repeat-containing domain showed interaction with Cul4A and the endogenous Ddb1 ().
Figure 7. ORCA is associated with Cul4A-Ddb1 E3 ligase. (A) ORCA immunoprecipitation in T7-ORCA and Myc-Cul4A transfected cells followed by immunoblots with ORCA and Myc. Rabbit IgG serves as the control. (B) T7 immunoprecipitation in cells transfected with T7-ORCA and Myc-Cul4A followed by immunoblots with T7 and Myc. Note that ORCA associates with Cul4A. Mouse IgG serves as the control. (C) ORCA immunoprecipitation in cells transfected with T7-ORCA and Myc-Ddb1 followed by immunoblots with ORCA and Myc. Rabbit IgG serves as the control. (D) Myc immunoprecipitations in cells transfected with T7-ORCA and Myc-Cul4A or Myc-Ddb1 followed by immunoblots with Myc and ORCA. Mouse IgG serves as the control. (E) Immunoprecipitations in cells expressing various T7-ORCA mutants and Myc-Cul4A were performed using the T7 antibody, and Cul4A and the endogenous Ddb1 were analyzed by immunoblots. Note that only the WD-containing constructs interact with Cul4A-Ddb1. (F) Depletion of Ddb1 or in combination with Orc2 followed by immunoblot analysis of ORCA.
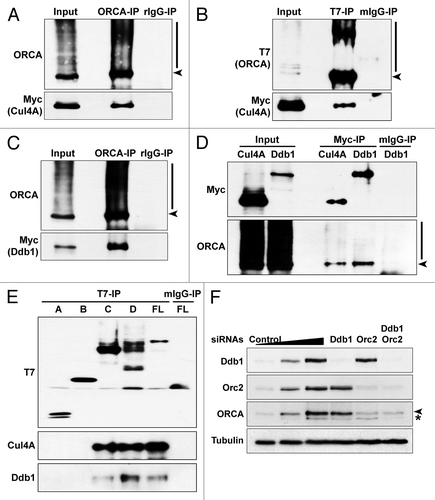
To address if Cul4A-Ddb1 mediates the ubiquitination and degradation of ORCA, we depleted Cul4A and Ddb1 from human cells as well as in combination with Orc2 ( and data not shown). We assessed if ORCA levels could be stabilized in the absence of Cul4A-Ddb1 individually or in combination with Orc2. No significant stabilization of ORCA was found under these conditions, suggesting that multiple E3 ligases may be targeting ORCA for ubiquitination (). It is equally likely that ORCA is a DCAF (Ddb1- and Cul4-associated factor) that along with Cul4A-Ddb1 provides the substrate specificity for this E3 ligase. Future work will test these possibilities.
Discussion
In eukaryotes, initiation of DNA replication requires the assembly of pre-RC in late mitosis and G1, with the sequential loading of the ORC, Cdc6, Cdt1 and MCM2–7 onto replication origins.Citation1 In order to prevent re-replication and control replication licensing, several factors, including Orc1, Cdt1 and its inhibitor Geminin, exhibit cell cycle-regulated preotein levels via ubiquitination-mediated proteasome degradation. We have shown that ORCA modulates ORC loading, binds to Cdt1 and Geminin, shows maximum levels during G1 and gets sequentially released from most sites, presumably by ubiquitination-mediated degradation, except at heterochromatic sites during the post-G1 part of the cell cycle.Citation29 ORCA binds to Cdt1 and Geminin, and the timely association of ORCA with ORC, Cdt1 and Geminin and the binding of each of these components to chromatin dictated by ORCA ensures “once per cell cycle” replication.Citation58
In the present study, we demonstrate that ORCA is polyubiquitinated in vivo with elevated ubiquitination and degradation at the G1/S boundary and is stabilized when cells are treated with the proteasome inhibitor MG132. Orc2, the direct binding partner of ORCA, stabilizes ORCA by preventing its proteasomal degradation. The WD domain of ORCA is responsible for ORC binding as well as chromatin association.Citation29 The WD repeats are organized to form a β-propeller fold of 6–8 blades, that together form an interaction scaffold.Citation62-Citation65 Recent work has shown that WD repeat propellers are a novel class of ubiquitin-binding domains.Citation49 We have observed that ORCA gets ubiquitinated at its WD repeat-containing domain. Interestingly, the WD repeat domain is also required for the association of ORCA to Cdt1 and Geminin, as well as to the E3 ligase Cul4A-Ddb1. Smaller truncation mutants of the ORCA WD domain have failed to provide any useful information, since the entire β-propeller structure seems to be required for association of ORCA to ORC as well as to chromatin. Determination of the crystal structure of ORCA is critical to understanding how ORCA can interact with several different proteins, as well as to chromatin utilizing its WD domain. However, one prediction would be that binding of Orc2 to ORCA prevents ubiquitin linkage formation on the WD domain. It is also equally likely that binding of Orc2 causes steric hindrance and does not allow the association of E3 ligases like Cul4A-Ddb1, thereby stabilizing ORCA. Future structure-function analyses would answer these questions.
