Abstract
Human embryonic stem cells have shown tremendous potential in regenerative medicine, and the recent progress in haploid embryonic stem cells provides new insights for future applications of embryonic stem cells. Disruption of normal fertilized embryos remains controversial; thus, the development of a new source for human embryonic stem cells is important for their usefulness. Here, we investigated the feasibility of haploid and diploid embryo reconstruction and embryonic stem cell derivation using microsurgically repaired tripronuclear human zygotes. Diploid and haploid zygotes were successfully reconstructed, but a large proportion of them still had a tripolar spindle assembly. The reconstructed embryos developed to the blastocyst stage, although the loss of chromosomes was observed in these zygotes. Finally, triploid and diploid human embryonic stem cells were derived from tripronuclear and reconstructed zygotes (from which only one pronucleus was removed), but haploid human embryonic stem cells were not successfully derived from the reconstructed zygotes when two pronuclei were removed. Both triploid and diploid human embryonic stem cells showed the general characteristics of human embryonic stem cells. These results indicate that the lower embryo quality resulting from abnormal spindle assembly contributed to the failure of the haploid embryonic stem cell derivation. However, the successful derivation of diploid embryonic stem cells demonstrated that microsurgical tripronuclear zygotes are an alternative source of human embryonic stem cells. In the future, improving spindle assembly will facilitate the application of triploid zygotes to the field of haploid embryonic stem cells.
Introduction
Embryonic stem cells (ES cells) have displayed tremendous potential in regenerative medicine and have been successfully derived from mice, rats, monkeys and humans.Citation1-Citation4 A typical characteristic of ES cells is the preservation of diploid karyotyping during long-term propagation. ES cells have shown the ability to self-renew during long-term propagation, and they maintain the normal karyotyping and differentiation both in vitro and in vivo. More importantly, ES cells have demonstrated pluripotency under specific conditions, differentiating into cell types, including neuronlineage cells, insulin-producing cells and even germ cells.Citation1,Citation4 In addition to cells, tissues have been generated from ES cells, demonstrating the importance of ES cells in clinical medicine.
ES cells containing only one set of chromosomes with characteristics similar to diploid ES cells have recently been derived from mice.Citation5,Citation6 Moreover, Yang et al. recently demonstrated that mouse haploid androgenic ES cells could function as sperm, and that the “fertilized” embryos created from MII oocytes and haploid androgeneic ES cells developed to term and resulted in live mice.Citation7 Therefore, haploid androgeneic ES cells provide a potential method to resolve infertility caused by azoospermia and could also be used as a transgenic tool. Several labs have reported haploid mouse ES cells,Citation5-Citation7 but not haploid human ES cells, thereby limiting further application in humans. One key limitation lies in obtaining haploid embryos or blastocysts for ES derivation. In mice, two approaches have been used to derive haploid embryos, including injecting one sperm while removing the oocyte’s chromosomes or removing the female pronucleus from the zygote.Citation7 However, in humans, both approaches require MII oocytes, which are difficult to obtain due to ethical issues.
Polyspermic zygotes may present a potential method to resolve this predicament. Polyspermy occurs when more than one sperm enters one oocyte, forming a zygote with more than two pronuclei. Polyspermy is regarded as invariably pathological, and the early embryo either fails to develop or develops abnormally.Citation8 In polyspermic zygotes, tripronuclear (3PN) zygotes with two sperm nuclei and one oocyte nucleus have been commonly found; approximately 2–5% of zygotes will become polyspermic during the in vitro fertilization process.
The developmental competence of polyspermic zygotes has been extensively studied. Balakier observed that 3PN zygotes were capable of substantial in vitro development, and 6% of such zygotes could develop to the blastocyst stage.Citation9 In a porcine study, Han et al. indicated that at least 40% of 3PN zygotes could develop to the blastocyst stage, but the cell number in the inner cell mass (ICM) was decreased when compared with diploid blastocysts.Citation10 Recently, Chen et al. derived one triploid ES cell line and three diploid ES cell lines from 12 blastocysts that were developed from 130 3PN zygotes, suggesting that polyspermic embryos could serve as an alternative source for normal euploid human embryonic stem cell (hES cell) lines.Citation11 In 1989, Malter et al. tried to reconstruct a diploid zygote using microsurgery; 1 blastocyst was obtained using microsurgery from seven tripronuclear human zygotes.Citation12 Ivakhnenko et al. observed that triploid and diploid embryos underwent their first cleavage division at similar times, indicating autonomous cytoplasmic activity in human zygotes.Citation13 This microsurgical repair method has also been used in clinical settings. Kattera et al. transferred repaired embryos to a 38-y-old woman, resulting in a normal, healthy baby boy.Citation14 Kattera’s study proved that repaired 3PN embryos are competent to develop to term, but the developmental competence of repaired 3PN embryos with two pronuclei removed has not yet been established.
In the present study, we investigated the developmental competence of diploid and haploid zygotes that were reconstructed from 3PN embryos and the feasibility of ES cell derivation using the resulting blastocysts. The resulting diploid hES cells from embryos with one PN removed had characteristics similar to those of hES cells from normal diploid fertilized embryos. However, no ES cells could be derived from haploid embryos, although the haploid zygotes successfully developed to the blastocyst stage.
Results
Effects of PN removal on the developmental competence and quality of the reconstructed embryos
The r2PN and r1PN embryos were reconstructed from the tripronuclear embryos by removing one or two pronuclei, respectively. The developmental competence results showed that the cleavage efficiency was not different among the 3PN, r2PN or r1PN groups (p > 0.05). However, the eight-cell and blastocyst efficiency of the reconstructed embryos was significantly decreased when compared with 3PN embryos, regardless of the number of PNs that were removed (p < 0.05) (). Embryo quality was evaluated using the eight-cell and blastocyst scoring methods. For eight-cell scoring in the r1PN group, the 8G1 ratio was significantly lower than in the other two groups (p < 0.05), and the 8G4 ratio was significantly higher when compared with the other two groups (p < 0.05). In the r2PN group, the 8G1 ratio was also significantly decreased compared with the 3PN group; the 8G3 ratio was higher than in the 3PN group, and the 8G4 ratio was not different between the groups. Moreover, there was no difference in 8G2 distribution between all three of the groups (). For blastocyst scoring in the r1PN group, two blastocysts were scored as BG3, the lowest quality in blastocyst grading, and only one belonged to the BG2 grade. In the r2PN group, one blastocyst was in the BG1 grade, and two blastocysts belonged to the BG2 grade, though there were still two blastocysts in the BG3 grade. In the 3PN group, three blastocysts were classified as BG1, and six blastocysts as BG2, with only one blastocyst in the BG3 grade (). The representative developmental images from the three groups are shown in .
