 ?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.Abstract
To date, a range of ion channels have been identified in chondrocytes using a number of different techniques, predominantly electrophysiological and/or biomolecular; each of these has its advantages and disadvantages. Here we aim to compare and contrast the data available from biophysical and microarray experiments. This letter analyses recent transcriptomics datasets from chondrocytes, accessible from the European Bioinformatics Institute (EBI). We discuss whether such bioinformatic analysis of microarray datasets can potentially accelerate identification and discovery of ion channels in chondrocytes. The ion channels which appear most frequently across these microarray datasets are discussed, along with their possible functions. We discuss whether functional or protein data exist which support the microarray data. A microarray experiment comparing gene expression in osteoarthritis and healthy cartilage is also discussed and we verify the differential expression of 2 of these genes, namely the genes encoding large calcium-activated potassium (BK) and aquaporin channels.
Chondrocytes are the resident cells of cartilage which produce, maintain, and degrade the extracellular matrix (ECM). Whilst the cells are non-excitable, they have already been shown to express a rich complement of ion channels.Citation1 Chondrocyte ion channels are involved in several critical functions including mechanotransductionCitation2 and apoptosis.Citation3 Many other functions in chondrocytes, and their precursors, have all been shown to directly involve ion channels.Citation4
Expression data to date have largely been obtained using traditional biochemical and physiological techniques such as flux studies, electrophysiology, and immunohistochemistry. More recently reverse transcription and real time (quantitative) polymerase chain reaction (PCR) methods have also identified novel ion channels in chondrocytes.Citation5 Over the past few years, microarray analysis or “transcriptomics” has added a new dimension to the study of gene expression. Whereas, in the past, one studied ion channels on a cell-by-cell, or antibody-by-antibody basis, this new “transcriptomic” or “expression profiling” technology potentially allows one to examine thousands of transcripts simultaneously. The technique is clearly very powerful but there are drawbacks too, since every scientific technique has its limitations. However, by combining modern omics strategies and conventional biomolecular techniques one can gain a more thorough and subtle understanding of chondrocyte biology.
The most widely used method for functional study of ion channels is patch-clamp electrophysiology. However, despite the prominence, historical dominance and power of this approach, it is a very slow method and in real cell systems (as opposed to heterologous expression systems) definitive identification of a specific ion channel is difficult. The identification of ion channels by patch clamp electrophysiology usually depends on the availability of a specific pharmacological ligand or a very distinct biophysical profile. Both of these are actually far rarer than one may think. For example, some of the most important channels for the control of chondrocyte function are the transient receptor potential (TRP) channels.Citation6 These are difficult to identify electrophysiologically because they show weak voltage sensitivity and there are relatively few selective ligands.
Traditionally, one has backed up the electrophysiology with immunohistochemistry or PCR studies, but even the strongest advocates of this approach would have to concede it is a slow process. Expression profiling with a microarray speeds up the characterization of ion channel transcription but, in addition to concerns over probe specificity, statistical validity and detection accuracy, there is a more fundamental issue. Even if particular ion channels are detected, and one accepts that this detection approach is valid, alone it gives no information on the role of that channel. Does the channel control membrane potential, volume, secretion, or have little role in that cell at all? The 2 data sets could be complementary to each other, however, if one makes the assumption that both experimental techniques are equally valid. The microarray data could direct electrophysiological experiments, by generating targets for study, and these biophysical studies could examine the role of the expressed genes. It is well known that there is a statistically significant, but weak, correlation between total RNA abundance and protein expression. This has been particularly well studied in yeastCitation7 and E. coli where, from gene to gene and cell to cell, ratio of mRNA copy number to protein number can vary from 1:100 to 1:10000.Citation8 The degree of correlation also depends on the gene ontology and can be as high as R2=0.5 or so.Citation9,Citation10 In the following text, we will compare the results of recent transcriptomic studies with those of traditional methods and ask; do the data match each other and what does each dataset add to the other?
In this letter we analyse and discuss the data from the following 10 chondrocyte Affymetrix microarray datasets openly available on the European Bioinformatics Institute (EBI) expression profiling database (EBI accession codes): E-GEOD-6119,Citation11 E-GEOD-10024,Citation12 E-GEOD-10556,Citation13 E-GEOD-1277,Citation14 E-GEOD-14402,Citation15 E-GEOD-16464,Citation16 E-GEOD-18052,Citation17 E-GEOD-18394,Citation18 E-GEOD-7683,Citation19 E-GEOD-8077.Citation20 These datasets were found by searching for “chondrocyte(s)” OR “cartilage and chondrocyte(s)”. These datasets have been produced from expression profiling experiments of extracted or primary cultures of chondrocytes from “healthy” cartilage from a range of human, bovine, rat, and mouse tissue.
A total of over 40,000 gene probe sets (reporters) have been used and these include those targeted at 200 or 300 ion channel, or related, plasma membrane proteins. In this paper, we have accepted the author’s detection limits and analysis where they have uploaded these to the European Bioinformatics Database. Where these are not given, we have processed the raw data with a Mas5 transformCitation21 using the open source Affymetrix Power Tools (Affymetrix). Probe annotations were derived from the Affymetrix annotations database files using custom Perl scripts (revisions “na31” except where stated). We have not analyzed data from other microarray platforms.