We have observed that the polyubiquitination of ORCA occurred on chromatin, as ORCA was resistant to detergent extraction (). It has been suggested that the ubiquitn-proteasome machinery is active in nuclear domains, and this, in turn, governs crucial cellular processes including DNA repair and gene regulation.Citation66 Fibrillarin immunostaining showed ubiquitinated ORCA localized adjacent to nucleoli; however, the functional relevance of this localization remains unclear.
The chromatin-mediated polyubiquitination of ORCA is similar to what has been reported for mammalian Orc1, Xenopus Cdt1 and Xenopus p27.Citation20,Citation67,Citation68 It has been proposed that Orc1 polyubiquitination acts as a dual-switch, whereby ubiquitination first serves as a signal to initiate DNA replication, following which the protein is targeted for degradation.Citation20 Interestingly, Cdt1 is also ubiquitinated on chromatin and requires DNA replication to be initiated.Citation67 Just like the pre-RC components Orc1 and Cdt1, ORCA shows cell cycle regulation with maximum levels at G1 and a significant reduction during G1/S boundary. It is possible that Orc2, by interacting with ORCA, prevents the proteasome-mediated degradation of ORCA, presumably at origins during G1. Loss of Orc2 from origins and retention only at heterochromatic structures in post-G1 cells suggest that ORCA may be degraded from origins in post-G1 cells by ubiquitin-mediated proteolysis, which is consistent with the elevated ORCA ubiquitination at the G1/S boundary. ORCA accomplishes the loading of ORC during G1. At the end of G1, Orc2 is released from origins; the residual ORCA without Orc2 (especially from origins) during the G1/S boundary is degraded by ubiquitination. This suggests that when signals dictate ORC to come off the chromatin or undergo destruction, cells also instruct ORCA to undergo rapid degradation to prevent ORC from being re-stabilized on the chromatin. This could be yet another mechanism ensuring that replication occurs only once during each cell division cycle. We have previously demonstrated that Orc2 is highly dynamic on chromatin,Citation69 and, therefore, the prediction is that every time Orc2 is released from chromatin, ORCA is targeted for degradation.
Our results demonstrate that ORCA interacts with Cul4A-Ddb1, and its proteolysis could be mediated by the Cul4A-Ddb1 pathway. Cul4 associates with Ddb1 to form an E3 ubiquitin ligase that constitutes a large family of the Cullin-RING ligases (CRLs) and targets substrate proteins for proteasome-mediated degradation.Citation70 The Cul4-Ddb1 is involved in various cellular events, including DNA replication and repair,Citation45,Citation46,Citation70-Citation78 histone methylationCitation50 and transcription.Citation79 It has also been shown that Cul4-Ddb1 interacts with WD-containing proteins including WDR5, EED and RBBP5, and associates with methylated H3. Further, inactivation of Cul4 and Ddb1 abolishes H3K4 mono- and tri-methylation.Citation50 Cul4-Ddb1 may also function via the WD-containing adaptors to affect histone methylation. ORCA has been shown to bind to repressive histone marks.Citation57,Citation80,Citation81 The interaction of ORCA with Cul4A-Ddb1 may not be solely for its degradation, but may also mediate other important regulatory pathways.
Several studies have shown that WD-containing proteins serve as the substrate-recognition subunits of the Cul4-Ddb1 ubiquitin ligase.Citation50,Citation82 Recently, a set of WD repeats containing proteins that interact with Cul4-Ddb1 ligase has been identified based on the structural studies, and these have been termed DCAFs (Ddb1- and Cul4-associated factors), or CDWs (Cul4- and Ddb1-associated WD repeat proteins). Ddb1, comprising three β-propeller structures, uses one β-propeller to interact with Cul4A and uses the other double β-propellers to bind the WD-containing protein.Citation50,Citation60,Citation61,Citation83 From this view, the WD-containing proteins serve as the substrate-specifying and recruiting adaptors for downstream effectors or the substrates themselves to fit into the structure. We have demonstrated that ORCA associates with Cul4-Ddb1, and it is the WD domain of ORCA that mediates this interaction. When we knockdown both Cul4 and Ddb1 in cells, we do not observe any significant stabilization of ORCA protein levels (). This implies that other E3 ligases might be involved in ORCA degradation. It is also likely that ORCA serves as an adaptor to recruit substrates to the Cul4A-Ddb1 E3 ligase. Future work on the identification of ORCA-interacting partners will provide more insights into this.