Table 1. Effects of PN removal on the development competence of reconstructed zygotes
Figure 1. Effects of PN removal on the quality of reconstructed eight-cell embryos and the representative image of embryos in 3PN, r2PN and r1PN groups. (A) The quality of embryos at the eight-cell stage tended to decrease with the increasing number of removed PN. The ratio of high-quality embryos was significantly decreased in the r1PN group. Means with “a” or “b” are significantly different (p < 0.05). (B) No significant morphological differences were observed among the three groups before blastocyst formation; however, no high-quality blastocysts were formed in the r1PN group.
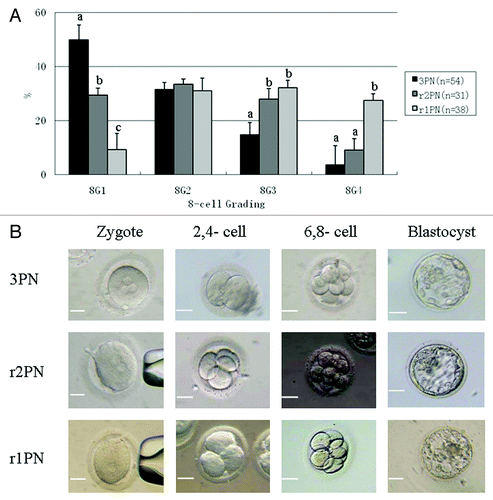
Table 2. Summary of quality scoring and ES derivation in 3PN-, r2PN- and r1PN-blastocysts
Assessment of the spindle assembly in the first mitotic cycle among the 3PN, r2PN and r1PN groups
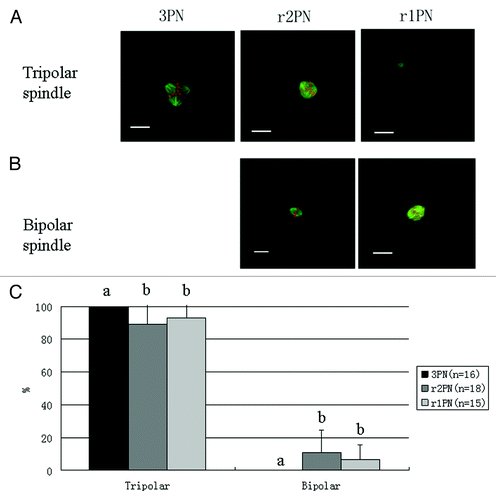
Spindle morphology and chromosome alignment were identified using immunofluorescence. Representative images of the spindle assembly from 3PN zygotes are shown in . In the 3PN group, the spindle morphology was tripolar, but in the r2PN and r1PN groups, two types of spindle morphologies were found: tripolar and dipolar. The distribution of dipolar and tripolar spindles in the r2PN and r1PN groups is shown in . There was no significant difference among the three groups (p > 0.05), but no bipolar spindle morphology was observed in the 3PN group.
Figure 2. Representative tripolar and bipolar spindle assembly among the 3PN, r2PN and r1PN groups. (A) Tripolar spindle morphology among the three groups; (B) bipolar spindle morphology in both the r2PN and r1PN groups; (C) all of the tested zygotes in the 3PN group had a tripolar spindle assembly, whereas 88.9% (n = 18) of the tested zygotes in the r2PN group and 93.3% (n = 15) of the tested zygotes in the r1PN group had a tripolar spindle assembly.
Haploid embryo identification by fluorescence in situ hybridization
The sets of chromosomes in the 3PN, r2PN and r1PN groups were identified using fluorescence in situ hybridization (FISH). Here, the numbers of chromosome 18 and the sex chromosomes were used as markers for embryo chromosome ploidy. shows representative images of female embryos from the three groups. In , it can be observed that the embryos have three sets of chromosome 18 and chromosome X, which means they are triploid. In , it can be observed that the embryos have two sets of chromosome 18 and chromosome X, which means they are diploid. Finally, in , it can be observed that only one set of chromosome 18 and chromosome X was observed, which means the embryo is haploid.
Figure 3. Analysis of chromosome ploidy using fluorescence in situ hybridization. (A) Triploid in 3PN zygote; (B) diploid in r2PN zygote; (C) haploid in r1PN zygote; (D) the loss of chromosome 18 was found in the r2PN and r1PN groups.
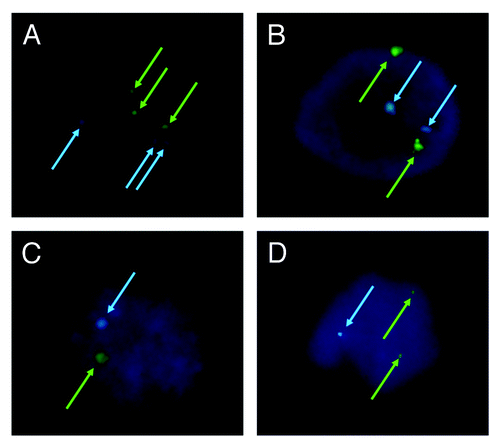
Moreover, chromosome loss was found in embryos belonging to the r2PN and r1PN groups (). Of the 10 single blastomeres tested that derived from r2PN eight-cell embryos, three embryos displayed the loss of chromosome 18. Of the eight single blastomeres tested that derived from r1PN eight-cell embryos, three embryos also displayed the loss of chromosome 18. However, no loss of chromosome 18 was observed in the 3PN group. Almost all of the embryos in which chromosome 18 could not be identified were in the 8G3 and 8G4 groups (Table S1). It is notable that the blastomere biopsy did not decrease the blastocyst formation efficiency of the eight-cell embryos from each group (). Moreover, the loss of chromosomes 16 and 21 was also tested by FISH, and those results were similar to the loss of chromosome 18 (Table S2).
