Abstract
Ubiquitin adducts surrounding DNA double-strand breaks (DSBs) have emerged as molecular platforms important for the assembly of DNA damage mediator and repair proteins. Central to these chromatin modifications lies the E2 UBC13, which has been implicated in a bipartite role in priming and amplifying lys63-linked ubiquitin chains on histone molecules through coupling with the E3 RNF8 and RNF168. However, unlike the RNF8-UBC13 holoenyzme, exactly how RNF168 work in concert with UBC13 remains obscure. To provide a structural perspective for the RNF168-UBC13 complex, we solved the crystal structure of the RNF168 RING domain. Interestingly, while the RNF168 RING adopts a typical RING finger fold with two zinc ions coordinated by several conserved cystine and histine residues arranged in a C3HC4 “cross-brace” manner, structural superimposition of RNF168 RING with other UBC13-binding E3 ubiquitin ligases revealed substantial differences at its corresponding UBC13-binding interface. Consistently, and in stark contrast to that between RNF8 and UBC13, RNF168 did not stably associate with UBC13 in vitro or in vivo. Moreover, domain-swapping experiments indicated that the RNF8 and RNF168 RING domains are not functionally interchangeable. We propose that RNF8 and RNF168 operate in different modes with their cognate E2 UBC13 at DSBs.
Introduction
Like many other covalent modification systems such as methylation, acetylation and phosphorylation, protein ubiquitylation has emerged as a critically important mechanism for spatiotemporal regulation of a repertoire of biological processes. A ubiquitylation reaction generally involves the concerted effort of three families of enzymes, namely ubiquitin-activating enzyme (E1), ubiquitin-conjugating enzyme (E2) and ubiquitin ligase (E3).Citation1 Apart from attaching to its substrates as monomers (mono-ubiquitylation), ubiquitin moieties can polymerise via each of its seven lysine residues to produce polyubiquitin chains. More recent studies also indicated that ubiquitin chains can be orientated in a head-to-tail manner (linear ubiquitin chains). Among these linkage-specific entities, K48-linked ubiquitin chains are associated with proteasome targeting, whereas ubiquitin polymers linked via K63-linkages play critical roles in protein kinase activation, protein trafficking and DNA damage response.Citation2-Citation5
DNA double-strand break (DSB) is one of the most lethal types of DNA lesions. Failure to repair DSBs contributes to genome instability and a host of human diseases, including cancer and immunodeficiency.Citation6,Citation7 To counter the deleterious effects of DSBs on genome integrity, cells have evolved elaborate protein networks that halt cell cycle progression and initiate DNA repair in response to genotoxic stress.Citation8 To this end, cells signal for DSB repair by initiating an H2AX-dependent signaling cascade that culminates into the assembly of a cohort of DNA damage mediator and repair proteins.Citation9 Accordingly, DSB signals are transmitted and amplified through a series of post-translational modifications, and recent studies have highlighted on usage of non-degradative ubiquitin chains catalyzed by the E3 ubiquitin ligases RNF8 and RNF168.Citation10
The ring finger protein RNF8 is tethered to DSBs via its N-terminal FHA domain that targets phosphorylated MDC1. Through its RING-dependent interaction with its cognate E2 UBC13, RNF8 conjugates ubiquitin adducts onto H2A-type histonesCitation11-Citation14 and allows recruitment of the second E3 ubiquitin ligase, namely the RIDDLE syndrome protein RNF168, to further amplify the signals.Citation15,Citation16 The current working model envisions that RNF168, in concert with UBC13, extends and spreads the RNF8-primed ubiquitin signals and, as such, represents the primary K63-ubiquitylating activity at DSBs important for assembly of downstream components, including tumor suppressors BRCA1 and 53BP1.Citation17,Citation18 While recent reports have identified a number of negative regulators of the RNF8/RNF168-dependent ubiquitin signaling pathway,Citation19-Citation23 mechanistically how the RNF168-UBC13 complex amplifies ubiquitin signals has remained obscure. What are the fundamental differences between RNF8-UBC13 and RNF168-UBC13?
In an attempt to dissect roles of RNF168, we solved the crystal structure of the RNF168 RING domain (residues 1–111, referred to as RNF168N-111). We found that the core RING domain of RNF168 adopted a typical RING finger fold comprising one central α helix, two antiparallel β strands and two long loops that are stabilized by the coordination of two zinc ions. Intriguingly, by comparing the structure of RNF168N-111 to the complex structures of TRAF6-UBC13, CHIP-UBC13 and RNF8-UBC13-MMS2, we identified a number of structural determinants on RNF168N-111 that may preclude its binding to UBC13. Our in vitro and in vivo data suggest that the RING domains of RNF8 and RNF168 may operate in different modes in the ubiquitin-dependent DNA damage signaling pathway.
Results
Consistent with a previous study reporting RNF168 as the primary ubiquitylating activity at DSBs,Citation18 we found that ectopic expression of RNF168, but not RNF8, promoted formation of ubiquitin adducts at DSBs, as indicated by FK2 and ubiquitylated-H2A (uH2A) foci (Fig. S1A and B). Moreover, depletion of RNF168 in HeLa cells resulted in drastic reduction of damage-induced ubiquitylation of γH2AX (Fig. S1C).Citation17 While these data support the general consensus that RNF168 amplifies the RNF8-primed ubiquitin signals at DSBs, it is not clear why UBC13, the same E2 that function with both RNF8 and RNF168, seems to exhibit much higher activity when paired with RNF168.