Materials and Methods
Cell culture and RNA interference
U2OS and WI38 cells were grown in Dulbecco’s modified Eagle medium (DMEM) containing high glucose, supplemented with penicillin-streptomycin and 10% fetal bovine serum (FBS) (Hyclone). When indicated, cells were incubated with DMEM + 20 µM MG132 for proteasome inhibition. Lipofectamine 2000 (Invitrogen) was used for transient transfections as per the manufacturer’s protocols. Orc2 siRNAsCitation21 and Ddb1 siRNAsCitation45 were delivered by lipofectamine RNAiMAX (Invitrogen) as a final concentration of 100 nM for three times at a gap of 24 h.
Plasmids and antibodies
Human ORCA cDNA was cloned into pCGT, pCGN and pEYFP-C1 vectors with CMV promoter (Clontech) to generate T7-ORCA, HA-ORCA and YFP-ORCA. Truncated mutants were obtained using PCR from the ORCA cDNA and also cloned into the pCGT vector. Site-directed mutagenesis (Stratagene) was performed to make the lysine (K) to arginine (R) mutants using standard protocols. Flag-Ub and His-Ub were kind gifts from Dr. Jie Chen, and Cul4A-Ddb1 constructs were from Dr. Yue Xiong.
The following antibodies (pAb: polyclonal antibodies; mAb: monoclonal antibodies) were used for immunoprecipitations and immunoblots as indicated: ORCA pAb (2853–2, 2854–1), Orc2 pAb (205), Orc3 pAb (Abcam), Myc mAb (9E10), Lys48-Ub mAb (Millipore), Lys63-Ub mAb (Millipore), α-Tubulin mAb (Sigma-Aldrich), T7 mAb (Novagen), GFP mAb (Roche and Covance).
Immunoprecipitation and immunofluorescence
Nuclear extracts were prepared first in hypotonic buffer (10 mM HEPES-NaOH pH 7.9, 10 mM KCl, 2 mM MgCl2, 0.34 M sucrose, 10% glycerol, 0.1% Triton X-100) plus a supplement consisting of 1 mM DTT, protein phosphatase inhibitors (10 mM NaF, 1 mM Na3VO4, 1 mM Na2H2P2O7), protease-inhibitor cocktail tablets (Roche), followed by nuclear extraction buffer (20 mM HEPES-NaOH pH 7.9, 2 mM MgCl2, 1 mM EGTA, 25% glycerol, 0.1% Triton X-100) plus a supplement consisting of 1 mM DTT, protein phosphatase inhibitors, protease inhibitors. And whole-cell lysates were made in the RIPA buffer (without DNase I or MNase treatment). Extracts were first pre-cleared by GammaBind G Sepharose resin (Amersham) and then incubated with antibodies at 4°C overnight. Resin was then added into extracts for 1.5 h, washed three times and resuspended in Laemmli buffer for immunoblot analysis. To make cell lysates for immunoprecipitations under the denaturing condition, cells were precipitated with 10% trichloroacetic acid (TCA) and washed twice with acetone. The dried pellets were then incubated with urea cracking buffer (50 mM Tris pH7.5, 8 M Urea, 1% SDS) for 10 min at 65°C, and diluted with 10 volumns of Tween-20 buffer (50 mM Tris pH7.5, 150 mM NaCl, 0.5% Tween-20, 0.1 mM EDTA).
Immunofluorescence studies were performed as described previously.Citation84 The YFP-ORCA-expressing cell line was either fixed in 2% paraformaldehyde for 15 min at room temperature and then permeabilized by PBS + 0.5% Triton X-100 for 7 min on ice, or pre-extracted in CSK buffer (10 mM PIPES pH 7.0, 100 mM NaCl, 300 mM sucrose, 3 mM MgCl2) + 0.5% Triton X-100 for 5 min on ice and then fixed with 2% paraformaldehyde. Cells were then blocked in PBS + 1% normal goat serum (NGS) and incubated with primary antibodies for 1 h and secondary antibodies for 45 min. DNA was stained with DAPI. Cells were examined on the DeltaVision optical sectioning deconvolution instrument (Applied Precision) on an Olympus microscope.
FRAP experiments were performed on the DeltaVision microscope (Applied Precision) using a 63 × NA 1.4 Planapochromat oil-immersion objective (Olympus) with a 488-nm laser line. Required regions were photobleached for 250–300 ms with laser; four pre-bleach and 50 post-bleach frames were recorded for each series. Quantification of fluorescence intensities were conducted in 30 nuclei for each condition. T (1/2) times of recovery and maximum recovery fraction were calculated from the curves.
Synchronization
To synchronize U2OS cells at the G1/S boundary, 2 mM thymidine was added. After 24 h, cells were washed three times with fresh medium, grown for 12 h and incubated with 2 mM thymidine again for 24 h. A portion of thymidine-released cells were further treated with 50 ng/ml nocodazole to be arrested in mitosis, and were then released into G1 phase. For proteasome-inhibition treatments, MG132 was added to double-thymidine blocked cells in the presence of 2 mM thymidine (G1/S) for 6 h, or added to 3 h post nocodazole release cells (G1) for 6 h. Equal amount of DMSO was used as the control.