Table 3. Effects of blastomere biopsy on the blastocyst formation among three groups
hES cell line derivation from blastocysts derived from the 3PN, r2PN and r1PN groups
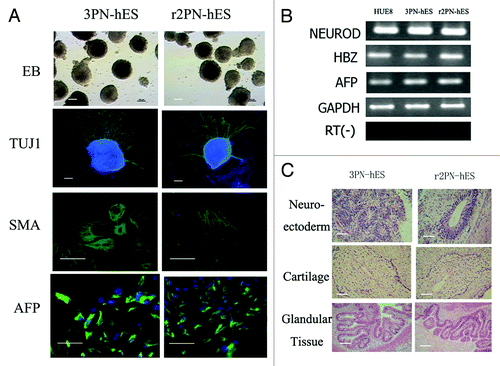
Blastocysts from the three groups were used to derive ES cells. From the 3PN group, a total of 10 blastocysts were put onto feeder layers, and six blastocysts attached and formed small cell colonies by the fifth day. After 1 wk, the growing cell colonies were divided into four or five clumps, from which three hES cell lines that could be propagated were successfully derived. From the r2PN group, five blastocysts were put on feeder layers, and two small colonies were formed after 5 d, resulting in the successful derivation of one hES cell line. In the r1PN group, the three blastocysts that were put onto the feeder layers could not attach to the feeder layers and were dead in the culture media after 1 wk (). All of the established hES cell lines displayed normal morphology, including distinct cell colony boundaries, high nuclear/cytoplasm ratio, tightly packed colonies and positive AP staining. The pluripotency marker (OCT-4) and cell surface markers (Tra-1-60 and SSEA-3) were positively expressed in all of the hES cell lines (). The karyotyping analysis showed that the 3PN-hES cells from the 3PN blastocysts were triploid (two cell lines are 69XXX; one cell line is 69 XXY), and that the r2PN-hES cells were diploid (). These hES cells differentiated to EBs in vitro that positively expressed markers for the three germ layers: SMA (mesoderm), TUJ-1 (ectoderm) and AFP (endoderm) (), and the RT-PCR results again showed the expression of marker genes for the three germ layers (AFP, endoderm; HBZ, mesoderm; NEUROD, ectoderm) (). Typical tissues from the three germ layers [neuroectoderm (ectoderm), cartilage (mesoderm), glandular tissue (endoderm)] were identified using HE staining two months after the hES cells had been injected into SCID mice ().
Figure 4. General characteristics of hES cells from 3PN and r2PN blastocysts. (A) The inner cell mass from the blastocyst could attach and form the primary colony, which then propagated by the mechanical method in both the 3PN and r2PN groups. The distinct cell colony boundary, high nuclear/cytoplasm ratio and tightly packed colonies could be observed, and these colonies showed positive AP activity. The ES cells positively expressed a pluripotency marker (OCT4) and ES cell surface markers (SSEA3 and TRA-1–60). (B) Triploid karyotyping (69, XXY and 69, XXX) was observed in 3PN-hES cells, and diploid karyotyping (46, XY) was observed in r2PN-hES cells.
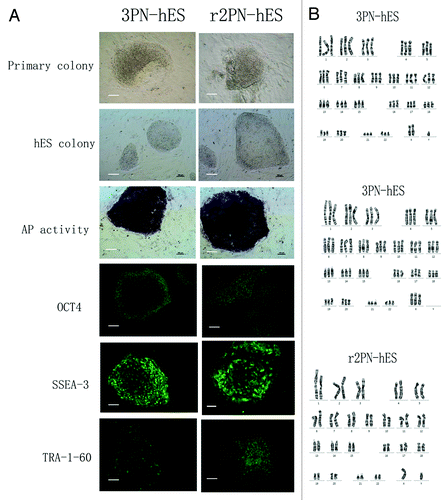
Figure 5. Differentiation ability in hES cells from the 3PN and r2PN groups. (A) EBs were formed in both types of ES cells, and TUJ-1 (ectoderm), SMA (mesoderm) and AFP (endoderm) were positively expressed in the differentiated EBs. (B) Genes from the endoderm (AFP), mesoderm (HBZ) and ectoderm (NEUROD) were identified in differentiated EB clumps. (C) Tissues from the three embryonic germ layers were identified in teratomas: neuroectoderm (ectoderm), cartilage (mesoderm), glandular tissue (endoderm).
Discussion
Polyspermy is a common phenomenon in some species when the oocytes are fertilized in vitro;Citation8 the resulting triploid zygotes have been used as an important resource in clinical settings (reconstructing diploid embryos for further development) and in basic research (reprogramming somatic cells).Citation14-Citation16
The present study investigated the feasibility of creating diploid and haploid ES cells by removing the PN from polyspermic zygotes. Our results indicate that the developmental competence of haploid r1PN was significantly decreased, and the resulting blastocysts failed to transform into ES cells. This failure was attributed to the lower quality of the blastocysts, which was most likely caused by abnormal spindle assembly. The increased loss of chromosomes was also observed in these lower-quality embryos.
In previous studies, 40% of porcine triploid zygotes developed to the blastocyst stage in vitro, but fewer than 6% of human triploid zygotes successfully developed to the blastocyst stage.Citation9,Citation10 In our study, the blastocyst development efficiency of the 3PN group was 10.8%, which is accordance with our previous report.Citation11 The clinical treatment and culture conditions might have contributed to this difference. The removal of PN to repair triploid zygotes has been used clinically, resulting in a live baby;Citation14 however, the developmental competence of r2PN zygotes is still unclear. Two studies indicate that approximately 15% of embryos could develop to the blastocyst stage, but only 6.8% developed to this stage in our study.Citation12,Citation17 This decreased developmental competence might be attributed to different micromanipulation and culture systems. In our study, we removed the pronuclei with the assistance of a Piezo apparatus and cultured the reconstructed embryos under 5% CO2 and 20% O2 conditions, and such r1PN embryos have poor developmental competence at the pre-implantation stage. Until now, no studies have been reported regarding haploid human embryos. In the mouse study, approximately 16–21% of haploid zygotes developed to the blastocyst stage, which was significantly lower than was observed for the diploid zygotes.Citation7 This trend is in accordance with our results.
Although Gu et al. suggested that PN removal could increase the developmental competence under low-oxygen conditions, which is contrary to our results, they also found that abnormal patterns in the first mitosis are not corrected by this removal.Citation12,Citation17 Exposure to atmospheric oxygen most likely has detrimental effects on blastocyst formation rates and on cell proliferation within individual blastocysts, which has been proven in mouse, sheep, cow, goat and pig.Citation18-Citation20 The study indicated that the detrimental effects could be primarily attributed to the oxidative stress that would result in apoptosis.Citation21 Therefore, some studies suggested that the viability of pre-implantation embryos and the quality of blastocysts were all improved significantly when the embryos were cultured under conditions of low oxygen concentration.Citation22,Citation23 More importantly, Lengner et al. demonstrated that the human blastocysts contained pre-X-inactivation cells, and that this state was preserved in vitro through culture under 5% oxygen, which suggested that not only blastocyst formation but also ES cell derivation from these blastocysts was improved under hypoxic conditions.Citation24 Therefore, the hypoxic conditions would contribute to the improvement of the developmental competence of embryos and should be tested in the culture process of r1PN embryos in further study.