Overall structure of RNF168N-111
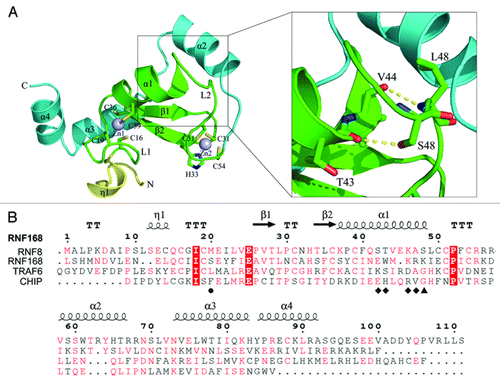
To provide a structural basis for its robust ubiquitylating activity, we solved the crystal structure of RNF168 RING domain (RNF168N-111). The overall structure of RNF168N-111 contains ordered residues from L3 to S93. The residues from G94 to S111 could not be observed in the electron density map, indicating high flexibility of this region. The structure of RNF168N-111 contains a core RING domain (residues 16–55) followed by three α helices flanking on one side of the RING domain. The core RING domain of RNF168 adopts the typical RING fold comprising of one central α helix (α1), two antiparallel β strands (β1 and β2) and two long loops (L1 and L2). The two β strands and one of the two long loops L1 are located on the N-terminal side of the central α helix, while loop L2 was found on the C-terminal side of the central α helix (). Similar to other RING domain structures, the long loops L1 and L2 are stabilized by coordination of two zinc ions. Generally, the conserved motif CX2CX(9–39)CX(1–3)HX(2–3)CX2CX(4–48)CX2C (where C is Cys, H is His and X is any residues) is observed to coordinate the two zinc ions in the RING domain structures.Citation24 Correspondingly, the signature motif comprising C16, C19, C36, C39, C31, H33, C51 and C54 was also identified in RNF168 RING domain structure. These defined cystine and histidine residues are arranged in a C3HC4 “cross-brace” manner to chelate the two zinc ions that stabilizes the conformations of loops L1 and L2 and enables correct folding of the RING domain ().Citation25 It is interesting to find that the last turn of helix α1 is distorted in the structure of RNF168N-111. In this region, the side chain of S48 forms a hydrogen bond with the main chain carbonyl group of T43, leading to a bent conformation of the central α helix. In addition, the main chain carbonyl group of V44 forms a hydrogen bond with the amide group of L49, which further stabilizes the distorted conformation of helix α1 (). Sequence alignment of the RING domain of RNF168 with several other E3 ubiquitin ligases capable of binding UBC13 with known structures shows that the serine residue is not conserved, indicating that the distorted central helix is a novel structural feature of the RNF168 RING domain ().
Figure 1. Overall structure of RNF8N-111. (A) Ribbon representation of the structure of RNF168N-111. The enlarged view of the interaction of helix α1 with the followed loop is shown on the right side. Amino acid residues are shown in a stick mode. Zinc ions are shown in a sphere mode. The core RING domain, the fragments preceding and following the RING domain are colored green, yellow and cyan, respectively. (B) Structure-based sequence alignment of RNF168 RING domain, TRAF6 RING domain, CHIP U-box domain and RNF8 RING domain. The conserved residues are in red and strictly conserved residues are highlighted red. The residues involved in the interaction with UBC13 on the central α helix are labeled by diamond. M20 and S48 of RNF168 and corresponding residues of TRAF6, CHIP and RNF8 are labeled by circle and triangle, respectively.
Comparison of the structure of RNF168N-111 with other E3 ubiquitin ligases capable of binding to UBC13
The interactions between E3 ligases and E2 enzymes play important roles in promoting ubiquitin transfer from E2-ubiquitin complex to its substrates. RNF168 was previously reported to interact with UBC13, and in concert with UBC13, catalyze K63-linked polyubiquitin chains on H2A-type histones.Citation15,Citation16 To gain further insight into its binding to UBC13, the structure of RNF168N-111 was compared with the structures of E3 ligases in complex with UBC13, including the structures of TRAF6 (tumor necrosis factor receptor-associated factor 6)-UBC13 and CHIP (C-terminal of Hsp70-interacting protein)-UBC13. TRAF6 is a RING domain E3 ligase while CHIP is an E3 ligase containing a C-terminal U-box domain that is structurally similar to RING domain lacking the metal chelating residues conserved in RING domains.Citation26,Citation27
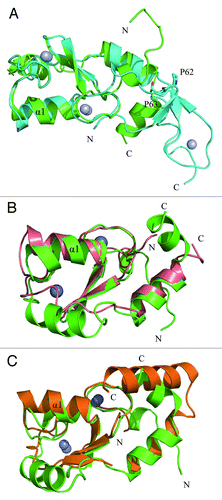
Superimposition of the RING domain structure of RNF168N-111 to the RING domain of TRAF6 and the U-box domain of CHIP showed that these structures are similar with the RMSD (root-mean-square deviation) values of 2.6 Å and 2.2 Å over aligned Cα atoms of TRAF6 (68 Cα atoms) and CHIP (69 Cα atoms). However, the conformations of the N-terminal fragment preceding the RING domain in these structures are different. There are two sequential proline residues in the sequence of TRAF6 and a sharp turn occurs in the region. Residues 59–61 in this fragment of TRAF6 form a β strand to interact with the β hairpin in the zinc finger region further stabilizing the U-shaped conformation. Consequently, the N-terminal fragment of TRAF6 folds back to interact with the RING domain and the zinc finger domain (). In RNF168N-111 and CHIP, the N-terminal sequence preceding the RING domain does not carry two sequential proline residues, so that the fragment adopts more extended conformation, protruding outside the RING and U-Box domains (). As described above, the last turn of helix α1 in RNF168N-111 is distorted leading to a bent conformation of the central α helix. The position of the last turn of the central α helix in RNF168N-111 shifts away from the metal binding sites that are adjacent to the interface between E2 and E3 enzyme complex compared with TRAF6 and CHIP (). In addition, a complex structure of RNF8-UBC13-MMS2 at 4.8 Å resolution was reported recently.Citation28 In the RING domain structure of RNF8, the central α helix adopts normal and unbent conformation in comparison to RNF168 ().