Acknowledgments
We thank members of the Prasanth laboratory for discussions and suggestions. We thank Drs. J. Cook, J. Chen, A. Dutta, M. Pagano, B. Stillman and Y. Xiong for providing reagents and suggestions. We would also like to thank Drs. K. Prasanth and S. Ceman for critical reading of the paper. This work was supported by NSF (0843604) and NIH (1RO1GM099669–01A1) awards to S.G.P.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- Bell SP, Dutta A. DNA replication in eukaryotic cells. Annu Rev Biochem 2002; 71:333 - 74; http://dx.doi.org/10.1146/annurev.biochem.71.110601.135425; PMID: 12045100
- Arias EE, Walter JC. Strength in numbers: preventing rereplication via multiple mechanisms in eukaryotic cells. Genes Dev 2007; 21:497 - 518; http://dx.doi.org/10.1101/gad.1508907; PMID: 17344412
- Drury LS, Diffley JF. Factors affecting the diversity of DNA replication licensing control in eukaryotes. Curr Biol 2009; 19:530 - 5; http://dx.doi.org/10.1016/j.cub.2009.02.034; PMID: 19285403
- Truong LN, Wu X. Prevention of DNA re-replication in eukaryotic cells. J Mol Cell Biol 2011; 3:13 - 22; http://dx.doi.org/10.1093/jmcb/mjq052; PMID: 21278447
- DePamphilis ML. Cell cycle dependent regulation of the origin recognition complex. Cell Cycle 2005; 4:70 - 9; http://dx.doi.org/10.4161/cc.4.1.1333; PMID: 15611627
- Woo RA, Poon RY. Cyclin-dependent kinases and S phase control in mammalian cells. Cell Cycle 2003; 2:316 - 24; http://dx.doi.org/10.4161/cc.2.4.468; PMID: 12851482
- Feng H, Kipreos ET. Preventing DNA re-replication--divergent safeguards in yeast and metazoa. Cell Cycle 2003; 2:431 - 4; http://dx.doi.org/10.4161/cc.2.5.527; PMID: 12963835
- Diffley JF, Cocker JH, Dowell SJ, Rowley A. Two steps in the assembly of complexes at yeast replication origins in vivo. Cell 1994; 78:303 - 16; http://dx.doi.org/10.1016/0092-8674(94)90299-2; PMID: 8044842
- Liang C, Stillman B. Persistent initiation of DNA replication and chromatin-bound MCM proteins during the cell cycle in cdc6 mutants. Genes Dev 1997; 11:3375 - 86; http://dx.doi.org/10.1101/gad.11.24.3375; PMID: 9407030
- Nguyen VQ, Co C, Li JJ. Cyclin-dependent kinases prevent DNA re-replication through multiple mechanisms. Nature 2001; 411:1068 - 73; http://dx.doi.org/10.1038/35082600; PMID: 11429609
- Wu PY, Nurse P. Establishing the program of origin firing during S phase in fission Yeast. Cell 2009; 136:852 - 64; http://dx.doi.org/10.1016/j.cell.2009.01.017; PMID: 19269364
- Hua XH, Newport J. Identification of a preinitiation step in DNA replication that is independent of origin recognition complex and cdc6, but dependent on cdk2. J Cell Biol 1998; 140:271 - 81; http://dx.doi.org/10.1083/jcb.140.2.271; PMID: 9442103
- Romanowski P, Madine MA, Rowles A, Blow JJ, Laskey RA. The Xenopus origin recognition complex is essential for DNA replication and MCM binding to chromatin. Curr Biol 1996; 6:1416 - 25; http://dx.doi.org/10.1016/S0960-9822(96)00746-4; PMID: 8939603
- Sun WH, Coleman TR, DePamphilis ML. Cell cycle-dependent regulation of the association between origin recognition proteins and somatic cell chromatin. EMBO J 2002; 21:1437 - 46; http://dx.doi.org/10.1093/emboj/21.6.1437; PMID: 11889049
- Rowles A, Tada S, Blow JJ. Changes in association of the Xenopus origin recognition complex with chromatin on licensing of replication origins. J Cell Sci 1999; 112:2011 - 8; PMID: 10341218
- Araki M, Wharton RP, Tang Z, Yu H, Asano M. Degradation of origin recognition complex large subunit by the anaphase-promoting complex in Drosophila. EMBO J 2003; 22:6115 - 26; http://dx.doi.org/10.1093/emboj/cdg573; PMID: 14609957