Abnormal spindle assemblies were observed in 3PN zygotes. A tripolar spindle was commonly observed in 3PN zygotes, but, interestingly, a large part of the spindle morphology was still tripolar when the zygotes entered into metaphase, even after the removal of one or two pronuclei during interphase. The relationship between spindle morphology and the developmental competence of oocytes has been previously studied. Chatzimeletiou et al. investigated the spindle morphology of developing and arrested embryos and observed that abnormal spindle formation is one of the most important factors affecting human embryonic development.Citation25 Moreover, spindle assembly is influenced by many factors, including maternal age, whether the oocyte was matured in vitro, cryopreservation and gamete manipulation in vitro.Citation26 In the present study, polyspermy also induced abnormal spindle assembly after fertilization. More importantly, the induced tripolar spindle morphology was not significantly improved or repaired after the removal of one or two pronuclei, which is in accordance with the previous study.Citation17
However, a few zygotes with normal bipolar spindles were observed in the present study. One possible reason for this finding is that the centriole from the sperm still existed in the 3PN zygotes. In humans, the centriole is located at the neck of the sperm, just beneath the basal plate and proximal to its nucleus; the configuration of the centriole is essential for ensuring the duplication and functionality of the centrosome in organizing embryonic spindles.Citation27 Sathananthan et al. demonstrated that the centriole is paternally derived in human zygotes, whereas it is maternally derived in mice.Citation28 Colombero et al. found that the female and male pronuclei could join even when the isolated human sperm head was injected into the oocyte, but only one out of five such zygotes had a normal diploid constitution.Citation29 Colombero’s study provides a reasonable explanation for our results, in which a functioning centrosome occasionally remained with the head nucleus. This centriole could be removed during the PN removal process, resulting in a bipolar spindle in tripronuclear human zygotes; however, if the centriole were kept in the cytoplasm after microsurgery, then a tripolar spindle would form.
Abnormal spindle assembly in embryos induces aneuploidy during the process of mitosis. Reverte et al. observed that abnormal spindle positioning and movements might also interfere with cytokinesis and lead to the accumulation of tetraploid cells.Citation30 FISH has been the preferred method for identifying the aneuploidy of human embryos, and chromosomes 13, 16, 18, 21, 22, X and Y are usually selected for testing in clinics.Citation31 Here, chromosomes 18, X and Y were tested. The identification of these three chromosomes by FISH was a feasible method for screening for aneuploidy in gametes.Citation32,Citation33 In our study, some embryos were triploid, diploid and haploid among the 3PN, r2PN and r1PN groups, respectively. However, some embryos among these three groups lost chromosomes, which resulted in the loss of chromosomes 16, 18 and 21 in the r2PN and r1PN embryos, but no embryos with such a loss were found in the control group. This result indicated that the PN removal could contribute to the loss of some chromosomes. The failure to identify these chromosomes by FISH was most likely due to the poor quality of the embryos, and a relationship between morphology and chromosomal constitution in human pre-implantation embryos has been suggested.Citation34 Moreover, it has been reported that aneuploid chromosomes affect developmental competence and embryo quality, increase cell fragments and induce cell apoptosis,Citation35-Citation37 which is in accordance with our results; furthermore, aneuploid chromosomes most likely also affected blastocyst formation and hES cell line derivation.
hES cell lines were derived from 3PN and r2PN blastocysts but not from r1PN blastocysts. This failure was attributed to the poor quality of the blastocysts from the r1PN group. In our study, the proportion of 8G1 embryos in the r1PN group was no more than 10%, which was significantly lower than in the other two groups. It has been demonstrated that poor quality blastocysts could be used as a resource to derive hES cell lines,Citation38,Citation39 but it is clear that high quality blastocysts are still more effective for establishing hES cell lines. Triploid hES cells have been derived from diploid or triploid blastocysts.Citation11,Citation40 These hES cells share characteristics such as self-renewal and multi-differentiation competence with diploid hES cells, which is in accordance with our results.
Except for blastocyst quality, signaling pathways and epigenetic instability were the most common causes of the failure of ES derivation. Haploid ES cells have been derived using mouse haploid blastocysts,Citation5-Citation7 but mouse and human ES cells differ importantly in the signaling pathways required to maintain pluripotency and self-renewal and for X chromosome inactivation. In mouse ES cells, these functions are regulated by LIF and BMP, or GSK3 and MAPK signal pathways,Citation41,Citation42 but in human ES cells, the combination of bFGF, Activin A or TGF-β and the activation of Wnt signaling pathways is dominant in maintaining pluripotency.Citation43-Citation45 X-chromosome inactivation was one of the most important epigenetic changes during the ES derivation. ICM-derived mouse ES cells have two active X chromosomes (XaXa) and will randomly inactivate one X chromosome upon differentiation (XaXi) through XistRNA coating the inactive X in cis;Citation46 however, in human, the X chromosome inactivation was pre-determined before blastocyst formation. Recent evidence suggested that the X chromosome inactivation was highly conserved in placental animals, and that the single XIST gene started to express with the zygotic genomic activation at the six or eight-cell stage.Citation47 Therefore, the distinct differences in signaling pathway dependence and epigenetic status suggest that new derivation systems or methods should be addressed in further studies of human haploid ES cells.