Figure 2. Comparison of the structure of RNF168N-111 with TRAF6, CHIP and RNF8. Superposition the structure of RNF168N-111 to (A) TRAF6 (PDB code: 3HCT), (B) CHIP (PDB code: 2C2V), (C) RNF8 (PDB code: 4EPO). Residues are shown in a stick mode. Metal ions are shown in a sphere mode. RNF168, TRAF6, CHIP and RNF8 are colored green, cyan, pink and brown, respectively.
Structure of the binding site of RNF168N-111 to UBC13
Since the overall structure of the RING domain of RNF168 is similar to the RING domain of TRAF6 and the U-box domain of CHIP, we speculated that the RNF168 RING domain should interact with UBC13 via the same binding site. We then conducted further analysis to compare the binding interface between UBC13 and its E3 ligases TRAF6, CHIP or RNF168N-111. Several residues within the RING domain of TRAF6, including E69, P71, I72, L74, M75, A101 and P106, are involved in the interaction with UBC13 on the interface between these two proteins.Citation26 Among these residues, L74 in loop L1 is entirely buried in a hydrophobic pocket on the UBC13 surface (, left panel). The corresponding residue of CHIP U-box domain is F238, which interacts with UBC13 in a similar manner (, middle panel). In RNF168, the residue is substituted by M20 ( and ). The side chain of M20 is longer than the side chains of L74 in TRAF6 and F238 in CHIP, respectively. The longer side chain of M20 of RNF168N-111 might cause steric clash, disrupting the interaction between the RNF168N-111 and UBC13 (, left and middle panel). Comparison of the RING domain of RNF168 to RNF8 in the complex structure of RNF8-UBC13-MMS2 revealed that the corresponding residue of RNF8 ring domain to M20 is S407 with much shorter side chain. The longer side chain of M20 is unable to fit in the binding pocket well and prevent the binding of RNF168 to UBC13 (, right panel).
Figure 3. Structural basis for the lack of interaction between RNF168N-111 and UBC13. (A) Comparison of the binding of loop L1 of TRAF6 (left panel), CHIP (middle panel) and RNF8 (right panel) to the hydrophobic biding pocket on UBC13 surface with RNF168N-111. (B) Comparison of the binding of the C-terminal end of the central α helix of TRAF6 (left panel), CHIP (middle panel) and RNF8 (right panel) to the hydrophobic pocket on UBC13 surface with RNF168N-111. (C) Comparison of the interaction between UBC13 and TRAF6 (left panel), CHIP (middle panel) and RNF8 (right panel) to RNF168N-111. RNF168, TRAF6, CHIP and RNF8 are shown in a ribbon mode and colored green, cyan, pink and brown, respectively. Residues are shown in a stick mode. UBC13 is shown in surface representation in (A and B). White, blue and red regions indicate neutral areas, positively charged areas and negatively charged areas, respectively. UBC13 is shown in a ribbon mode and colored yellow in (C). The PDB codes of TRAF6, CHIP and RNF8 in complex with UBC13 are 3HCT, 2C2V and 4EPO, respectively.
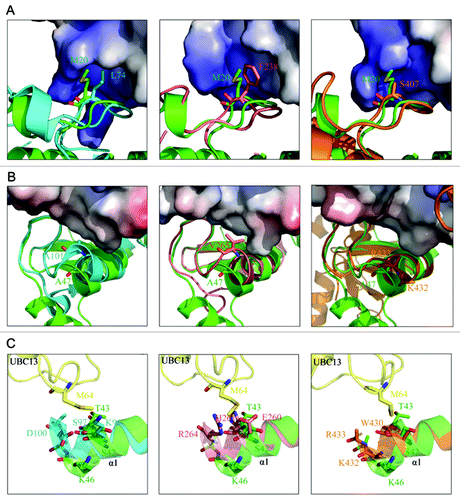
The central helix α1 of RNF168 is bent compared with TRAF6, CHIP and RNF8 because of the hydrogen bonds formed between S48 and T43 and V44 and L49 ( and ). The hydrogen bond between S48 and T43 is considered to be the critical driving force for the formation of the bent central α helix of RNF168 RING domain. Because the sequence alignment result showed that corresponding residues of TRAF6, CHIP and RNF8 are glycine and lysine, respectively, lacking the side chain to form the same hydrogen bond (), the central α helices of TRAF6, CHIP and RNF8 adopt normal conformation (). Consequently, the C-terminal end of the central α helix of RNF168 RING domain shifts away from the binding interface of the E2 and E3 enzyme complexes (). When the structures of these proteins are superposed, the central helix of RNF168 rotates about 21°, 21° and 19°, and the C terminus shifts 4.1 Å, 4.7 Å and 6.1 Å relative to TRAF6, CHIP and RNF8, respectively (Fig. S2A–C). At this binding site, A101 of TRF6 and V265 of CHIP at the end of the central α helix are buried in a hydrophobic pocket on the surface of UBC13, respectively. A47 of RNF168 at the corresponding site is located farther away from the binding pocket because of the bent central α helix (, left and middle panel), suggesting that RNF168N-111 may exhibit reduced affinity for UBC13. In the structure of RNF8-UBC13-MMS2 complex, two residues, K432 and R433 with positive charges, are located at this site. Although the side chains of K432 and R433 are invisible, they may interact with UBC13 by salt bridges since UBC13 bears negative charges nearby (, right panel). The interactions at this site between RNF8 and UBC13 are different from those observed in TRAF6-UBC13 and CHIP-UBC13 complexes. However, the bent conformation of central α helix of RNF168 causes A47 shifting away from the interacting interface of UBC13 in a similar way when compared with RNF8 (, right panel).
