Abstract
The kidney and brain protein (KIBRA) is a scaffold or an adaptor-like protein with WW, C2-like and atypical protein kinase C (aPKC)-binding domains. Genetic studies in Drosophila revealed that KIBRA is an upstream regulator of the conserved Hippo pathway, which is implicated in organ size determination. In addition, genome-wide studies revealed an association between the single nucleotide polymorphism in the KIBRA gene locus and human episodic memory performance. However, the mechanism of action through which KIBRA is linked to these functions remains poorly understood. Recent studies on the biochemical and cellular properties of KIBRA reveal the role of KIBRA as a regulator of membrane trafficking. Further, KIBRA directly inhibits the activity of the cell polarity regulator, aPKC, which is required for apical protein exocytosis. Here, we discuss how this KIBRA-aPKC connection, a potential regulator of membrane trafficking and cell polarity, can contribute to the recently discovered functions of KIBRA.
KIBRA is a scaffold or adaptor-like protein implicated in a variety of cellular processes and functions that include regulation of the cytoskeleton,Citation1,Citation2 mitogen-activated protein kinase (MAPK) activation,Citation3 and directional cell migration.Citation2,Citation4 Recent genetic studies in Drosophila revealed an unexpected role of KIBRA in the control of body organ size; KIBRA was reported to be involved in cell growth and apoptosis, acting as an upstream regulator of the Hippo pathwayCitation5-Citation7 (For a review see refs. Citation8, Citation9). Importantly, this function of KIBRA seems to be conserved in mammals.Citation10,Citation11 In a completely different context, a whole-genome association study using single nucleotide polymorphisms (SNPs) has established an association of the KIBRA gene locus with human episodic memory performance.Citation12 The T-allele of the C/T polymorphism in the ninth intron of the KIBRA gene (rs17070145) has been linked to increased episodic memory (For a review see ref. Citation13). Although the mechanism by which the SNPs influence the memory formation remains unclear, a recent study using KIBRA-knockout mice showed that KIBRA is necessary for the contextual and trace fear memory in adult mice.Citation14 At the cellular level, accumulating evidence suggests that the involvement of KIBRA in plasma membrane trafficking in several contexts.Citation14-Citation16 Furthermore, at the molecular level, KIBRA associates with aPKC and inhibits its kinase activity.Citation15 In this minireview, we briefly summarize our current knowledge on molecular and cellular functions of KIBRA and discuss how these functions of KIBRA are linked to each other, especially from the point of view of the role played by the KIBRA-aPKC interaction in membrane trafficking and cell polarity.
Genes, Protein Sequences and Binding Partners
Before discussing cellular functions, it is important to overview the basic properties of KIBRA. KIBRA is a member of the WWC family of proteins, which contain two WW domains and a C2-like domain (). The WW domains are composed of 34 amino acids including two tryptophans (WW) and can recognize proline-rich motifs, such as PPxY and PPxP. The C2 domain is composed of approximately 130 amino acid residues that bind to two or three calcium ions and to phospholipids for targeting proteins to the plasma membrane. In addition to the WW and C2-like domains, WWC proteins share potential coiled-coil domains, an aPKC-binding region and a PDZ-binding motif (). There are two paralogs of KIBRA in humans: WWC2, which has a 49% total sequence identity with KIBRA, and WWC3, which has a 39% total sequence identity with KIBRA, but lacks the WW domains. Not all species have all three paralogs of KIBRA: no ortholog of KIBRA has been identified in Caenorhabditis elegans, only one ortholog of KIBRA (Kibra) has been identified in Drosophila, only KIBRA and WWC3 are present in fish, and only KIBRA and WWC2 in mice.
Figure 1. KIBRA homologs and binding partners. The sequence similarity of each domain to human KIBRA is shown as a percentage. The arrowhead indicates the site of the serine residue that can be phosphorylated by aurora kinase.
