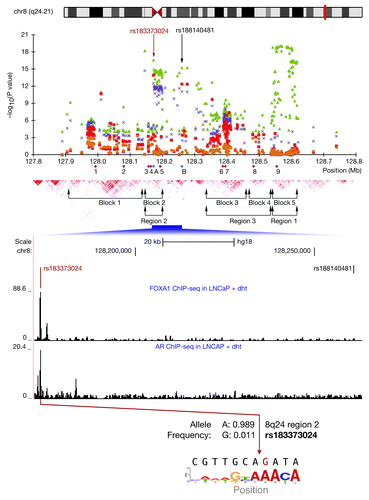
The chromosome 8q24 region is dense with single-nucleotide-polymorphism (SNP) variants associated with risk for prostate and other cancers. Many of the SNPs lie within an apparent gene “desert,” meaning that there are no annotated coding genes within 1 MB, a sufficiently large distance that assignment of viable hypotheses of SNP functionality is difficult even for other SNPs correlated by linkage disequilibrium (LD). The region of cancer risk falls centromeric of the notorious proto-oncogene cMyc and telomeric of the FAM84B gene; the latter encodes a DNA-repair enzyme known to be upregulated in breast cancers. In October 2012, Gudmundsson and colleagues reported the discovery of a novel and rare index SNP, rs188140481, which falls within the 8q24 region and confers a relatively high odds-ratio (2.9) of highly significant risk (p = 6.2 × 10−34) for prostate cancer.Citation1 It is unclear what function can be ascribed to rs188140481 due to lack of evidence for chromatin biological features at its genomic location. We used FunciSNP,Citation2 an R/Bioconductor software package developed by us, to analyze this novel index SNP for correlated SNPs within known biological/chromatin features from recently published high-throughput data. Our analysis resulted in the identification of a single correlated SNP, rs183373024, which was also identified by Gudmundsson et al.,Citation1 but rejected as a candidate for further study on the basis that it is highly correlated with the index SNP rs188140481 (r2 = 0.87), while having slightly weaker combined replication results. However, of the two SNPs, which both were reported to be highly significant risk SNPs for prostate cancer, rs183373024 alone maps to transcription factor-occupied regions of biological interest (). Thus, significant ChIP-seq peaks for binding of the androgen receptor (AR) and FoxA1 have been reported for this region in prostate cancer cells (LNCaP).Citation3,Citation4 Since this is a region of open chromatin,Citation1 to which AR and FoxA1 are bound, it may be implicated in prostate cancer tumorigenesis. Furthermore, upon closer analysis of the sequence, the rs183373024 minor allele G that carries risk replaces the middle A of an adenine triplet in the major allele, which is plausibly bound by FoxA1; the reference sequence (CGTTGCAAATAT) scores high (0.8) when aligned against public models. Even more provocatively, the predicted binding specificity for FoxA1 is greatly reduced by the introduction of the risk allele G in that position, leading us to hypothesize that the effect of the minor allele of rs183373024, associated with prostate cancer risk, is to reduce (even totally destroy) binding of FoxA1 to this putative enhancer region. Based on the above compelling evidence, we propose here that the lesion created by the minor (G) allele at rs183373024 may result in the downregulation of an as-yet-unidentified tumor suppressor-like gene target of the FoxA1 enhancer. Since the gene target(s) of an enhancer are difficult in principle to predict, the nearest protein-coding oncogene, cMYC is not necessarily the primary target of this enhancer. We note that there are other candidates located more proximal to rs183373024. The first, PCAT1, encodes a ~70 kb distant lincRNA recently shown to be a novel prostate-specific repressor transcript, which is also highly upregulated in a subset of prostate cancers.Citation5 PCAT1 was proposed to promote cell proliferation, but its role in cancer predisposition remains unclear. The second, POU5F1P1, is a pseudogene ~200 kb distant from the SNP, with significant homology to the POU5F1 proto-oncogeneCitation6 on chromosome 6. These genes are all candidates to be affected by the rs183373024 SNP. Recent evidence has pointed to the proposition that the 8q24 gene desert region may be a fertile ground for the evolutionary development of enhancers, which, if modified by chance via SNPs, have effects on risk for complex diseases such as cancers. By adding the FoxA1 binding site containing rs183373024 to the list of affected risk elements at this locus, we can now account for functionality in all three regions of prostate cancer risk, namely rs11986220 (FoxA1 binding) in region 1,Citation7 rs6983267 (TCF7L2 binding) in region 3Citation8,Citation9 and now the one reported here, rs183373024 (FoxA1 binding) in region 2. The entire 8q24 can thus be viewed as a nursery of enhancers affecting cancer risk via the regulation of distant gene expression.
Figure 1. Functional annotation of a region of chromosome 8q24. Figure 1 from reference Citation1 was modified to add annotations of potential functionality by incorporating raw signal tracks of FoxA1 and AR CHIP-seq data. The SNP (rs183373024) is as indicated and shows how the G allele may destroy the FoxA1 site. The consensus FoxA1 sequence is shown. The position of index SNP (rs188140481) is as indicated.
Acknowledgments
This work was supported by the National Institutes of Health (R01 CA136924 and U19 CA148537 to G.A.C.) and from a subcontract (to G.A.C. from Harvard) of a David Mazzone award. The scientific development and funding of this project were in part supported by the Genetic Associations and Mechanisms in Oncology (GAME-ON), a NCI Cancer Post-GWAS Initiative.
Author Contributions
All three authors were involved in all aspects of the design, analyses and writing of the manuscript.
References
- Gudmundsson J, et al. Nat Genet 2012; 44:1326 - 9; http://dx.doi.org/10.1038/ng.2437; PMID: 23104005
- Coetzee SG, et al. Nucleic Acids Res 2012; 40:e139; http://dx.doi.org/10.1093/nar/gks542; PMID: 22684628
- Wang D, et al. Nature 2011; 474:390 - 4; http://dx.doi.org/10.1038/nature10006; PMID: 21572438
- Yu J, et al. Cancer Cell 2007; 12:419 - 31; http://dx.doi.org/10.1016/j.ccr.2007.10.016; PMID: 17996646
- Prensner JR, et al. Nat Biotechnol 2011; 29:742 - 9; http://dx.doi.org/10.1038/nbt.1914; PMID: 21804560
- Campbell PA, et al. PLoS One 2007; 2:e553; http://dx.doi.org/10.1371/journal.pone.0000553; PMID: 17579724
- Jia L, et al. PLoS Genet 2009; 5:e1000597; http://dx.doi.org/10.1371/journal.pgen.1000597; PMID: 19680443
- Pomerantz MM, et al. Nat Genet 2009; 41:882 - 4; http://dx.doi.org/10.1038/ng.403; PMID: 19561607
- Ting MC, et al. Dis Model Mech 2012; 5:366 - 74; http://dx.doi.org/10.1242/dmm.008458; PMID: 22279083