Commonality between Datasets
Typically, using standard assessment criteria, approximately 50 to 100 channel targets are detected in chondrocytes. Not surprisingly the dataset from each study are rather different to each other, but there is a common set of 7 probes that are detected in all 10 of these studies. These are given in . It is important to note that this analysis does not select particular ion channel genes to study, but reveals all the genes commonly expressed in the above microarray datasets (note that there are over 300 ion channel gene probes encoded on each of the cited Affymetrix chips; accessory proteins such as the channel “tetramerization domain” proteins are excluded). With such a large number of ion channel transcripts on the Affymetrix chips one would expect the random co-detection of transcripts across datasets. To quantify this we used the following statistical analysis. The probability of more than n transcripts being co-localised in d datasets is given by:
Table 1: Ion channels detected in all 10 of the microarray studies considered in this report.
where p is the probability of a given gene appearing in a datasets, and there are a total of g genes on each array. This gives a p-value for 7 (or more) transcripts appearing in all 10 datasets by chance as p <1e-14.
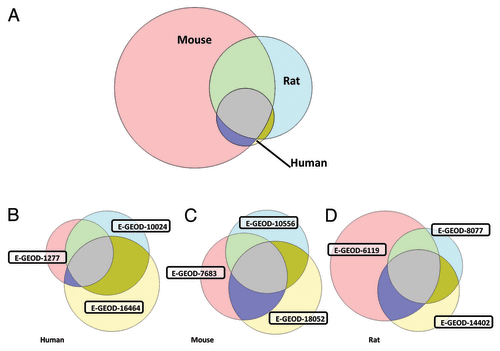
It should be noted that these microarray datasets were derived from different species (3 rat, 3 mouse, 3 human and 1 bovine) and there are potential differences in chondrocyte isolation protocols. Constraining analysis to just rodent (6 datasets) returns a set of 23 commonly expressed ion channel genes (). quantitatively illustrates both the overlap of genes commonly expressed between species () and the overlap between each of the transcripts from human, mouse and rat (, respectively). It is evident that far more transcripts were detected in all 3 of the mouse datasets than in all 3 of the human datasets. This could be for three reasons; firstly, it is possible that the sensitivity of the mouse chips is greater, but we have seen no specific evidence for this. Secondly, each of the protocols requires manual dissection and separation of chondrocytes from the subchondral bone and adnexa. It is possible that mouse “chondrocyte” samples are inherently contaminated with non-chondrocyte tissue. In an electrophysiological or immunohistochemical study such contamination would be relatively easy to detect, but in a biochemical protocol, where harvested tissue is macerated and then processed, it could be missed. Thirdly, it is conceivable that there are genuine phenotypic differences between chondrocytes in mice and other animals. Such differences have been discussed elsewhere.Citation22,Citation23
Table 2: Ion channel gene IDs from all those gene transcripts detected in all 6 rodent microarray studies.
Figure 1. Commonality of gene transcript expression between datasets. (A) Commonality between species. These represent transcripts present in each of the 3 datasets for each of the human, mouse, and rat datasets (i.e., includes 9 datasets total). Far more are observed in rodent datasets (mouse especially) than human. (B) Commonality between human derived datasets. The human studies used tissue harvested from either adolescents receiving limb length correction surgery (E-GEOD-1277), adults receiving ACL surgery (E-GEOD-16464), or post mortem (E-GEOD-10024). Samples were taken from knee (E-GEOD-16464), distal femur (E-GEOD-1277), or shoulder (E-GEOD-10024) and focussed on articular (E-GEOD-10024), mixed (E-GEOD-16464), or growth plate (E-GEOD-1277) chondrocytes. Chip Ids; E-GEOD-10024 used HG-U133A and E-GEOD-16464 used the slightly newer HG-U133A_Plus_2, but E-GEOD-1277 used the U95AV2 GeneChip. All 3 human studies used expanded chondrocytes, but E-GEOD-10024 and E-GEOD-16464 re-constituted those into 3D cultures. Extraction enzymes were collagenase P (E-GEOD-10024), clostridial collagenase and deoxyribonuclease I (E-GEOD-16464) and trypsin (E-GEOD-1277). (C) Commonality between mouse derived datasets. Rodent studies suffer from inherent difficulties in extraction of tissue since cartilage is thinner than larger animals. Tissue used from the microarray studies analysed in this letter came from a variety of joints from immature mice and are likely to include mixed chondrocyte phenotypes. Where stated explicitly, chondrocytes were expanded in monolayer cultures following collagenase based isolation (E-GEOD-8052 and E-GEOD-7683). All 3 studies (E-GEOD-10556, E-GEOD-18052 and E-GEOD-7683) used the same Affymetrix Mouse430_2 chips. (D) Commonality between rat derived datasets. The rat femoral head (E-GEOD-6119, E-GEOD-14402) or knee (E-GEOD-8077) tissue was harvested from a range of ages from one day old neonates (from which “only the outer two-thirds of cartilage” was used to select for articular chondrocytes, E-GEOD-14402) to several month old rats (300-320g, E-GEOD-8077). Strain was either Wistar (E-GEOD-6119), Sprague-Dawley (E-GEOD-8077) or not stated. E-GEOD-6119 and E-GEOD-14402 both used monolayer expanded chondrocytes following collagenase II based isolation. E-GEOD-6119 also included pronase, but E-GEOD-8077 used direct RNA extraction from macerated tissue. All the included rat studies used the Affymetrix Rat230_2 chips. One bovine dataset derived from chondrocytes 3D cultured from carpal bones of 3 to 6 mo old calves was also analyzed (E-GEOD-18394, Affymetrix Bovine chip, annotated with version “na29”). Since there was only 1 bovine chondrocyte dataset on EBI (albeit including a number of replicates) this is not included in the Venn diagrams. All other datasets were annotated with revision Affymetrix annotation version “na31”. Note that each of the 3 species sets in (A) is equivalent to the commonly expressed regions of the Venn diagrams in (B, C, and D).