In addition, M64 of UBC13 formed van der Waals interactions with the side chains of residues K96, S97 and D100 and E260, H261 and R264 on the central α helix of TRAF6 and CHIP, respectively. In the structure of RNF8-UBC13-MMS2 complex, this binding pocket is formed by E429, W430, K432 and R433 of RNF8. The corresponding residues of RNF168 are S42, T43 and K46, whose side chains are capable of forming van der Waals, contacts with M64 of UBC13. Because of the bent conformation of the central α helix, the distance between this binding pocket of RNF168 to M64 of UBC13 is longer in comparison to TRAF6, CHIP and RNF8 ().
RNF168 does not stably associate with UBC13
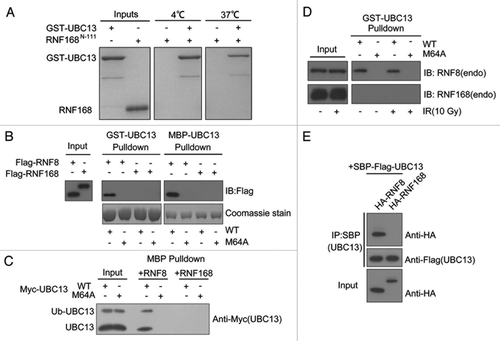
RNF168 is a RING domain-containing factor that cooperates with RNF8 to catalyze K63-linked ubiquitin chains on histone H2A and H2AX in response to DSB.Citation15,Citation29 Generally, the RING domain of E3 ubiquitin ligase promotes ubiquitin transfer by bringing charged E2 enzymes into close proximity with its substrates. In this regard, we examined the interaction between RNF168N-111 and UBC13 by GST pull-down assay. Consistent with our prediction based on structural analysis, we were unable to detect any interaction between RNF168N-111 and UBC13 either at 4°C or 37°C (). To more rigorously examine the interaction between RNF168 and UBC13, we performed further interaction studies using RNF8 as control, which has been shown extensively to interact with UBC13.Citation30-Citation32 Bacterially expressed and purified GST-UBC13 or MBP-UBC13 proteins, and their respective M64A mutants, were incubated with lysates derived from cells overexpressing RNF8 or RNF168. Pull-down experiments demonstrated robust RNF8-UBC13 interactions, which is dependent on the UBC13 M64 residue. By contrast, UBC13 proteins did not detectably associate with RNF168 (). The same was true when bacterially expressed and purified MBP-RNF8 or RNF168 fusion proteins were incubated with lysates derived from cells expressing either wild-type UBC13 or its M64A mutant (). In addition, DNA damage did not affect the binding between RNF8 and UBC13, and failed to promote any stable interaction between RNF168 and UBC13 (). Finally, by use of co-immunoprecipitation, we found that RNF8, but not RNF168, associated with UBC13 (). Together, we concluded that RNF168 does not stably associate with UBC13.
Figure 4. RNF168 does not stably associate with UBC13. (A) The interaction between RNF168N-111 and UBC13 analyzed by GST pull-down assay at 4°C and 37°C. (B) Agarose conjugated with bacterially expressed and purified GST-UBC13 or MBP-UBC13 proteins, and their corresponding M64A mutants, were incubated with lysates derived from cells expressing Flag-RNF8 or Flag-RNF168 for 4 h at 4°C with gentle agitation. Thereafter, agarose beads were washed three times with NETN buffer and proteins were separated by SDS-PAGE. Western blotting experiments were done using indicated antibodies. (C) MBP-RNF8 or MBP-RNF168 protein were incubated with lysates derived from cells expressing myc-tagged UBC13 wild-type (WT) or its M64A mutant. Bound complexes were washed and analyzed using anti-myc (9E10) antibodies. (D) GST-UBC13 proteins were incubated with lysates derived from irradiated cells or untreated cells. Bound proteins were analyzed using indicated antibodies. (E) 293T cells were co-transfected with indicated constructs. Cells were subsequently lysed using NETN buffer, and streptavidin beads were used to precipitate UBC13 protein complexes. Proteins were separated by SDS-PAGE and were analyzed by western blotting using indicated antibodies.
The RING domains of RNF168 and RNF8 are not functionally interchangeable
Our observations that RNF168 does not stably associate with UBC13 suggested that RNF8 and RNF168 may have evolved to interact with UBC13 in different modes. To strengthen this possibility, we performed domain-swapping experiments to examine whether the RNF8 and RNF168 RING domains were interchangeable in the DNA damage signaling pathway. We generated chimeric constructs where the RNF8 RING deletion mutant was fused with the RNF168 RING (), and reconstituted RNF8-deficient MEF cells to assess whether these ectopically expressed proteins may restore RNF8 function in promoting FK2 and 53BP1 foci formation. Expression of RNF8, but not its RING deletion mutant, restored FK2 and 53BP1 foci (). This was also true in cells expressing RNF8ΔRING-ChfrRING.Citation32 By contrast, replacing the RNF8 RING with that of RNF168 did not restore RNF8 functions in vivo, supporting our hypothesis in via inherent differences exists between the RNF8 and RNF168 RING domains. Consistently, we found that GST-UBC13 proteins precipitated wild-type RNF8 and RNF8ΔRING-ChfrRING, but not RNF8ΔRING nor RNF8ΔRING-RNF168RING ().