Although ES cell derivation from r1PN embryos failed, there were still significant findings for ES derivation from r2PN embryos in this study. The result of successful derivation of diploid ES cell lines from modified triploid embryos provides an alternative method for ES cell derivation. In the previous study, ES cell derivation was highly controversial, as the ICM must be isolated from normal fertilized embryos that have the ability to develop further, even into live babies. More studies have been performed to try to derive ES cells without destroying the fertilized embryos, including the use of poor-quality embryos, developmentally arrested embryos and even the blastomere biopsied from eight-cell embryos; however, the derivation efficiency was poor.Citation39,Citation48,Citation49 Here, we showed a much higher efficiency of ES derivation from r2PN embryos, which would be helpful for deriving more human ES cell lines. Moreover, this approach would allow researchers more opportunities to derive human ES cells with certain diseases, such as genetic mutations and metabolic and endocrine diseases. It has been difficult to obtain such embryos, and the pathological studies of ES cells were limited. In pre-implantation genetic diagnosis (PGD) procedures, some embryos with genetic mutations were identified and, thus, discarded, which could most likely be applied in ES derivation and further studies. However, because many diseases cannot be identified by PGD testing, the production of ES cells derived from these embryos becomes nearly impossible. Therefore, the modification of triploid zygotes provides an opportunity to obtain these ES cells.
In summary, human triploid polyspermic zygotes were successfully used to derive diploid hES cells after removing one pronucleus, but haploid hES cells failed to survive after the removal of two pronuclei because of the lower quality of the blastocysts. This failure could be attributed to the abnormal spindle assembly, causing the loss of chromosomes, whereas aneuploid chromosomes are considered to be one of the key factors that decrease embryo quality. Thus, more studies are needed to improve the spindle assembly and make polyspermic zygotes a reliable source of haploid hES cells.
Methods and Materials
All chemicals were purchased from Sigma Aldrich Co. unless otherwise indicated.
Experimental design
The present study investigated the feasibility of haploid ES deviation using reconstructed polyspermic zygotes. In experiment 1, one or two pronuclei were removed from 3PN zygotes by micromanipulation, resulting in reconstructed diploid zygotes (r2PN) and reconstructed haploid zygotes (r1PN), respectively. The developmental competence was compared, and embryo quality scores were recorded at the eight-cell and blastocyst stages among the 3PN, r2PN and r1PN groups. In experiment 2, the spindle assembly and tubulin protein location were identified by immunostaining the three groups. In experiment 3, chromosome 18, chromosome X and chromosome Y were detected using FISH in the three groups. In experiment 4, the blastocysts that were developed from the three groups were implanted onto the feeder layer, and ES cell derivation was attempted.
Ethical statement
The present study was approved by the ethics committee of the Third Affiliated Hospital of Guangzhou Medical University and the Peking University Third Hospital. The patients involved in this study knew and comprehended the usage of the polyspermic zygotes and voluntarily discarded them after signing the informed consents.
Collection of polyspermic zygotes
Overall, 367 polyspermic zygotes were collected from 162 patients in this study. The patients were treated with IVF due to female oviduct factors. After IVF treatment, fertilization was observed the next day, and the polyspermic zygotes were selected and transferred into a new culture dish for further micromanipulation.
Pronucleus removal by micromanipulation
The polyspermic zygotes were transferred into GMOPS media. The male pronucleus was selected for removal. The criteria used to identify the male pronuclei were (1) pronucleus-associated sperm tails, (2) increased pronucleus size and (3) a greater distance (relative to the female pronuclei) from the second polar body. For the r2PN group, one pronucleus, which was away from the polar body I (PB I), was identified clearly in the eyepiece. One hole was made in the zona pellucida using a 12–15 μm diameter injector needle, with the assistance of a Piezo apparatus; the pronucleus was then aspirated out of the oocyte using the injector needle. The reconstructed embryos were transferred and cultured in Gm media until the blastocyst stage. For the r1PN group, two pronuclei, which were away from the polar body I (PB I), were identified clearly in the eyepiece. One hole was made in the zona pellucida using a 12–15 μm diameter injector needle, and then the two pronuclei were aspirated out of the oocyte together using the injector needle. The reconstructed embryos were transferred and cultured in Gm media until the blastocyst stage.
Blastomere biopsy using micromanipulation
One blastomere was removed from 3PN, r2PN and r1PN embryos at the six- or eight-cell stage, as previously described.Citation50,Citation51 The embryos were transferred to GMOPS media, and then one hole was made in the zona pellucida using a 12–15 μm diameter injector needle, with the assistance of a Piezo apparatus. Then, one blastomere was aspirated out of the oocyte using the injector needle. The embryos were transferred and cultured in Gm media until the blastocyst stage. The blastomere was transferred into another drop for chromosome detection.
Embryo grading
The method of assessing the quality of embryos at the eight-cell stage was modified from the criteria published by Xia.Citation52 The scores were as follows: for 8G1 scoring, the embryos must have eight blastomeres with equal volume and with fragments that are less than 5%; for 8G2 scoring, the embryos must have eight blastomeres with equal volume and with fragments that are less than 20%; for 8G3 scoring, the embryos must have 4–8 blastomeres with unequal volume and with fragments less than 50%; and for 8G4 scoring, the embryos must have almost no blastomeres and consist nearly entirely of fragments. The method of assessing the quality of embryos at the blastocyst stage was referenced in Dokras’s study.Citation53
Immunostaining
Fifteen zygotes from each group were fixed in 4% paraformaldehyde for 30 min and then permeabilized with 0.2% Triton X-100 for 30 min, followed by blocking in 3% BSA in PBS for 2 h. The zygotes were incubated with a fluorescein isothiocyanate (FITC)-conjugated mouse monoclonal antibody against α-tubulin (Sigma F2168) diluted 1:100. The embryos were washed three times and stained with 10 μg/mL propidium iodide for 30 min. Finally, the embryos were mounted on glass slides and examined with an A1R Confocal Laser-Scanning Microscope (Nikon). hES cell pluripotency markers were stained as described in our previous study, and the ES cell colonies were fixed and treated with primary antibodies, including Oct4, SSEA-3 and Tra-1-60. Nuclei were stained with DAPI.
FISH analysis
The single blastomere was treated as described in our previous study. FISH for chromosomes 18, X and Y was performed on 20 samples. A standard panel of commercially available probes for chromosomes 18, X and Y was used (FISH kit for the detection of chromosome numerical anomalies in prenatal diagnosis®, Jinpujia Medical Technology Co. Ltd.). The X, Y and 18 probes were labeled with a green fluorochrome (CSP spectrum green), a red fluorochrome (CSP spectrum orange) and a blue fluorochrome (CSP spectrum blue), respectively. The slides were examined on a fluorescence microscope, and the results were analyzed after 48 h.
hES cell derivation
The blastocysts were used for ES derivation at day 5.5. The immunosurgery method was applied for ICM isolation, as described in our previous work.Citation54 After 5–7 d, the human ICM formed a small colony, and the culture media were changed every 2 d. The small colony was allowed to grow for 7 more days and was mechanically dissociated into three to four small clumps using a micropipette. After five passages, the human ES cell colonies were propagated using 1 mg/ml of type IV collagenase every 4–7 d.