The remainder of this letter focuses on whether the microarray data provides useful clues as to which channels are expressed in chondrocytes, and whether, therefore, initial processing of microarray data will improve the rate of channel detection in chondrocytes, or potentially other tissues.
Chloride Channels
The chloride channel superfamilyi is huge and includes the large ClC family, CFTR protein, calcium-activated, volume-activated, P64 related chloride channels (Clns), and intracellular chloride channels (CLIC). In many systems chloride channels have been less well studied than cation channels, although they were some of the earliest ion channels identified in chondrocytes.Citation24,Citation25 Analysis of the current datasets reveals that both p64-related (CLIC1, CLIC4) and ClNS1A ion channels were detected in all 10 microarray experiments. The function of the corresponding channels are unknown, complex, controversial, or a combination of all these. CLIC1, CLIC4, and other members of the CLIC family of proteins appear to be legitimate anion channels.Citation26 They are often referred to as “p64-related” simply because their earliest characterization appeared to be of a 64kDa protein.Citation27 They appear to be of relatively low conductance for a chloride channel (8-40pS, depending on experimental conditionsCitation28-Citation30). Whilst, as their name implies, these channels can localize to intracellular compartments,Citation31,Citation32 in some cell types they also appear to be in the plasma membrane and could serve a role in secretion.Citation32,Citation33 One possibility is that the channel shuttles to and from the membrane in a cell cycle dependent way.Citation32 The pCln channel (sometimes referred to as P-glycoprotein, pICln), also detected in 10 out of 10 datasets, was first identified by Paulmichl et alCitation34 as a putative rather ubiquitous volume-sensitive chloride channel. More recently, this volume regulator role has been refutedCitation35,Citation36 and roles in gene-regulation and development have been proposed.Citation37,Citation38 The controversy surrounding the nature of this channel is discussed in detail by StrangeCitation39 and Furst.Citation40 The particular issue is that it cannot be clearly determined whether this is a volume-sensitive channel, another type of channel, or is a regulator of a channel endogenous in the various expression systems in which it has been studied.Citation39 Even the fundamental property of ion selectivity is controversial, since recent studies have shown a rather higher permeability of pCln to cations than would be expected for a chloride channel.Citation41 It is possible that one or other of these channels does constitute the chloride channel identified by Tsuga et alCitation25 in chondrocytes, but it is unlikely since that channel is more typical of a classical maxi-type chloride channel in electrophysiological and pharmacological terms.Citation26,Citation42
The Voltage Dependent Anion Channels
The detection of both VDAC1 and VDAC2 is particularly interesting. The associated channels are thought of, generally, as mitochondrial ion channels, found in the outer membrane of this organelle. The proteins have also been detected in the plasma membrane, where they exhibit voltage-gated anion channel activity (based on data in the NCBInr and UniProt databases). VDAC channels are also implicated in apoptosis and, as such, they will be of profound importance to all cells in which they are expressed. It has also now been suggested by a number of authors that some VDAC protein expression is also of plasma membrane ion channels.Citation43,Citation44 The phenotype of this channel is that of a maxi-chloride channel. This would be very much in line with the original chondrocyte chloride channel work of TsugaCitation25 and Sugimoto.Citation24 The channel identified in these studies was the maxi-chloride channel, which is remarkably similar to the maxi-Cl/VDAC channel. In our own unpublished work, we see clear expression of a large conductance, niflumic acid and SITS-sensitive chloride channel which appears likely to be maxi-chloride.
The Large Calcium-Activated Potassium Channel
It is no surprise that KCNMA1 (BK) has been detected in all of the 10 datasets analyzed here. Currents either broadly, or specifically, identified as being through BK channels have been described in a number of papers. In our own work we have shown not only that BK currents are present by electrophysiology, but also that the KCNMA1 was detectable by immunohistochemical methods.Citation45 Interestingly, whilst the BK function modifying β-subunit KCNMB1 was detected in our own immunohistochemical studies it was not detected in any of the 10 microarray studies discussed here. There was detection of KCNMB2 and KCNB4 in 2 and 3 out of the 10 studies respectively. Activation of BK results in such large currents that it is likely to be involved with regulation of intracellular osmolarity and volume. This is important, since it known to be activated by stretch.Citation45 Early work did not show whether this activation was direct or indirect via an increase of intracellular calcium ions, but our more recent studies show rather convincingly that TRPV4 is activated by stretch and that this results in the opening of a potassium channel. BK is, so far, the only candidate for this role.
The Sodium Channel β-Subunit
The detection of the sodium β1-subunit SCN1B in all 10 studies is interesting, since the α-subunits were undetectable in most studies. This subunit has been shown, in neurons to convey subtle changes to expression patterns and functional properties of voltage-gated sodium channels.Citation46 There has been 1 electrophysiological study, which identified voltage-gated sodium channels in chondrocytes.Citation24 This work has not been followed up on and, since chondrocytes do not fire action potentials, it is difficult to see what the role of a voltage-gated sodium channel might be. For completeness: SCN7A (Nav2.1), a somewhat atypical Na+ channel was detected in 4/10 experiments, and the classical sodium channel SCN2A1 in 3/10, and the transient type sodium channel SCN11A and SCN10A in 2/10 datasets. SCN5A and 3A were detected in 1/10.