Figure 5. RNF8 and RNF168 RING domains are not functionally interchangeable. (A and D) Schematic illustrations of RNF8 and RNF168 chimeric constructs. (B and E) RNF8−/− MEFs or RIDDLE cells (RNF168 mutant cells) reconstituted with indicated constructs were irradiated (10 Gy) and were processed for immunofluorescent experiments to assess FK2 and 53BP1 foci formation. (C) GST-UBC13 interacted with RNF8 and Chfr RING but not RNF168 RING. Agarose beads conjugated with GST-UBC13 proteins were incubated with lysates derived from cells expressing indicated proteins. Bound protein complexes were separated by SDS-PAGE and analyzed by western blotting using indicated antibodies.
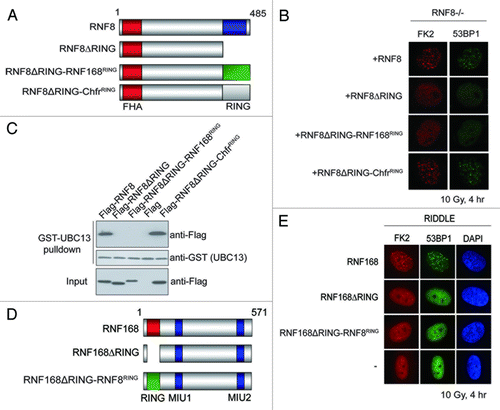
We also performed reconstitution experiments in RNF168 mutant RIDDLE cellsCitation33 using an RNF168-expressing construct in which its RING domain was replaced with that from RNF8 (). While expression of wild-type RNF168 restored FK2 and 53BP1 foci formation, reconstitution of RNF168ΔRING or RNF168ΔRING-RNF8RING did not ().
Discussion
In concert with its cognate E2 UBC13, the E3 ubiquitin ligase RNF168 has been implicated in DNA signal propagation by catalyzing K63-linked ubiquitin chains onto histone molecules at the vicinity of DSBs.Citation15,Citation16 To provide a structural perspective into its ubiquitylating property, we solved the crystal structure of the RNF168 RING domain. Although the overall structure of RNF168 RING domain is similar to the reported structures of RING domain and U-box domain of E3 ligases (), several unique structural features have been identified by comparing the structure of RING domain of RNF168 to TRAF6 CHIP and RNF8.Citation26-Citation28 It was shown previously that several residues of TRAF6, including E69, P71, I72, L74, M75, A101 and P106, are important for the binding of TRAF6 to UBC13.Citation26 Among these residues, L74 is buried in a hydrophobic pocket on the surface of UBC13 (, left panel). The corresponding residues are M20 of RNF168, S407 of RNF8 and F238 of CHIP, respectively ( and ). The side chain of M20 is longer than S407, L74 and F238 in RNF8, TRAF6 and CHIP, respectively, which might disrupt the interaction between RNF168 and UBC13 because of the steric hindrance effect. Interestingly, mutations of L74 in TRAF6 to glutamic acid or lysine residues with longer side chains totally abolish the binding of TRAF6 to UBC13.Citation26 Although the glutamic acid and lysine residues are able to disrupt the hydrophobic interaction between TRAF6 and UBC13, the steric hindrance effect of the longer side chains may also play an important role in regulating the interaction. Another peculiar structural feature on the RNF168 RING domain is that the hydrogen bonds formed between S48 and T43 and V44 and L48 result in a bent conformation of the central α helix ( and ). Consequently, the residue A47 located at the end of the central α helix shifts away from the predicted interaction interface, leading to weaker association between RNF168 and UBC13 (). In addition, several residues of the central α helix in TRAF6, CHIP and RNF8 are involved in the formation of hydrophobic interaction with M64 of UBC13 ().Citation26-Citation28 The bent conformation of the central α helix of RNF168 might disrupt its interaction with M64 of UBC13, further weakening the association between RNF168 and UBC13.
RNF168 and UBC13 co-localize at DSBs to catalyze ubiquitin conjugates onto H2A-type histones in response to DNA damage. Since our data indicated that RNF168 does not stably bind to UBC13, the interaction between these two proteins may be transient in vivo. Another possibility is that some unknown proteins may exist to stabilize the association of RNF168 with UBC13. As shown before, HERC2 facilitates the assembly of RNF8-UBC13 complex to promote RNF8-mediated ubiquitylation at sites of DSBs.Citation34 Since HERC2 binds and stabilizes RNF168,Citation34 it may have a similar role in promoting the assembly of RNF168-UBC13 complex in vivo. A recent report reveal that RNF168 is unable to promote the formation of a ubiquitin chain on histone H2A in the presence of UBC13, indicating that RNF168 may recruit E2 enzymes other than UBC13 to propagate the signals at DSBs.Citation35 Together, with our observations that RNF168 is unable to associate with UBC13 stably, suggests an alternative possibility that RNF168 may recruit other E2 enzymes to ubiquitinate histone H2A at DSBs.
Both RNF8 and RNF168 have been reported to promote histone H2A ubiquitylation in concert with UBC13 in vivo.Citation11-Citation13,Citation15,Citation16 Deletion of the RING domain of RNF168 impaired the formation of ubiquitin adducts and compromised the accumulation of DNA damage repair proteins at DSB sites.Citation11,Citation28 While genetic studies suggest that RNF8 and RNF168 are recruited to DSBs sequentially to catalyze a bipartite mode of histone ubiquitylation events, a recent report from the Sixma lab provided compelling biochemical evidence to challenge this view, implicating RNF168 as the “initiator” in the step-wise amplification of H2A-type histones at DSBs.Citation35 Regardless, our observations that RNF168 RING cannot compensate RNF8 RING (vice versa) in the DNA damage signaling pathway support our hypothesis in which the two E3 ubiquitin ligases operate in different modes, with UBC13 in the ubiquitin-dependent DNA damage signaling pathway.