- Kreitz S, Ritzi M, Baack M, Knippers R. The human origin recognition complex protein 1 dissociates from chromatin during S phase in HeLa cells. J Biol Chem 2001; 276:6337 - 42; http://dx.doi.org/10.1074/jbc.M009473200; PMID: 11102449
- Siddiqui K, Stillman B. ATP-dependent assembly of the human origin recognition complex. J Biol Chem 2007; 282:32370 - 83; http://dx.doi.org/10.1074/jbc.M705905200; PMID: 17716973
- Tatsumi Y, Ohta S, Kimura H, Tsurimoto T, Obuse C. The ORC1 cycle in human cells: I. cell cycle-regulated oscillation of human ORC1. J Biol Chem 2003; 278:41528 - 34; http://dx.doi.org/10.1074/jbc.M307534200; PMID: 12909627
- Méndez J, Zou-Yang XH, Kim SY, Hidaka M, Tansey WP, Stillman B. Human origin recognition complex large subunit is degraded by ubiquitin-mediated proteolysis after initiation of DNA replication. Mol Cell 2002; 9:481 - 91; http://dx.doi.org/10.1016/S1097-2765(02)00467-7; PMID: 11931757
- Prasanth SG, Prasanth KV, Siddiqui K, Spector DL, Stillman B. Human Orc2 localizes to centrosomes, centromeres and heterochromatin during chromosome inheritance. EMBO J 2004; 23:2651 - 63; http://dx.doi.org/10.1038/sj.emboj.7600255; PMID: 15215892
- Fujita M. Cdt1 revisited: complex and tight regulation during the cell cycle and consequences of deregulation in mammalian cells. Cell Div 2006; 1:22; http://dx.doi.org/10.1186/1747-1028-1-22; PMID: 17042960
- Tada S. Cdt1 and geminin: role during cell cycle progression and DNA damage in higher eukaryotes. Front Biosci 2007; 12:1629 - 41; http://dx.doi.org/10.2741/2175; PMID: 17127409
- McGarry TJ, Kirschner MW. Geminin, an inhibitor of DNA replication, is degraded during mitosis. Cell 1998; 93:1043 - 53; http://dx.doi.org/10.1016/S0092-8674(00)81209-X; PMID: 9635433
- Wohlschlegel JA, Dwyer BT, Dhar SK, Cvetic C, Walter JC, Dutta A. Inhibition of eukaryotic DNA replication by geminin binding to Cdt1. Science 2000; 290:2309 - 12; http://dx.doi.org/10.1126/science.290.5500.2309; PMID: 11125146
- Saxena S, Dutta A. Geminin and p53: deterrents to rereplication in human cancer cells. Cell Cycle 2003; 2:283 - 6; http://dx.doi.org/10.4161/cc.2.4.443; PMID: 12851473
- Xouri G, Dimaki M, Bastiaens PI, Lygerou Z. Cdt1 interactions in the licensing process: a model for dynamic spatiotemporal control of licensing. Cell Cycle 2007; 6:1549 - 52; http://dx.doi.org/10.4161/cc.6.13.4455; PMID: 17598984
- Diffley JF. Regulation of early events in chromosome replication. Curr Biol 2004; 14:R778 - 86; http://dx.doi.org/10.1016/j.cub.2004.09.019; PMID: 15380092
- Shen Z, Sathyan KM, Geng Y, Zheng R, Chakraborty A, Freeman B, et al. A WD-repeat protein stabilizes ORC binding to chromatin. Mol Cell 2010; 40:99 - 111; http://dx.doi.org/10.1016/j.molcel.2010.09.021; PMID: 20932478
- Chen ZJ, Sun LJ. Nonproteolytic functions of ubiquitin in cell signaling. Mol Cell 2009; 33:275 - 86; http://dx.doi.org/10.1016/j.molcel.2009.01.014; PMID: 19217402
- Kirkin V, McEwan DG, Novak I, Dikic I. A role for ubiquitin in selective autophagy. Mol Cell 2009; 34:259 - 69; http://dx.doi.org/10.1016/j.molcel.2009.04.026; PMID: 19450525
- Weake VM, Workman JL. Histone ubiquitination: triggering gene activity. Mol Cell 2008; 29:653 - 63; http://dx.doi.org/10.1016/j.molcel.2008.02.014; PMID: 18374642
- Cardozo T, Pagano M. The SCF ubiquitin ligase: insights into a molecular machine. Nat Rev Mol Cell Biol 2004; 5:739 - 51; http://dx.doi.org/10.1038/nrm1471; PMID: 15340381
- Petroski MD, Deshaies RJ. Function and regulation of cullin-RING ubiquitin ligases. Nat Rev Mol Cell Biol 2005; 6:9 - 20; http://dx.doi.org/10.1038/nrm1547; PMID: 15688063