Transient Receptor Potential (TRP) Channels
The next most commonly identified ion channel transcripts are given in . This list includes TRPC1, of the canonical TRP, and TRPV4, a vanilloid channel, both of which were identified in 9/10 microarray studies. In traditional protein and pharmacological studies, however, three TRP channels have been identified in chondrocytes TRPV4,Citation47,Citation48 TRPV5,Citation6,Citation48 and TRPV6Citation48 but not TRPC1. We have not investigated TRPC1 ourselves, however. The TRP channels identified pharmacologically in chondrocytes have different, but related roles. TRPV5 appears important for setting the membrane potential, crucial to maintenance of cell volume in chondrocytes.Citation6 TRPV4, however, appears to be critical for allowing entry of Ca2+ and activation of BK channels during imposed volume increase, and thus the process of regulatory volume decrease in epithelial cells.Citation49
Table 3. Ion channels detected in 9/10 of the microarray studies considered in this report.
Changes of Transcript Levels with Onset of Osteoarthritis
To use traditional methods to identify changes in channel expression between populations of cells from different tissues is probably not feasible unless high throughput automated ion channel recording equipment can be adapted for this purpose. However, observation of changes in cellular properties and mathematical models may provide clues as to which channels have changed in OA. An alternative approach would be to use microarray comparisons between normal and OA tissue, detect changes in channel mRNA abundance and then follow-up with functional or histochemical experiments. Generally, differential expression patterns from microarray studies are used to identify changed pathways, however, this process could miss changes which take place in individual ion channels not associated with an established pathway. The next microarray study considered in this review specifically compared transcript abundance from chondrocytes in human normal and OA cartilage.Citation50 The authors focused on (and verified) changes in many cartilage phenotypic markers, but did not specifically consider changes in other proteins such as ion channels. Probing this list for ion channels and porins significantly changed (greater than 2-fold change) produces the data in . The data clearly show an approximately 3-fold change in ENaC, TMEM16A and BK (KCNMA1) transcript abundance and a 38-fold change in the aquaporin AQP1 channel transcript abundance. Interestingly each of these channels are important for chondrocyte cell volume control.Citation2, Citation55 From this, by way of proof of principle we verified BK channel changes with immunohistochemistry and aquaporin channel changes with a functional assay.
Table 4. Ion channels and porins significantly changed in OA.
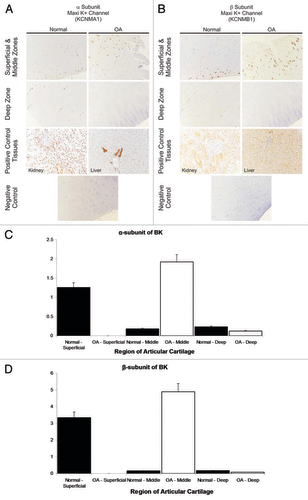
We analysed, using immunohistochemistry, whether protein expression of BK is increased. Tissue was taken from stifle joints of horses with and without OA (see for representative example). We investigated the expression of both α- and β-subunits of BK (only KCNMA1 was included in Karlsson 2010,Citation50 there were no probe sets for KCNMB1 on the chip). Semi-quantitative analysis of protein expression density shows that both BK subunits were significantly increased in OA, in the middle zone ().
Figure 2. Immunohistochemical identification of KCNMA1 (BK α-subunit) and KCNB1 (BK β-subunit) in sections of healthy and OA equine cartilage. The data from normal equines is reproduced with permission from Mobasheri et al.Citation45 Macroscopically normal articular cartilage samples were obtained from weight-bearing regions of the metacarpophalangeal joints of horses of mixed breed, age, and sex. Joint tissues were sourced from an abattoir in Nantwich, Cheshire and Taunton Devon. Animals were euthanized for non-research purposes having been stunned before slaughter for meat in accordance with Welfare of Animals (Slaughter or Killing) Regulations 1995. Sections of normal (n=6) and OA (n=3) equine cartilage were probed for channel expression by immunohistochemistry essentially as previously described.Citation45 Sections were incubated overnight at 4°C with rabbit polyclonal antibodies to the KCNMA1 and KCNB1. Antibody dilutions used ranged from 1:200 to 1:1500 in tris-buffered saline containing 1% bovine serum albumin. Slides were incubated with horseradish peroxidase-labelled polymer conjugated to affinity-purified goat anti-rabbit immunoglobulins. Cell nuclei were counterstained by incubation with aqueous haematoxylin (code no. S3309; Dako). Positive control samples were included from liver and kidney. Omission of primary antibody served as negative controls. Photomicrographs of immunostained tissue sections captured using Nikon Digital Sight DS-5M camera driven by Nikon Eclipsenet image capture software (Nikon). Positive staining is indicated by brown staining and particular evident at middle/superficial zones. (C and D) Semi-quantified protein expression density. The largest increase in expression density (from data such as that illustrated in ) is in the middle zone, for both KCNMA1 (C) and KCNMB1 (D) (α- and β- subunit respectively). Note that in OA tissue there was insufficient superficial data to quantify expression levels.