Karyotyping
Karyotyping identification was performed following our previous study. hES cells were passaged onto matrigel without a feeder layer, and these cells were collected and trypsinized after incubation for 3 d. Then, the cells were incubated in 0.075 mol/L potassium chloride for 10 min at 37°C after being rinsed in phosphate-buffered saline (PBS) solution. Finally, the cells were fixed with methanol/glacial acetic acid and dropped onto glass slides. Chromosome spreads were Giemsa-banded and photographed. The karyotype of hES cells was determined at passage 20.
Assessment of the differentiation capacity in vitro and in vivo
The embryoid body (EB) was used to assess the differentiation ability in vitro of hES cells via specific gene expression. hES cells were cultured in suspension, and then EB growth was observed after 2 wk. After collection, the EB was analyzed by RT-PCR to analyze the specific gene expression for the three embryonic germ layers: AFP (endoderm), NEUROD1 (ectoderm) and HBZ (mesoderm). Moreover, after 7 d cultured in suspension, EBs were transferred to a gelatin-coated plate and cultured in the same medium for another 7 d. The differentiated EBs were immunostained with AFP (1:100, Human Germ Layer Marker Kit, Chemicon), α-SMA (1:200, Human Germ Layer Marker Kit, Chemicon) and Tuj1 (1:200, Human Germ Layer Marker Kit, Chemicon). The secondary antibodies used were Alexa 488-conjugated goat anti-mouse IgG (1:500, Invitrogen) and Alexa 488-conjugated goat anti-rabbit IgG (1:500, Invitrogen). Nuclei were stained with DAPI.
Teratoma production was used to assess the in vivo differentiation ability of hES cells. The hES cells from passage 10 or beyond were treated with a 1 mg/ml type IV collagenase solution for 10–15 min and then dispersed into 300–400 small hES colony suspensions. The colonies were collected and subcutaneously injected into the inguinal grooves of 6-wk-old male severe combined immunodeficiency (SCID) mice. Eight weeks later, the resultant tumors were removed, fixed for 4–8 h in 4% paraformaldehyde and embedded in paraffin. After staining with hematoxylin and eosin, the sections were examined under a light microscope for the presence of tissues derived from the three germ layers.
Statistical analysis
The ratios of normal or abnormal spindle morphologies among the tripronuclear, r2PN and r1PN groups were analyzed using the chi-square test. Eight-cell embryo grading and the developmental efficiency of fertilized and cloned embryos were analyzed using a one-way ANOVA test with SPSS13.0 software. p < 0.05 was regarded as a significant difference.
Additional material
Download Zip (108.5 KB)Acknowledgments
This work was supported in part by the Ministry of Science and Technology of China Grants (973 program; 2011CB944504), the Program for Changjiang Scholars and Innovative Research Team in University of Ministry of Education of China (30825038), the National Natural Science Funds for Young Scholar (31000661), the National Natural Science Foundation of China (81100473, U1132005), Zhujiang Science and Technology Star Project of Guangzhou (2012J2200006) and Guangdong education fund (2012KJCX0087).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- Evans MJ, Kaufman MH. Establishment in culture of pluripotential cells from mouse embryos. Nature 1981; 292:154 - 6; http://dx.doi.org/10.1038/292154a0; PMID: 7242681
- Li P, Tong C, Mehrian-Shai R, Jia L, Wu N, Yan Y, et al. Germline competent embryonic stem cells derived from rat blastocysts. Cell 2008; 135:1299 - 310; http://dx.doi.org/10.1016/j.cell.2008.12.006; PMID: 19109898
- Thomson JA, Kalishman J, Golos TG, Durning M, Harris CP, Becker RA, et al. Isolation of a primate embryonic stem cell line. Proc Natl Acad Sci USA 1995; 92:7844 - 8; http://dx.doi.org/10.1073/pnas.92.17.7844; PMID: 7544005
- Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, et al. Embryonic stem cell lines derived from human blastocysts. Science 1998; 282:1145 - 7; http://dx.doi.org/10.1126/science.282.5391.1145; PMID: 9804556
- Elling U, Taubenschmid J, Wirnsberger G, O’Malley R, Demers SP, Vanhaelen Q, et al. Forward and reverse genetics through derivation of haploid mouse embryonic stem cells. Cell Stem Cell 2011; 9:563 - 74; http://dx.doi.org/10.1016/j.stem.2011.10.012; PMID: 22136931
- Leeb M, Wutz A. Derivation of haploid embryonic stem cells from mouse embryos. Nature 2011; 479:131 - 4; http://dx.doi.org/10.1038/nature10448; PMID: 21900896
- Yang H, Shi L, Wang BA, Liang D, Zhong C, Liu W, et al. Generation of genetically modified mice by oocyte injection of androgenetic haploid embryonic stem cells. Cell 2012; 149:605 - 17; http://dx.doi.org/10.1016/j.cell.2012.04.002; PMID: 22541431
- Hunter RH. Sperm-egg interactions in the pig: monospermy, extensive polyspermy, and the formation of chromatin aggregates. J Anat 1976; 122:43 - 59; PMID: 988013
- Balakier H. Tripronuclear human zygotes: the first cell cycle and subsequent development. Hum Reprod 1993; 8:1892 - 7; PMID: 8288756
- Han YM, Abeydeera LR, Kim JH, Moon HB, Cabot RA, Day BN, et al. Growth retardation of inner cell mass cells in polyspermic porcine embryos produced in vitro. Biol Reprod 1999; 60:1110 - 3; http://dx.doi.org/10.1095/biolreprod60.5.1110; PMID: 10208971