During the preparation of this manuscript, Glover and colleagues reported their structural study on the RNF168 RING domain.Citation28 Our study not only compliments their work, but considering the emerging evidence that reinstates the lys63-ub-promoting activity to the RNF8-UBC13 holoenzyme, it is tempting to speculate that ubcH5c, or a yet-to-be-identified E2, may participate in the early stages of DSB signaling. Further work will be necessary to fully elucidate the differences between RNF8 and RNF168 in catalyzing DSB-associated ubiquitylation events, and how they contribute to the maturing of the ubiquitin landscape at DSBs.
Materials and Methods
Cloning, overexpression and purification
The DNA fragment encoding RNF168N-111 was amplified by PCR using human fetal brain cDNA library as template. The PCR product was digested with enzymes of BamHI/XhoI and ligated into the pET-22b vector with a 6 × His-tag fused to its N terminus. The resulting plasmid was confirmed by DNA sequencing. The expression plasmid containing the RING domain of RNF168 was transformed into E. coli Rosetta 2 (DE3) cells. The E. coli cells were grown at 37°C in LB media supplemented with 100 μg/ml ampicillin until OD600nm reached 0.8. One-tenth mM IPTG was added into the media and the cells were further incubated for 20 h at 16°C.
The cells were harvested and resuspended with lysis buffer (50 mM Tris-HCl, pH 7.5, 500 mM NaCl, 10 mM imidazole). The cells were lysed using ultrasonication on ice and the lysate was centrifuged at 12,000 rpm for 30 min. The supernatant was loaded onto a Ni-NTA nickel-chelating column (QIAGEN) pre-equilibrated with lysis buffer. The column was washed by 50 column volumes of washing buffer (50 mM Tris-HCl, pH 7.5, 500 mM NaCl and 50 mM imidazole) to remove contaminant. Target protein was eluted by elution buffer (50 mM Tris-HCl, pH 7.5, 500 mM NaCl, 250 mM imidazole). The eluted protein was subjected to a HiLoad 16/60 superdex 75 column (GE Healthcare) pre-equilibrated with the column buffer (20 mM Tris-HCl, pH 7.5, 200 mM NaCl, 1 mM DTT). The eluted fraction containing RNF168N-111 was concentrated to 3 mg/ml for further use. All steps of protein purification were performed at 4°C.
Crystallization and data collection
The purified RNF168N-111 was crystallized using sitting drop vapor diffusion method at 18°C. Initial crystal trials were performed using Crystal Screen, Crystal Screen II, Index (Hampton Research), Wizard I and Wizard II (Emerald Biosystems). Finally, the diffraction quality crystals were grown in the buffer containing 1.6 M sodium malonate, pH 7.0. The crystals of RNF168N-111 were cryoprotected by the buffer containing 1.6 M sodium malonate and 10% glycerol. The crystal was flash cooled in liquid nitrogen and subjected to X-ray diffraction data collection at 100K on synchrotron beamline BL17U1 of SSRF (Shanghai). Diffraction data were processed with HKL2000 software package.Citation36
Structure determination and refinement
The structure of RNF168N-111 was solved by using single wavelength anomalous diffraction of the intrinsic zinc atoms. The zinc ions were located and the initial phases were calculated using the PHENIX program package. Initial structure model was built automatically using AUTOBUILD in the PHENIX program package.Citation37 The model was refined to 2.6 Å using program REFMAC5 and COOT alternatively resulting in the final model with an R-factor of 22.28% (R-free = 27.61%).Citation38,Citation39 The quality of the final structure model was analyzed with the PROCHECK program.Citation40 The coordinates and structure factors are deposited in the Protein Data Bank under the accession code 4GB0. The data processing and structure determination statistics are listed in .
Table 1. Data collection and structure refinement statistics
Pulldown assay
UBC13 (Homo sapiens) was cloned into pGEX-6P-1 vector with a GST-tag fused to its N terminus and overexpressed in E. coli Rosetta2 (DE3) cells. GST-UBC13 was purified with Glutathione Sepharose beads (GE Healthcare) in PBS buffer. Purified GST-UBC13 was incubated with 20 μl Glutathione Sepharose beads pre-equilibrated with buffer A containing 137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 2 mM KH2PO4, 1 mM DTT, pH 7.4. GST-UBC13 immobilized with Glutathione Sepharose beads was incubated with purified RNF168N-111. After incubation at 4°C or 37°C for 60 min, the beads were washed with buffer A three times. Beads were boiled with SDS-sample buffer and proteins retained on the Glutathione Sepharose beads were analyzed on SDS-PAGE.
Immunofluorescence studies
Cells grown on coverslips were irradiated (10 Gy) and were subsequently fixed with 3% paraformaldehyde for 15 min at room temperature. Cells were washed with PBS and permeabilised with 0.5% triton solution for 1 min. After washing with PBS, cells were immunostained with primary and secondary antibodies for 30 min, respectively. Nuclei were visualized by staining with DAPI. Coverslips were mounted and IR-induced foci were visualized using an Olympus BX51 fluorescence microscope.