- Hochstrasser M. Ubiquitin-dependent protein degradation. Annu Rev Genet 1996; 30:405 - 39; http://dx.doi.org/10.1146/annurev.genet.30.1.405; PMID: 8982460
- Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem 1998; 67:425 - 79; http://dx.doi.org/10.1146/annurev.biochem.67.1.425; PMID: 9759494
- Pickart CM. Mechanisms underlying ubiquitination. Annu Rev Biochem 2001; 70:503 - 33; http://dx.doi.org/10.1146/annurev.biochem.70.1.503; PMID: 11395416
- Li X, Zhao Q, Liao R, Sun P, Wu X. The SCF(Skp2) ubiquitin ligase complex interacts with the human replication licensing factor Cdt1 and regulates Cdt1 degradation. J Biol Chem 2003; 278:30854 - 8; http://dx.doi.org/10.1074/jbc.C300251200; PMID: 12840033
- Liu E, Li X, Yan F, Zhao Q, Wu X. Cyclin-dependent kinases phosphorylate human Cdt1 and induce its degradation. J Biol Chem 2004; 279:17283 - 8; http://dx.doi.org/10.1074/jbc.C300549200; PMID: 15004027
- Sugimoto N, Tatsumi Y, Tsurumi T, Matsukage A, Kiyono T, Nishitani H, et al. Cdt1 phosphorylation by cyclin A-dependent kinases negatively regulates its function without affecting geminin binding. J Biol Chem 2004; 279:19691 - 7; http://dx.doi.org/10.1074/jbc.M313175200; PMID: 14993212
- Nishitani H, Sugimoto N, Roukos V, Nakanishi Y, Saijo M, Obuse C, et al. Two E3 ubiquitin ligases, SCF-Skp2 and DDB1-Cul4, target human Cdt1 for proteolysis. EMBO J 2006; 25:1126 - 36; http://dx.doi.org/10.1038/sj.emboj.7601002; PMID: 16482215
- Zhong W, Feng H, Santiago FE, Kipreos ET. CUL-4 ubiquitin ligase maintains genome stability by restraining DNA-replication licensing. Nature 2003; 423:885 - 9; http://dx.doi.org/10.1038/nature01747; PMID: 12815436
- Arias EE, Walter JC. PCNA functions as a molecular platform to trigger Cdt1 destruction and prevent re-replication. Nat Cell Biol 2006; 8:84 - 90; http://dx.doi.org/10.1038/ncb1346; PMID: 16362051
- Ralph E, Boye E, Kearsey SE. DNA damage induces Cdt1 proteolysis in fission yeast through a pathway dependent on Cdt2 and Ddb1. EMBO Rep 2006; 7:1134 - 9; http://dx.doi.org/10.1038/sj.embor.7400827; PMID: 17039252
- Hu J, McCall CM, Ohta T, Xiong Y. Targeted ubiquitination of CDT1 by the DDB1-CUL4A-ROC1 ligase in response to DNA damage. Nat Cell Biol 2004; 6:1003 - 9; http://dx.doi.org/10.1038/ncb1172; PMID: 15448697
- Higa LA, Banks D, Wu M, Kobayashi R, Sun H, Zhang H. L2DTL/CDT2 interacts with the CUL4/DDB1 complex and PCNA and regulates CDT1 proteolysis in response to DNA damage. Cell Cycle 2006; 5:1675 - 80; http://dx.doi.org/10.4161/cc.5.15.3149; PMID: 16861906
- Komander D. The emerging complexity of protein ubiquitination. Biochem Soc Trans 2009; 37:937 - 53; http://dx.doi.org/10.1042/BST0370937; PMID: 19754430
- Hicke L, Schubert HL, Hill CP. Ubiquitin-binding domains. Nat Rev Mol Cell Biol 2005; 6:610 - 21; http://dx.doi.org/10.1038/nrm1701; PMID: 16064137
- Pashkova N, Gakhar L, Winistorfer SC, Yu L, Ramaswamy S, Piper RC. WD40 repeat propellers define a ubiquitin-binding domain that regulates turnover of F box proteins. Mol Cell 2010; 40:433 - 43; http://dx.doi.org/10.1016/j.molcel.2010.10.018; PMID: 21070969
- Higa LA, Wu M, Ye T, Kobayashi R, Sun H, Zhang H. CUL4-DDB1 ubiquitin ligase interacts with multiple WD40-repeat proteins and regulates histone methylation. Nat Cell Biol 2006; 8:1277 - 83; http://dx.doi.org/10.1038/ncb1490; PMID: 17041588
- Shimizu Y, Okuda-Shimizu Y, Hendershot LM. Ubiquitylation of an ERAD substrate occurs on multiple types of amino acids. Mol Cell 2010; 40:917 - 26; http://dx.doi.org/10.1016/j.molcel.2010.11.033; PMID: 21172657
- Woelk T, Sigismund S, Penengo L, Polo S. The ubiquitination code: a signalling problem. Cell Div 2007; 2:11; http://dx.doi.org/10.1186/1747-1028-2-11; PMID: 17355622