- Chen X, Luo Y, Fan Y, Yue L, Wu X, Chen Y, et al. Triploid and diploid embryonic stem cell lines derived from tripronuclear human zygotes. J Assist Reprod Genet 2012; 29:713 - 21; http://dx.doi.org/10.1007/s10815-012-9764-4; PMID: 22527897
- Malter HE, Cohen J. Embryonic development after microsurgical repair of polyspermic human zygotes. Fertil Steril 1989; 52:373 - 80; PMID: 2776891
- Ivakhnenko V, Cieslak J, Verlinsky Y. A microsurgical technique for enucleation of multipronuclear human zygotes. Hum Reprod 2000; 15:911 - 6; http://dx.doi.org/10.1093/humrep/15.4.911; PMID: 10739841
- Kattera S, Chen C. Normal birth after microsurgical enucleation of tripronuclear human zygotes: case report. Hum Reprod 2003; 18:1319 - 22; http://dx.doi.org/10.1093/humrep/deg262; PMID: 12773466
- Fan Y, Chen X, Luo Y, Chen X, Li S, Huang Y, et al. Developmental potential of human oocytes reconstructed by transferring somatic cell nuclei into polyspermic zygote cytoplasm. Biochem Biophys Res Commun 2009; 382:119 - 23; http://dx.doi.org/10.1016/j.bbrc.2009.02.143; PMID: 19265682
- Egli D, Rosains J, Birkhoff G, Eggan K. Developmental reprogramming after chromosome transfer into mitotic mouse zygotes. Nature 2007; 447:679 - 85; http://dx.doi.org/10.1038/nature05879; PMID: 17554301
- Gu YF, Lin G, Lu CF, Lu GX. Analysis of the first mitotic spindles in human in vitro fertilized tripronuclear zygotes after pronuclear removal. Reprod Biomed Online 2009; 19:745 - 54; http://dx.doi.org/10.1016/j.rbmo.2009.09.013; PMID: 20021725
- Wang F, Thirumangalathu S, Loeken MR. Establishment of new mouse embryonic stem cell lines is improved by physiological glucose and oxygen. Cloning Stem Cells 2006; 8:108 - 16; http://dx.doi.org/10.1089/clo.2006.8.108; PMID: 16776602
- Thompson JG, Simpson AC, Pugh PA, Donnelly PE, Tervit HR. Effect of oxygen concentration on in-vitro development of preimplantation sheep and cattle embryos. J Reprod Fertil 1990; 89:573 - 8; http://dx.doi.org/10.1530/jrf.0.0890573; PMID: 2401984
- Karja NW, Wongsrikeao P, Murakami M, Agung B, Fahrudin M, Nagai T, et al. Effects of oxygen tension on the development and quality of porcine in vitro fertilized embryos. Theriogenology 2004; 62:1585 - 95; http://dx.doi.org/10.1016/j.theriogenology.2004.03.012; PMID: 15511546
- Van Soom A, Yuan YQ, Peelman LJ, de Matos DG, Dewulf J, Laevens H, et al. Prevalence of apoptosis and inner cell allocation in bovine embryos cultured under different oxygen tensions with or without cysteine addition. Theriogenology 2002; 57:1453 - 65; http://dx.doi.org/10.1016/S0093-691X(01)00726-9; PMID: 12054204
- Dumoulin JC, Meijers CJ, Bras M, Coonen E, Geraedts JP, Evers JL. Effect of oxygen concentration on human in-vitro fertilization and embryo culture. Hum Reprod 1999; 14:465 - 9; http://dx.doi.org/10.1093/humrep/14.2.465; PMID: 10099995
- Kea B, Gebhardt J, Watt J, Westphal LM, Lathi RB, Milki AA, et al. Effect of reduced oxygen concentrations on the outcome of in vitro fertilization. Fertil Steril 2007; 87:213 - 6; http://dx.doi.org/10.1016/j.fertnstert.2006.05.066; PMID: 17081523
- Lengner CJ, Gimelbrant AA, Erwin JA, Cheng AW, Guenther MG, Welstead GG, et al. Derivation of pre-X inactivation human embryonic stem cells under physiological oxygen concentrations. Cell 2010; 141:872 - 83; http://dx.doi.org/10.1016/j.cell.2010.04.010; PMID: 20471072
- Chatzimeletiou K, Morrison EE, Prapas N, Prapas Y, Handyside AH. Spindle abnormalities in normally developing and arrested human preimplantation embryos in vitro identified by confocal laser scanning microscopy. Hum Reprod 2005; 20:672 - 82; http://dx.doi.org/10.1093/humrep/deh652; PMID: 15689349
- Dumoulin JM, Coonen E, Bras M, Bergers-Janssen JM, Ignoul-Vanvuchelen RC, van Wissen LC, et al. Embryo development and chromosomal anomalies after ICSI: effect of the injection procedure. Hum Reprod 2001; 16:306 - 12; http://dx.doi.org/10.1093/humrep/16.2.306; PMID: 11157825
- Palermo GD, Colombero LT, Rosenwaks Z. The human sperm centrosome is responsible for normal syngamy and early embryonic development. Rev Reprod 1997; 2:19 - 27; http://dx.doi.org/10.1530/ror.0.0020019; PMID: 9414462
- Sathananthan AH, Kola I, Osborne J, Trounson A, Ng SC, Bongso A, et al. Centrioles in the beginning of human development. Proc Natl Acad Sci USA 1991; 88:4806 - 10; http://dx.doi.org/10.1073/pnas.88.11.4806; PMID: 2052559
- Colombero LT. MM, Hariprashad J, Oquendo M, Rosenwaks Z and Palermo GD. Labelling and localization of the centriole/centrosome in intact and dissected spermatozoa. Fertil Steril 1996; S122
- Reverte CG, Benware A, Jones CW, LaFlamme SE. Perturbing integrin function inhibits microtubule growth from centrosomes, spindle assembly, and cytokinesis. J Cell Biol 2006; 174:491 - 7; http://dx.doi.org/10.1083/jcb.200603069; PMID: 16908668
- Ziebe S, Lundin K, Loft A, Bergh C, Nyboe Andersen A, Selleskog U, et al, CEMAS II and Study Group. FISH analysis for chromosomes 13, 16, 18, 21, 22, X and Y in all blastomeres of IVF pre-embryos from 144 randomly selected donated human oocytes and impact on pre-embryo morphology. Hum Reprod 2003; 18:2575 - 81; http://dx.doi.org/10.1093/humrep/deg489; PMID: 14645173
- Aran B, Blanco J, Vidal F, Vendrell JM, Egozcue S, Barri PN, et al. Screening for abnormalities of chromosomes X, Y, and 18 and for diploidy in spermatozoa from infertile men participating in an in vitro fertilization-intracytoplasmic sperm injection program. Fertil Steril 1999; 72:696 - 701; http://dx.doi.org/10.1016/S0015-0282(99)00307-6; PMID: 10521113