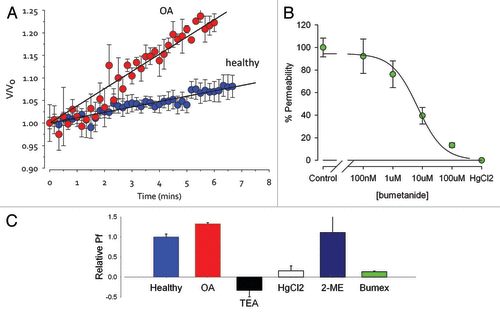
The accepted assay for aquaporin expression is that developed by Preston et al.Citation51 Cells are challenged with a hypotonic solution, causing them to swell, and the rate of swell can be measured to determine the aquaporin expression. The bioinformatics showed a 38-fold increase in AQP1 transcript abundance in OA and we see a significant increase in functional aquaporin expression in tissue from OA joints (), although the increase is much smaller than the change in AQP1 transcript abundance. Current pharmacological tools do not allow categorical determination of aquaporin subtype, however, here, water permeability of chondrocytes was blocked by concentrations of bumetanide, TEA, and HgCl2 consistent with that expected for AQP1Citation53,Citation54 ().
Figure 3. Water Permeability (Aquaporin) Assay. Water permeability can be calculated from the initial slope of the relative volume (V/Vo) against time curve. Where V is the volume at time t and Vo is the volume at time zero. This is the accepted physiological assay for aquaporin expression. (A) Permeability is 30±3% (p<0.05, n=4) greater in chondrocytes from dogs with osteoarthritis (OA). TEA (a blocker of AQP1Citation53,Citation54), bumetanideCitation54 (pIC50 5.17±0.11µM, n=6, “Bumex”, a blocker of AQP1 and 4), and mercuric chloride (HgCl2 a non-specific AQP blocker reversed by 2-mercaptoethanol) ME) are included to determine AQP type (B and C). Chondrocytes were harvested from canine clinical waste tissue with owner consent. Cells were placed in a “physiological saline” solution including 120 mM sucrose (osmolarity 300mOsm), then moved to an identical physiological saline without the sucrose. Cells at first swell as water enters the cell due to osmosis. Live cell imaging was achieved with a Nikon Diaphot microscope equipped with a Sony ICX098QB high sensitivity CCD. Images were analysed offline with ImageJVolume was calculated from the 2D surface area (A) of the cell disc by assuming the cell is approximately spherical as described previously,Citation6 using the following equation: (Equation 2). Except where stated, data are presented normalized for starting volume (V0) as V/V0, where V is the volume at time t. Visual data were analyzed with ImageJ and ANOVA performed with SPSS (SPSS Inc.). Note that canine tissue was harvested from clinical waste tissue with Local Ethical Approval, no dogs were harmed for the study.
In summary, there appears to be considerable agreement between transcriptomic studies and physiological or immunohistochemical studies. It would seem that most channels common to all 10 datasets can be identified by these other techniques. There are examples, however, of proteins which have been identified in chondrocytes yet show up in few datasets. For example, the ASIC channel (ACCN2 gene) has been shown by immunohistochemistry and rt-PCR yet shows up in only one of the three rat datasets discussed here.Citation52 Therefore, combining these approaches should massively speed up the rate of discovery of ion channels in cell types which can be isolated in sufficient quantities to perform such studies. There are tissues, such as the brain, where cell types are too intermingled to allow identification of unique cell types, but for many tissues in the musculoskeletal system (or cell lines), the combination of transcriptomics and protein studies seems ideal. With regard to OA, this strategy has allowed us to very rapidly pinpoint some phenotypic changes in the expression of two important channels in OA: an aquaporin and the BK channel. Further protein and functional experiments will be necessary to establish whether the other KCa channels are also altered, and in particular whether these changes contribute to or result from progression of OA.
| Abbreviations: | ||
| EBI | = | European Bioinformatics Institute |
| ECM | = | extracellular matrix |
| BK | = | large conductance calcium-activated potassium channel |
| PCR | = | polymerase chain reaction |
| TRP | = | transient receptor potential |
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
The authors thank Andy Jones for assistance with the bioinformatics, Peter Cripps for assistance with the statistical analysis and Prof John Innes for supply of the canine tissue.
Funding
The research leading to these results has received full funding from the European Union Seventh Framework Programme (FP7/2007-2013) under grant agreement no. 305815 (http://cordis.europa.eu/projects/rcn/105314_en.html) (http://ec.europa.eu/research/health/medical-research/severe-chronich-diseases/projects/d-board_en.html).
Author Contributions
All authors have made substantial intellectual contributions to the conception and design of the study, data acquisition, analysis and interpretation. RBJ conceived the study. All authors contributed to data collection, interpretation and analysis. All authors contributed to data interpretation and manuscript preparation and approved the final version submitted.
References
- Barrett-Jolley R, Lewis R, Fallman R, Mobasheri A. The emerging chondrocyte channelome. Frontiers in Membrane Physiol Biophys 2010; 1.