- Spence J, Gali RR, Dittmar G, Sherman F, Karin M, Finley D. Cell cycle-regulated modification of the ribosome by a variant multiubiquitin chain. Cell 2000; 102:67 - 76; http://dx.doi.org/10.1016/S0092-8674(00)00011-8; PMID: 10929714
- Hoege C, Pfander B, Moldovan GL, Pyrowolakis G, Jentsch S. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature 2002; 419:135 - 41; http://dx.doi.org/10.1038/nature00991; PMID: 12226657
- Hofmann RM, Pickart CM. Noncanonical MMS2-encoded ubiquitin-conjugating enzyme functions in assembly of novel polyubiquitin chains for DNA repair. Cell 1999; 96:645 - 53; http://dx.doi.org/10.1016/S0092-8674(00)80575-9; PMID: 10089880
- Vong QP, Cao K, Li HY, Iglesias PA, Zheng Y. Chromosome alignment and segregation regulated by ubiquitination of survivin. Science 2005; 310:1499 - 504; http://dx.doi.org/10.1126/science.1120160; PMID: 16322459
- Bartke T, Vermeulen M, Xhemalce B, Robson SC, Mann M, Kouzarides T. Nucleosome-interacting proteins regulated by DNA and histone methylation. Cell 2010; 143:470 - 84; http://dx.doi.org/10.1016/j.cell.2010.10.012; PMID: 21029866
- Shen Z, Chakraborty A, Jain A, Giri S, Ha T, Prasanth KV, et al. Dynamic association of ORCA with prereplicative complex components regulates DNA replication initiation. Mol Cell Biol 2012; 32:3107 - 20; http://dx.doi.org/10.1128/MCB.00362-12; PMID: 22645314
- Shcherbik N, Pestov DG. Ubiquitin and ubiquitin-like proteins in the nucleolus: multitasking tools for a ribosome factory. Genes Cancer 2010; 1:681 - 9; http://dx.doi.org/10.1177/1947601910381382; PMID: 21113400
- Angers S, Li T, Yi X, MacCoss MJ, Moon RT, Zheng N. Molecular architecture and assembly of the DDB1-CUL4A ubiquitin ligase machinery. Nature 2006; 443:590 - 3; PMID: 16964240
- Jin J, Arias EE, Chen J, Harper JW, Walter JC. A family of diverse Cul4-Ddb1-interacting proteins includes Cdt2, which is required for S phase destruction of the replication factor Cdt1. Mol Cell 2006; 23:709 - 21; http://dx.doi.org/10.1016/j.molcel.2006.08.010; PMID: 16949367
- Li D, Roberts R. WD-repeat proteins: structure characteristics, biological function, and their involvement in human diseases. Cell Mol Life Sci 2001; 58:2085 - 97; http://dx.doi.org/10.1007/PL00000838; PMID: 11814058
- Neer EJ, Schmidt CJ, Nambudripad R, Smith TF. The ancient regulatory-protein family of WD-repeat proteins. Nature 1994; 371:297 - 300; http://dx.doi.org/10.1038/371297a0; PMID: 8090199
- Smith TF, Gaitatzes C, Saxena K, Neer EJ. The WD repeat: a common architecture for diverse functions. Trends Biochem Sci 1999; 24:181 - 5; http://dx.doi.org/10.1016/S0968-0004(99)01384-5; PMID: 10322433
- Migliori V, Mapelli M, Guccione E. On WD40 proteins: Propelling our knowledge of transcriptional control?. Epigenetics 2012; 7:8; http://dx.doi.org/10.4161/epi.21140; PMID: 22810296
- von Mikecz A. The nuclear ubiquitin-proteasome system. J Cell Sci 2006; 119:1977 - 84; http://dx.doi.org/10.1242/jcs.03008; PMID: 16687735
- Arias EE, Walter JC. Replication-dependent destruction of Cdt1 limits DNA replication to a single round per cell cycle in Xenopus egg extracts. Genes Dev 2005; 19:114 - 26; http://dx.doi.org/10.1101/gad.1255805; PMID: 15598982
- Furstenthal L, Swanson C, Kaiser BK, Eldridge AG, Jackson PK. Triggering ubiquitination of a CDK inhibitor at origins of DNA replication. Nat Cell Biol 2001; 3:715 - 22; http://dx.doi.org/10.1038/35087026; PMID: 11483956
- Prasanth SG, Shen Z, Prasanth KV, Stillman B. Human origin recognition complex is essential for HP1 binding to chromatin and heterochromatin organization. Proc Natl Acad Sci USA 2010; 107:15093 - 8; http://dx.doi.org/10.1073/pnas.1009945107; PMID: 20689044