Additional material
Download Zip (4.5 MB)Acknowledgments
We thank the staff at beamline BL17U1 of the Shanghai Synchrotron Radiation Facility for assistance with data collection, and Dr. Grant Stewart for the RIDDLE cells. This work was supported by the Chinese Ministry of Science and Technology (Project Nos. 2009CB825502 and 2012CB917202 to J.Z.), the Ministry of Education of China (Ph.D. Programs Foundation Project No. 20113402110033 to J.Z.), the National Natural Science Foundation of China (Project Nos. 31171241 and 30970576 to J.Z.), the Chinese Academy of Sciences (“100 Talents Program” to J.Z.) and the Research Grant Council Hong Kong (General Research Fund Project No. 767811 to M.S.Y.H.).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- Pickart CM, Eddins MJ. Ubiquitin: structures, functions, mechanisms. Biochim Biophys Acta 2004; 1695:55 - 72; http://dx.doi.org/10.1016/j.bbamcr.2004.09.019; PMID: 15571809
- Haglund K, Dikic I. Ubiquitylation and cell signaling. EMBO J 2005; 24:3353 - 9; http://dx.doi.org/10.1038/sj.emboj.7600808; PMID: 16148945
- Thrower JS, Hoffman L, Rechsteiner M, Pickart CM. Recognition of the polyubiquitin proteolytic signal. EMBO J 2000; 19:94 - 102; http://dx.doi.org/10.1093/emboj/19.1.94; PMID: 10619848
- Ulrich HD, Walden H. Ubiquitin signalling in DNA replication and repair. Nat Rev Mol Cell Biol 2010; 11:479 - 89; http://dx.doi.org/10.1038/nrm2921; PMID: 20551964
- Huang TT, D’Andrea AD. Regulation of DNA repair by ubiquitylation. Nat Rev Mol Cell Biol 2006; 7:323 - 34; http://dx.doi.org/10.1038/nrm1908; PMID: 16633336
- Negrini S, Gorgoulis VG, Halazonetis TD. Genomic instability--an evolving hallmark of cancer. Nat Rev Mol Cell Biol 2010; 11:220 - 8; http://dx.doi.org/10.1038/nrm2858; PMID: 20177397
- Jackson SP, Bartek J. The DNA-damage response in human biology and disease. Nature 2009; 461:1071 - 8; http://dx.doi.org/10.1038/nature08467; PMID: 19847258
- Ciccia A, Elledge SJ. The DNA damage response: making it safe to play with knives. Mol Cell 2010; 40:179 - 204; http://dx.doi.org/10.1016/j.molcel.2010.09.019; PMID: 20965415
- Huen MS, Chen J. Assembly of checkpoint and repair machineries at DNA damage sites. Trends Biochem Sci 2010; 35:101 - 8; http://dx.doi.org/10.1016/j.tibs.2009.09.001; PMID: 19875294
- Polo SE, Jackson SP. Dynamics of DNA damage response proteins at DNA breaks: a focus on protein modifications. Genes Dev 2011; 25:409 - 33; http://dx.doi.org/10.1101/gad.2021311; PMID: 21363960
- Huen MSY, Grant R, Manke I, Minn K, Yu XC, Yaffe MB, et al. RNF8 transduces the DNA-damage signal via histone ubiquitylation and checkpoint protein assembly. Cell 2007; 131:901 - 14; http://dx.doi.org/10.1016/j.cell.2007.09.041; PMID: 18001825
- Mailand N, Bekker-Jensen S, Faustrup H, Melander F, Bartek J, Lukas C, et al. RNF8 ubiquitylates histones at DNA double-strand breaks and promotes assembly of repair proteins. Cell 2007; 131:887 - 900; http://dx.doi.org/10.1016/j.cell.2007.09.040; PMID: 18001824
- Kolas NK, Chapman JR, Nakada S, Ylanko J, Chahwan R, Sweeney FD, et al. Orchestration of the DNA-damage response by the RNF8 ubiquitin ligase. Science 2007; 318:1637 - 40; http://dx.doi.org/10.1126/science.1150034; PMID: 18006705
- Wang B, Elledge SJ. Ubc13/Rnf8 ubiquitin ligases control foci formation of the Rap80/Abraxas/Brca1/Brcc36 complex in response to DNA damage. Proc Natl Acad Sci USA 2007; 104:20759 - 63; http://dx.doi.org/10.1073/pnas.0710061104; PMID: 18077395
- Doil C, Mailand N, Bekker-Jensen S, Menard P, Larsen DH, Pepperkok R, et al. RNF168 binds and amplifies ubiquitin conjugates on damaged chromosomes to allow accumulation of repair proteins. Cell 2009; 136:435 - 46; http://dx.doi.org/10.1016/j.cell.2008.12.041; PMID: 19203579
- Stewart GS, Panier S, Townsend K, Al-Hakim AK, Kolas NK, Miller ES, et al. The RIDDLE syndrome protein mediates a ubiquitin-dependent signaling cascade at sites of DNA damage. Cell 2009; 136:420 - 34; http://dx.doi.org/10.1016/j.cell.2008.12.042; PMID: 19203578
- Sy SM, Jiang J, Dong SS, Lok GT, Wu J, Cai H, et al. Critical roles of ring finger protein RNF8 in replication stress responses. J Biol Chem 2011; 286:22355 - 61; http://dx.doi.org/10.1074/jbc.M111.232041; PMID: 21558560
- Feng L, Chen J. The E3 ligase RNF8 regulates KU80 removal and NHEJ repair. Nat Struct Mol Biol 2012; 19:201 - 6; http://dx.doi.org/10.1038/nsmb.2211; PMID: 22266820
- Poulsen M, Lukas C, Lukas J, Bekker-Jensen S, Mailand N. Human RNF169 is a negative regulator of the ubiquitin-dependent response to DNA double-strand breaks. J Cell Biol 2012; 197:189 - 99; http://dx.doi.org/10.1083/jcb.201109100; PMID: 22492721