- Lewis R, Feetham CH, Barrett-Jolley R. Cell volume regulation in chondrocytes. Cell Physiol Biochem 2011; 28:1111 - 22; http://dx.doi.org/10.1159/000335847; PMID: 22179000
- Kumagai K, Imai S, Toyoda F, Okumura N, Isoya E, Matsuura H, et al. 17β-Estradiol inhibits the doxorubicin-induced apoptosis via block of volume-sensitive Cl+ current in rabbit articular chondrocytes. Br J Pharmacol 2011; 166:702 - 20; http://dx.doi.org/10.1111/j.1476-5381.2011.01802.x
- Mobasheri A, Lewis R, Ferreira-Mendes A, Rufino A, Dart C, Barrett-Jolley R. Potassium channels in articular chondrocytes. Channels (Austin) 2012; 6:416 - 25; http://dx.doi.org/10.4161/chan.22340; PMID: 23064164
- Funabashi K, Ohya S, Yamamura H, Hatano N, Muraki K, Giles W, Imaizumi Y. Accelerated Ca2+ entry by membrane hyperpolarization due to Ca2+-activated K+ channel activation in response to histamine in chondrocytes. Am J Physiol Cell Physiol 2010; 298:C786 - 97; http://dx.doi.org/10.1152/ajpcell.00469.2009; PMID: 20042729
- Lewis R, Asplin KE, Bruce G, Dart C, Mobasheri A, Barrett-Jolley R. The role of the membrane potential in chondrocyte volume regulation. J Cell Physiol 2011; 226:2979 - 86; http://dx.doi.org/10.1002/jcp.22646; PMID: 21328349
- Gygi SP, Rochon Y, Franza BR, Aebersold R. Correlation between protein and mRNA abundance in yeast. Mol Cell Biol 1999; 19:1720 - 30; PMID: 10022859
- Tyagi SE. Genomics. E. coli, what a noisy bug. Science 2010; 329:518 - 9; http://dx.doi.org/10.1126/science.1194036; PMID: 20671174
- Guo YF, Xiao P, Lei SF, Deng FY, Xiao GG, Liu YZ, Chen X, Li L, Wu S, Chen Y, et al. How is mRNA expression predictive for protein expression? A correlation study on human circulating monocytes. Acta Biochim Biophys Sin (Shanghai) 2008; 40:426 - 36; http://dx.doi.org/10.1111/j.1745-7270.2008.00418.x; PMID: 18465028
- Anderson L, Seilhamer J. A comparison of selected mRNA and protein abundances in human liver. Electrophoresis 1997; 18:533 - 7; http://dx.doi.org/10.1002/elps.1150180333; PMID: 9150937
- Gouze J-N, Gouze E, Popp MP, Bush ML, Dacanay EA, Kay JD, et al. Exogenous glucosamine globally protects chondrocytes from the arthritogenic effects of IL-1 beta. Arthritis Res Ther 2006; •••:8
- Andreas K, Lübke C, Häupl T, Dehne T, Morawietz L, Ringe J, Kaps C, Sittinger M. Key regulatory molecules of cartilage destruction in rheumatoid arthritis: an in vitro study. Arthritis Res Ther 2008; 10:R9; http://dx.doi.org/10.1186/ar2358; PMID: 18205922
- Fukada T, Civic N, Furuichi T, Shimoda S, Mishima K, Higashiyama H, Idaira Y, Asada Y, Kitamura H, Yamasaki S, et al. The zinc transporter SLC39A13/ZIP13 is required for connective tissue development; its involvement in BMP/TGF-beta signaling pathways. PLoS One 2008; 3:e3642; http://dx.doi.org/10.1371/journal.pone.0003642; PMID: 18985159
- Olney RC, Wang JW, Sylvester JE, Mougey EB. Growth factor regulation of human growth plate chondrocyte proliferation in vitro. Biochem Biophys Res Commun 2004; 317:1171 - 82; http://dx.doi.org/10.1016/j.bbrc.2004.03.170; PMID: 15094393
- Rockel JS, Bernier SM, Leask A. Egr-1 inhibits the expression of extracellular matrix genes in chondrocytes by TNF alpha-induced MEK/ERK signalling. Arthritis Res Ther 2009; •••:11
- Dehne T, Karlsson C, Ringe J, Sittinger M, Lindahl A. Chondrogenic differentiation potential of osteoarthritic chondrocytes and their possible use in matrix-associated autologous chondrocyte transplantation. Arthritis Res Ther 2009; 11:R133; http://dx.doi.org/10.1186/ar2800; PMID: 19723327
- Saito A, Hino S-i, Murakami T, Kanemoto S, Kondo S, Saitoh M, Nishimura R, Yoneda T, Furuichi T, Ikegawa S, et al. Regulation of endoplasmic reticulum stress response by a BBF2H7-mediated Sec23a pathway is essential for chondrogenesis. Nat Cell Biol 2009; 11:1197 - 204; http://dx.doi.org/10.1038/ncb1962; PMID: 19767744
- Huang AH, Stein A, Mauck RL. Evaluation of the complex transcriptional topography of mesenchymal stem cell chondrogenesis for cartilage tissue engineering. Tissue Eng Part A 2010; 16:2699 - 708; http://dx.doi.org/10.1089/ten.tea.2010.0042; PMID: 20367254