- Munné S, Grifo J, Cohen J, Weier HU. Chromosome abnormalities in human arrested preimplantation embryos: a multiple-probe FISH study. Am J Hum Genet 1994; 55:150 - 9; PMID: 8023843
- Pellestor F, Girardet A, Andréo B, Arnal F, Humeau C. Relationship between morphology and chromosomal constitution in human preimplantation embryo. Mol Reprod Dev 1994; 39:141 - 6; http://dx.doi.org/10.1002/mrd.1080390204; PMID: 7826614
- Findikli N, Kahraman S, Kumtepe Y, Donmez E, Benkhalifa M, Biricik A, et al. Assessment of DNA fragmentation and aneuploidy on poor quality human embryos. Reprod Biomed Online 2004; 8:196 - 206; http://dx.doi.org/10.1016/S1472-6483(10)60516-0; PMID: 14989798
- Liu L, Trimarchi JR, Keefe DL. Haploidy but not parthenogenetic activation leads to increased incidence of apoptosis in mouse embryos. Biol Reprod 2002; 66:204 - 10; http://dx.doi.org/10.1095/biolreprod66.1.204; PMID: 11751284
- Munné S. Chromosome abnormalities and their relationship to morphology and development of human embryos. Reprod Biomed Online 2006; 12:234 - 53; http://dx.doi.org/10.1016/S1472-6483(10)60866-8; PMID: 16478592
- Mitalipova M, Calhoun J, Shin S, Wininger D, Schulz T, Noggle S, et al. Human embryonic stem cell lines derived from discarded embryos. Stem Cells 2003; 21:521 - 6; http://dx.doi.org/10.1634/stemcells.21-5-521; PMID: 12968106
- Lerou PH, Yabuuchi A, Huo H, Takeuchi A, Shea J, Cimini T, et al. Human embryonic stem cell derivation from poor-quality embryos. Nat Biotechnol 2008; 26:212 - 4; http://dx.doi.org/10.1038/nbt1378; PMID: 18223642
- Baharvand H, Ashtiani SK, Taee A, Massumi M, Valojerdi MR, Yazdi PE, et al. Generation of new human embryonic stem cell lines with diploid and triploid karyotypes. Dev Growth Differ 2006; 48:117 - 28; http://dx.doi.org/10.1111/j.1440-169X.2006.00851.x; PMID: 16512855
- Ying QL, Nichols J, Chambers I, Smith A. BMP induction of Id proteins suppresses differentiation and sustains embryonic stem cell self-renewal in collaboration with STAT3. Cell 2003; 115:281 - 92; http://dx.doi.org/10.1016/S0092-8674(03)00847-X; PMID: 14636556
- Ying QL, Wray J, Nichols J, Batlle-Morera L, Doble B, Woodgett J, et al. The ground state of embryonic stem cell self-renewal. Nature 2008; 453:519 - 23; http://dx.doi.org/10.1038/nature06968; PMID: 18497825
- Brons IG, Smithers LE, Trotter MW, Rugg-Gunn P, Sun B, Chuva de Sousa Lopes SM, et al. Derivation of pluripotent epiblast stem cells from mammalian embryos. Nature 2007; 448:191 - 5; http://dx.doi.org/10.1038/nature05950; PMID: 17597762
- Chou YF, Chen HH, Eijpe M, Yabuuchi A, Chenoweth JG, Tesar P, et al. The growth factor environment defines distinct pluripotent ground states in novel blastocyst-derived stem cells. Cell 2008; 135:449 - 61; http://dx.doi.org/10.1016/j.cell.2008.08.035; PMID: 18984157
- James D, Levine AJ, Besser D, Hemmati-Brivanlou A. TGFbeta/activin/nodal signaling is necessary for the maintenance of pluripotency in human embryonic stem cells. Development 2005; 132:1273 - 82; http://dx.doi.org/10.1242/dev.01706; PMID: 15703277
- Monkhorst K, Jonkers I, Rentmeester E, Grosveld F, Gribnau J. X inactivation counting and choice is a stochastic process: evidence for involvement of an X-linked activator. Cell 2008; 132:410 - 21; http://dx.doi.org/10.1016/j.cell.2007.12.036; PMID: 18267073
- van den Berg IM, Laven JS, Stevens M, Jonkers I, Galjaard RJ, Gribnau J, et al. X chromosome inactivation is initiated in human preimplantation embryos. Am J Hum Genet 2009; 84:771 - 9; http://dx.doi.org/10.1016/j.ajhg.2009.05.003; PMID: 19481196
- Zhang X, Stojkovic P, Przyborski S, Cooke M, Armstrong L, Lako M, et al. Derivation of human embryonic stem cells from developing and arrested embryos. Stem Cells 2006; 24:2669 - 76; http://dx.doi.org/10.1634/stemcells.2006-0377; PMID: 16990582
- Mateizel I, De Temmerman N, Ullmann U, Cauffman G, Sermon K, Van de Velde H, et al. Derivation of human embryonic stem cell lines from embryos obtained after IVF and after PGD for monogenic disorders. Hum Reprod 2006; 21:503 - 11; http://dx.doi.org/10.1093/humrep/dei345; PMID: 16284066
- Sermon K, Van Steirteghem A, Liebaers I. Preimplantation genetic diagnosis. Lancet 2004; 363:1633 - 41; http://dx.doi.org/10.1016/S0140-6736(04)16209-0; PMID: 15145639
- Yu Y, Yan J, Li M, Yan L, Zhao Y, Lian Y, et al. Effects of combined epidermal growth factor, brain-derived neurotrophic factor and insulin-like growth factor-1 on human oocyte maturation and early fertilized and cloned embryo development. Hum Reprod 2012; 27:2146 - 59; http://dx.doi.org/10.1093/humrep/des099; PMID: 22532606
- Xia P. Intracytoplasmic sperm injection: correlation of oocyte grade based on polar body, perivitelline space and cytoplasmic inclusions with fertilization rate and embryo quality. Hum Reprod 1997; 12:1750 - 5; http://dx.doi.org/10.1093/humrep/12.8.1750; PMID: 9308806
- Dokras A, Sargent IL, Barlow DH. Human blastocyst grading: an indicator of developmental potential?. Hum Reprod 1993; 8:2119 - 27; PMID: 8150914
- Mai Q, Yu Y, Li T, Wang L, Chen MJ, Huang SZ, et al. Derivation of human embryonic stem cell lines from parthenogenetic blastocysts. Cell Res 2007; 17:1008 - 19; http://dx.doi.org/10.1038/cr.2007.102; PMID: 18071366