- Gudjonsson T, Altmeyer M, Savic V, Toledo L, Dinant C, Grøfte M, et al. TRIP12 and UBR5 suppress spreading of chromatin ubiquitylation at damaged chromosomes. Cell 2012; 150:697 - 709; http://dx.doi.org/10.1016/j.cell.2012.06.039; PMID: 22884692
- Chen J, Feng W, Jiang J, Deng Y, Huen MS. Ring finger protein RNF169 antagonizes the ubiquitin-dependent signaling cascade at sites of DNA damage. J Biol Chem 2012; 287:27715 - 22; http://dx.doi.org/10.1074/jbc.M112.373530; PMID: 22733822
- Panier S, Ichijima Y, Fradet-Turcotte A, Leung CC, Kaustov L, Arrowsmith CH, et al. Tandem protein interaction modules organize the ubiquitin-dependent response to DNA double-strand breaks. Mol Cell 2012; 47:383 - 95; http://dx.doi.org/10.1016/j.molcel.2012.05.045; PMID: 22742833
- Nakada S, Tai I, Panier S, Al-Hakim A, Iemura S, Juang YC, et al. Non-canonical inhibition of DNA damage-dependent ubiquitination by OTUB1. Nature 2010; 466:941 - 6; http://dx.doi.org/10.1038/nature09297; PMID: 20725033
- Borden KL, Freemont PS. The RING finger domain: a recent example of a sequence-structure family. Curr Opin Struct Biol 1996; 6:395 - 401; http://dx.doi.org/10.1016/S0959-440X(96)80060-1; PMID: 8804826
- Chasapis CT, Spyroulias GA. RING finger E(3) ubiquitin ligases: structure and drug discovery. Curr Pharm Des 2009; 15:3716 - 31; http://dx.doi.org/10.2174/138161209789271825; PMID: 19925422
- Yin Q, Lin SC, Lamothe B, Lu M, Lo YC, Hura G, et al. E2 interaction and dimerization in the crystal structure of TRAF6. Nat Struct Mol Biol 2009; 16:658 - 66; http://dx.doi.org/10.1038/nsmb.1605; PMID: 19465916
- Zhang M, Windheim M, Roe SM, Peggie M, Cohen P, Prodromou C, et al. Chaperoned ubiquitylation--crystal structures of the CHIP U box E3 ubiquitin ligase and a CHIP-Ubc13-Uev1a complex. Mol Cell 2005; 20:525 - 38; http://dx.doi.org/10.1016/j.molcel.2005.09.023; PMID: 16307917
- Campbell SJ, Edwards RA, Leung CC, Neculai D, Hodge CD, Dhe-Paganon S, et al. Molecular insights into the function of RING finger (RNF)-containing proteins hRNF8 and hRNF168 in Ubc13/Mms2-dependent ubiquitylation. J Biol Chem 2012; 287:23900 - 10; http://dx.doi.org/10.1074/jbc.M112.359653; PMID: 22589545
- Stewart GS, Panier S, Townsend K, Al-Hakim AK, Kolas NK, Miller ES, et al. The RIDDLE syndrome protein mediates a ubiquitin-dependent signaling cascade at sites of DNA damage. Cell 2009; 136:420 - 34; http://dx.doi.org/10.1016/j.cell.2008.12.042; PMID: 19203578
- Lok GT, Sy SM, Dong SS, Ching YP, Tsao SW, Thomson TM, et al. Differential regulation of RNF8-mediated Lys48- and Lys63-based poly-ubiquitylation. Nucleic Acids Res 2012; 40:196 - 205; http://dx.doi.org/10.1093/nar/gkr655; PMID: 21911360
- Plans V, Scheper J, Soler M, Loukili N, Okano Y, Thomson TM. The RING finger protein RNF8 recruits UBC13 for lysine 63-based self polyubiquitylation. J Cell Biochem 2006; 97:572 - 82; http://dx.doi.org/10.1002/jcb.20587; PMID: 16215985
- Huen MS, Huang J, Yuan J, Yamamoto M, Akira S, Ashley C, et al. Noncanonical E2 variant-independent function of UBC13 in promoting checkpoint protein assembly. Mol Cell Biol 2008; 28:6104 - 12; http://dx.doi.org/10.1128/MCB.00987-08; PMID: 18678647
- Stewart GS, Stankovic T, Byrd PJ, Wechsler T, Miller ES, Huissoon A, et al. RIDDLE immunodeficiency syndrome is linked to defects in 53BP1-mediated DNA damage signaling. Proc Natl Acad Sci USA 2007; 104:16910 - 5; http://dx.doi.org/10.1073/pnas.0708408104; PMID: 17940005
- Bekker-Jensen S, Rendtlew Danielsen J, Fugger K, Gromova I, Nerstedt A, Lukas C, et al. HERC2 coordinates ubiquitin-dependent assembly of DNA repair factors on damaged chromosomes. Nat Cell Biol 2010; 12:80-6; sup pp 1-12.
- Mattiroli F, Vissers JH, van Dijk WJ, Ikpa P, Citterio E, Vermeulen W, et al. RNF168 ubiquitinates K13-15 on H2A/H2AX to drive DNA damage signaling. Cell 2012; 150:1182 - 95; http://dx.doi.org/10.1016/j.cell.2012.08.005; PMID: 22980979
- Otwinowski Z, Minor W. Processing of X-Ray Diffraction Data Collected in Oscillation Mode. Methods Enzymol 1997; 276:307 - 26; http://dx.doi.org/10.1016/S0076-6879(97)76066-X
- Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr 2002; 58:1948 - 54; http://dx.doi.org/10.1107/S0907444902016657; PMID: 12393927
- Vagin AA, Steiner RA, Lebedev AA, Potterton L, McNicholas S, Long F, et al. REFMAC5 dictionary: organization of prior chemical knowledge and guidelines for its use. Acta Crystallogr D Biol Crystallogr 2004; 60:2184 - 95; http://dx.doi.org/10.1107/S0907444904023510; PMID: 15572771
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 2004; 60:2126 - 32; http://dx.doi.org/10.1107/S0907444904019158; PMID: 15572765
- Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Cryst 1993; 26:283 - 91; http://dx.doi.org/10.1107/S0021889892009944