- James CG, Ulici V, Tuckermann J, Underhill TM, Beier F. Expression profiling of Dexamethasone-treated primary chondrocytes identifies targets of glucocorticoid signalling in endochondral bone development. BMC Genomics 2007; 8:205; http://dx.doi.org/10.1186/1471-2164-8-205; PMID: 17603917
- Appleton CTG, Pitelka V, Henry J, Beier F. Global analyses of gene expression in early experimental osteoarthritis. Arthritis Rheum 2007; 56:1854 - 68; http://dx.doi.org/10.1002/art.22711; PMID: 17530714
- Hubbell E, Liu WM, Mei R. Robust estimators for expression analysis. Bioinformatics 2002; 18:1585 - 92; http://dx.doi.org/10.1093/bioinformatics/18.12.1585; PMID: 12490442
- Clark RB, Hatano N, Kondo C, Belke DD, Brown BS, Kumar S, Votta BJ, Giles WR. Voltage-gated K+ currents in mouse articular chondrocytes regulate membrane potential. Channels (Austin) 2010; 4:179 - 91; http://dx.doi.org/10.4161/chan.4.3.11629; PMID: 20372061
- Mobasheri A, Lewis R, Ferreira-Mendes A, Rufino A, Dart C, Barrett-Jolley R. Potassium channels in articular chondrocytes. Channels (Austin) 2012; 6:416 - 25; http://dx.doi.org/10.4161/chan.22340; PMID: 23064164
- Sugimoto T, Yoshino M, Nagao M, Ishii S, Yabu H. Voltage-gated ionic channels in cultured rabbit articular chondrocytes. Comp Biochem Physiol C Pharmacol Toxicol Endocrinol 1996; 115:223 - 32; http://dx.doi.org/10.1016/S0742-8413(96)00091-6; PMID: 9375360
- Tsuga K, Tohse N, Yoshino M, Sugimoto T, Yamashita T, Ishii S, Yabu H. Chloride conductance determining membrane potential of rabbit articular chondrocytes. J Membr Biol 2002; 185:75 - 81; http://dx.doi.org/10.1007/s00232-001-0112-3; PMID: 11891566
- Jentsch TJ, Stein V, Weinreich F, Zdebik AA. Molecular structure and physiological function of chloride channels. Physiol Rev 2002; 82:503 - 68; PMID: 11917096
- Redhead CR, Edelman AE, Brown D, Landry DW, al-Awqati Q. A ubiquitous 64-kDa protein is a component of a chloride channel of plasma and intracellular membranes. Proc Natl Acad Sci U S A 1992; 89:3716 - 20; http://dx.doi.org/10.1073/pnas.89.9.3716; PMID: 1315034
- Tonini R, Ferroni A, Valenzuela SM, Warton K, Campbell TJ, Breit SN, Mazzanti M. Functional characterization of the NCC27 nuclear protein in stable transfected CHO-K1 cells. FASEB J 2000; 14:1171 - 8; PMID: 10834939
- Edwards JC, Tulk B, Schlesinger PH. Functional expression of p64, an intracellular chloride channel protein. J Membr Biol 1998; 163:119 - 27; http://dx.doi.org/10.1007/s002329900376; PMID: 9592076
- Singh H. Two decades with dimorphic Chloride Intracellular Channels (CLICs). FEBS Lett 2010; 584:2112 - 21; http://dx.doi.org/10.1016/j.febslet.2010.03.013; PMID: 20226783
- Berryman M, Bretscher A. Identification of a novel member of the chloride intracellular channel gene family (CLIC5) that associates with the actin cytoskeleton of placental microvilli. Mol Biol Cell 2000; 11:1509 - 21; http://dx.doi.org/10.1091/mbc.11.5.1509; PMID: 10793131
- Valenzuela SM, Mazzanti M, Tonini R, Qiu MR, Warton K, Musgrove EA, Campbell TJ, Breit SN. The nuclear chloride ion channel NCC27 is involved in regulation of the cell cycle. J Physiol 2000; 529:541 - 52; http://dx.doi.org/10.1111/j.1469-7793.2000.00541.x; PMID: 11195932
- Ulmasov B, Bruno J, Woost PG, Edwards JC. Tissue and subcellular distribution of CLIC1. BMC Cell Biol 2007; 8:8; http://dx.doi.org/10.1186/1471-2121-8-8; PMID: 17326840
- Paulmichl M, Norris AA, Rainey DK. Role of chloride channel modulation in the mechanism of action of nedocromil sodium. Int Arch Allergy Immunol 1995; 107:416; http://dx.doi.org/10.1159/000237060; PMID: 7613196
- Voets T, Buyse G, Tytgat J, Droogmans G, Eggermont J, Nilius B. The chloride current induced by expression of the protein pICln in Xenopus oocytes differs from the endogenous volume-sensitive chloride current. J Physiol 1996; 495:441 - 7; PMID: 8887755
- Nilius B, Droogmans G. Amazing chloride channels: an overview. Blackwell Science Ltd, 2003:119-47.