- McCall CM, Hu J, Xiong Y. Recruiting substrates to cullin 4-dependent ubiquitin ligases by DDB1. Cell Cycle 2005; 4:27 - 9; http://dx.doi.org/10.4161/cc.4.1.1396; PMID: 15655366
- Nag A, Bondar T, Shiv S, Raychaudhuri P. The xeroderma pigmentosum group E gene product DDB2 is a specific target of cullin 4A in mammalian cells. Mol Cell Biol 2001; 21:6738 - 47; http://dx.doi.org/10.1128/MCB.21.20.6738-6747.2001; PMID: 11564859
- Chen X, Zhang Y, Douglas L, Zhou P. UV-damaged DNA-binding proteins are targets of CUL-4A-mediated ubiquitination and degradation. J Biol Chem 2001; 276:48175 - 82; PMID: 11673459
- Groisman R, Polanowska J, Kuraoka I, Sawada J, Saijo M, Drapkin R, et al. The ubiquitin ligase activity in the DDB2 and CSA complexes is differentially regulated by the COP9 signalosome in response to DNA damage. Cell 2003; 113:357 - 67; http://dx.doi.org/10.1016/S0092-8674(03)00316-7; PMID: 12732143
- Sugasawa K, Okuda Y, Saijo M, Nishi R, Matsuda N, Chu G, et al. UV-induced ubiquitylation of XPC protein mediated by UV-DDB-ubiquitin ligase complex. Cell 2005; 121:387 - 400; http://dx.doi.org/10.1016/j.cell.2005.02.035; PMID: 15882621
- Kapetanaki MG, Guerrero-Santoro J, Bisi DC, Hsieh CL, Rapić-Otrin V, Levine AS. The DDB1-CUL4ADDB2 ubiquitin ligase is deficient in xeroderma pigmentosum group E and targets histone H2A at UV-damaged DNA sites. Proc Natl Acad Sci USA 2006; 103:2588 - 93; http://dx.doi.org/10.1073/pnas.0511160103; PMID: 16473935
- Wang H, Zhai L, Xu J, Joo HY, Jackson S, Erdjument-Bromage H, et al. Histone H3 and H4 ubiquitylation by the CUL4-DDB-ROC1 ubiquitin ligase facilitates cellular response to DNA damage. Mol Cell 2006; 22:383 - 94; http://dx.doi.org/10.1016/j.molcel.2006.03.035; PMID: 16678110
- Higa LA, Mihaylov IS, Banks DP, Zheng J, Zhang H. Radiation-mediated proteolysis of CDT1 by CUL4-ROC1 and CSN complexes constitutes a new checkpoint. Nat Cell Biol 2003; 5:1008 - 15; http://dx.doi.org/10.1038/ncb1061; PMID: 14578910
- Bondar T, Ponomarev A, Raychaudhuri P. Ddb1 is required for the proteolysis of the Schizosaccharomyces pombe replication inhibitor Spd1 during S phase and after DNA damage. J Biol Chem 2004; 279:9937 - 43; http://dx.doi.org/10.1074/jbc.M312570200; PMID: 14701809
- Wertz IE, O’Rourke KM, Zhang Z, Dornan D, Arnott D, Deshaies RJ, et al. Human De-etiolated-1 regulates c-Jun by assembling a CUL4A ubiquitin ligase. Science 2004; 303:1371 - 4; http://dx.doi.org/10.1126/science.1093549; PMID: 14739464
- Vermeulen M, Eberl HC, Matarese F, Marks H, Denissov S, Butter F, et al. Quantitative interaction proteomics and genome-wide profiling of epigenetic histone marks and their readers. Cell 2010; 142:967 - 80; http://dx.doi.org/10.1016/j.cell.2010.08.020; PMID: 20850016
- Chan KM, Zhang Z. Leucine-rich repeat and WD repeat-containing protein 1 is recruited to pericentric heterochromatin by trimethylated lysine 9 of histone H3 and maintains heterochromatin silencing. J Biol Chem 2012; 287:15024 - 33; http://dx.doi.org/10.1074/jbc.M111.337980; PMID: 22427655
- Higa LA, Zhang H. Stealing the spotlight: CUL4-DDB1 ubiquitin ligase docks WD40-repeat proteins to destroy. Cell Div 2007; 2:5; http://dx.doi.org/10.1186/1747-1028-2-5; PMID: 17280619
- Lee J, Zhou P. DCAFs, the missing link of the CUL4-DDB1 ubiquitin ligase. Mol Cell 2007; 26:775 - 80; http://dx.doi.org/10.1016/j.molcel.2007.06.001; PMID: 17588513
- Sathyan KM, Shen Z, Tripathi V, Prasanth KV, Prasanth SG. A BEN-domain-containing protein associates with heterochromatin and represses transcription. J Cell Sci 2011; 124:3149 - 63; http://dx.doi.org/10.1242/jcs.086603; PMID: 21914818