- Krapivinsky G, Pu W, Wickman K, Krapivinsky L, Clapham DE. pICln binds to a mammalian homolog of a yeast protein involved in regulation of cell morphology. J Biol Chem 1998; 273:10811 - 4; http://dx.doi.org/10.1074/jbc.273.18.10811; PMID: 9556550
- Pu WT, Wickman K, Clapham DE. ICln is essential for cellular and early embryonic viability. J Biol Chem 2000; 275:12363 - 6; http://dx.doi.org/10.1074/jbc.275.17.12363; PMID: 10777517
- Strange K. Molecular identity of the outwardly rectifying, swelling-activated anion channel: time to reevaluate pICln. J Gen Physiol 1998; 111:617 - 22; http://dx.doi.org/10.1085/jgp.111.5.617; PMID: 9565399
- Fürst J, Gschwentner M, Ritter M, Bottà G, Jakab M, Mayer M, Garavaglia L, Bazzini C, Rodighiero S, Meyer G, et al. Molecular and functional aspects of anionic channels activated during regulatory volume decrease in mammalian cells. Pflugers Arch 2002; 444:1 - 25; http://dx.doi.org/10.1007/s00424-002-0805-1; PMID: 11976912
- Fürst J, Bazzini C, Jakab M, Meyer G, König M, Gschwentner M, Ritter M, Schmarda A, Bottà G, Benz R, et al. Functional reconstitution of ICln in lipid bilayers. Pflugers Arch 2000; 440:100 - 15; PMID: 10864003
- Jentsch TJ. Chloride channels are different. Nature 2002; 415:276 - 7; http://dx.doi.org/10.1038/415276a; PMID: 11796992
- Bahamonde MI, Valverde MA. Voltage-dependent anion channel localises to the plasma membrane and peripheral but not perinuclear mitochondria. Pflugers Arch 2003; 446:309 - 13; PMID: 12698369
- Kotake S, Yago T, Kawamoto M, Nanke Y. Voltage-dependent anion channels (VDACs, porin) expressed in the plasma membrane regulate the differentiation and function of human osteoclasts. Cell Biol Int 2013; 37:65 - 77; http://dx.doi.org/10.1002/cbin.10013; PMID: 23319323
- Mobasheri A, Lewis R, Maxwell JEJ, Hill C, Womack M, Barrett-Jolley R. Characterization of a stretch-activated potassium channel in chondrocytes. J Cell Physiol 2010; 223:511 - 8; http://dx.doi.org/10.1002/jcp.22075; PMID: 20162564
- Meadows LS, Malhotra J, Loukas A, Thyagarajan V, Kazen-Gillespie KA, Koopman MC, Kriegler S, Isom LL, Ragsdale DS. Functional and biochemical analysis of a sodium channel β1 subunit mutation responsible for generalized epilepsy with febrile seizures plus type 1. J Neurosci 2002; 22:10699 - 709; PMID: 12486163
- Phan MN, Leddy HA, Votta BJ, Kumar S, Levy DS, Lipshutz DB, Lee SH, Liedtke W, Guilak F. Functional characterization of TRPV4 as an osmotically sensitive ion channel in porcine articular chondrocytes. Arthritis Rheum 2009; 60:3028 - 37; http://dx.doi.org/10.1002/art.24799; PMID: 19790068
- Hdud IM, El-Shafei AA, Loughna P, Barrett-Jolley R, Mobasheri A. Expression of Transient Receptor Potential Vanilloid (TRPV) Channels in Different Passages of Articular Chondrocytes. Int J Mol Sci 2012; 13:4433 - 45; http://dx.doi.org/10.3390/ijms13044433; PMID: 22605988
- Fernández-Fernández JM, Andrade YN, Arniges M, Fernandes J, Plata C, Rubio-Moscardo F, Vázquez E, Valverde MA. Functional coupling of TRPV4 cationic channel and large conductance, calcium-dependent potassium channel in human bronchial epithelial cell lines. Pflugers Arch 2008; 457:149 - 59; http://dx.doi.org/10.1007/s00424-008-0516-3; PMID: 18458941
- Karlsson C, Dehne T, Lindahl A, Brittberg M, Pruss A, Sittinger M, Ringe J. Genome-wide expression profiling reveals new candidate genes associated with osteoarthritis. Osteoarthritis Cartilage 2010; 18:581 - 92; http://dx.doi.org/10.1016/j.joca.2009.12.002; PMID: 20060954
- Preston GM, Carroll TP, Guggino WB, Agre P. Appearance of water channels in Xenopus oocytes expressing red cell CHIP28 protein. Science 1992; 256:385 - 7; http://dx.doi.org/10.1126/science.256.5055.385; PMID: 1373524
- Yuan F-L, Chen F-H, Lu W-G, Li X, Wu F-R, Li J-P, Li CW, Wang Y, Zhang TY, Hu W. Acid-sensing ion channel 1a mediates acid-induced increases in intracellular calcium in rat articular chondrocytes. Mol Cell Biochem 2010; 340:153 - 9; http://dx.doi.org/10.1007/s11010-010-0412-y; PMID: 20179994
- Müller EM, Hub JS, Grubmüller H, de Groot BL. Is TEA an inhibitor for human Aquaporin-1?. Pflugers Arch 2008; 456:663 - 9; http://dx.doi.org/10.1007/s00424-007-0422-0; PMID: 18196268
- Detmers FJM, de Groot BL, Müller EM, Hinton A, Konings IBM, Sze M, Flitsch SL, Grubmüller H, Deen PM. Quaternary ammonium compounds as water channel blockers. Specificity, potency, and site of action. J Biol Chem 2006; 281:14207 - 14; http://dx.doi.org/10.1074/jbc.M513072200; PMID: 16551622
- Lewis R, Feetham CH, Gentles L, Penny J, Tregilgas L, Tohami W, Mobasheri A, Barrett-Jolley R. Benzamil sensitive ion channels contribute to volume regulation in canine chondrocytes. Br J Pharmacol 2013; 168:1584 - 96; http://dx.doi.org/10.1111/j.1476-5381.2012.02185.x; PMID: 22928819