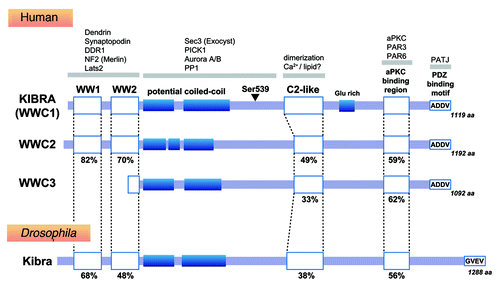
The WW domains in KIBRA can recognize the PPxY motifCitation1 and are highly conserved (> 50% similarity) from Drosophila to humans. The WW domains of KIBRA interact with components of the Hippo pathway, such as neurofibromin 2 (NF2)/Merlin (belonging to the FERM protein family) and the nuclear Dbf2-related kinase Lats2, thereby leading to KIBRA-induced activation of this pathway to suppress the transcriptional activity of Yes-associated protein (YAP).Citation5-Citation7,Citation11 KIBRA also binds to the discoidin domain receptor 1 (DDR1) through the WW domains to mediate MAPK activation in response to collagen in mammary epithelial cells.Citation3 The WW domains also bind to cytoskeletal-associated proteins, such as dendrinCitation1 and synaptopodin,Citation2 suggesting the importance of the interaction between KIBRA and the cytoskeleton. The C2-like domain of KIBRA can mediate its homodimerization.Citation17 Further, critical residues for lipid and calcium binding are conserved, and the crystal structure of the C2-like domain of KIBRA (DOI:10.2210/pdb2z0u/pdb) is very similar to the C2 domain of synaptotagmin, rabphilin and conventional protein kinase C (PKC)-β, suggesting that the C2-like domain of KIBRA may be involved in calcium/lipid-dependent signaling. Although the intermediate region between the WW and C2 domains is not so conserved, this region commonly contains potential coiled-coil sequences and has several important binding partners. Examples of these binding partners include a component of the exocyst complex, Sec3, to regulate directional cell migration,Citation4 and protein interacting with C kinase 1 (PICK1) to regulate the trafficking of α-amino-3-hydroxyl-5-methyl-4-isoxazole-propionate (AMPA) receptors, the major excitatory neurotransmitter receptors in the brain.Citation14 The intermediate sequence between the WW and C2 domains of KIBRA also contains a conserved motif with a serine residue (S539), whose phosphorylation status is regulated by aurora kinase and protein phosphatase 1.Citation10 Phosphorylation of S539 results in the dissociation of NF2/Merlin from KIBRA.Citation10 The C-terminal region of KIBRA seems to be important for the interaction of KIBRA with cell polarity regulators such as the partitioning defective (PAR) proteins: the aPKC-binding region associates with a core polarity regulator known as the PAR3-aPKC-PAR6 complex (PAR complex) to regulate apical vesicle trafficking,Citation15 and the C-terminal type III PDZ- binding motif (ADDV) binds to a cell polarity regulator called protein associated with tight junction (PATJ) to regulate the directional migration of podocytes.Citation2 In Drosophila, the C-terminal PDZ-binding motif seems to be conserved as a type II PDZ-binding motif (GVEV).Citation18 Considering that the aPKC-binding region and the PDZ binding motif are highly conserved between species, the interaction between KIBRA and the polarity complex may be of fundamental importance to these organisms.
Involvement of KIBRA in Membrane/Vesicular Trafficking through Early/Recycling Endosomes
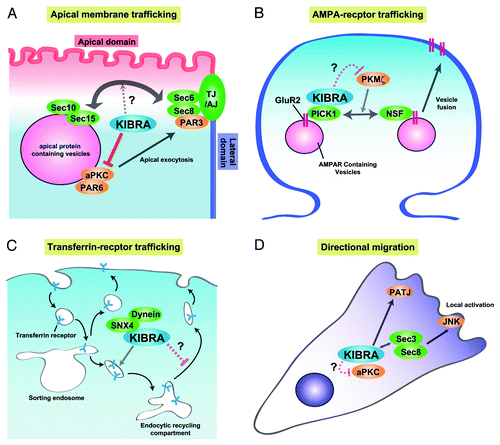
Recent discoveries about the cellular functions of KIBRA suggest that KIBRA plays an important role in membrane homeostasis via the regulation of vesicular transport. Analysis using the Madin-Darby canine kidney (MDCK) cells, which are cultured epithelial cells derived from the canine kidney, revealed the function of KIBRA in apical plasma membrane trafficking in epithelial cells.Citation15 Epithelial cells maintain their apical-basolateral membrane polarity by regulating the polarized trafficking machinery, which transports different plasma membrane proteins to the apical and basolateral membrane domains.Citation19 In the absence of cell-cell contact, large intracellular vacuoles called vacuolar apical compartments (VACs) appear,Citation20 which are thought to be normal intermediates in the biogenesis of apical surface and often result from the fusion of apical transport vesicles under conditions of reduced delivery form the apical recycling/early endosome to the apical plasma membrane.Citation19 Even in the absence of cell-cell contact, however, KIBRA-depleted MDCK cells did not exhibit the formation of VACs.Citation15 Furthermore, KIBRA knockdown accelerates the exocytosis of the apical protein, p75, to the cell surface under these conditions, suggesting that KIBRA exerts an inhibitory action on vesicular trafficking from the endosome to the plasma membrane (exocytosis) ().Citation15 The knockdown of KIBRA does not have any effect on apical protein internalization induced by collagen overlay, suggesting that the endocytic pathway is not affected by KIBRA.Citation15 The involvement of KIBRA in exocytosis, but not in endocytosis, seems to be a conserved property observed in a different biological context. In hippocampal neurons, KIBRA has been shown to be involved in the suppression of AMPA receptor trafficking to the cell surface; the knockdown of KIBRA did not affect internalization of the GluR2 subunit of AMPA receptor, but accelerated the recycling speed to the cell surfaceCitation14 (). In another study, Traer and colleaguesCitation16 showed that KIBRA is required for the sorting of the transferrin receptor (TfnR). Knockdown of KIBRA in HeLa cells disrupted the endosomal recycling compartment, possibly due to inhibition of the trafficking between the endosomal sorting compartment to the endosomal recycling compartment (). Importantly, there were no effects on the internalization of TfnR. Although this report did not examine whether the recycling of TfnR is affected by the knockdown of KIBRA, it supports the idea that KIBRA plays important roles in endosomal trafficking in various types of cells.
Figure 2. Models of the molecular mechanism of KIBRA in regulating membrane trafficking. (A) In epithelial cells, KIBRA suppresses the kinase activity of aPKC, resulting in suppression of the trafficking of apical-containing vesicles to the cell surface or cell-cell contact site. Association between KIBRA and the exocyst complex can influence this process. (B) In neurons, KIBRA associates with PICK1 and suppresses recycling of the AMPA receptor subunit, GluR2, to the cell surface. PKMζ enhances GluR2 surface expression via regulating the NSF-mediated release of GluR2 from PICK1. KIBRA may inhibit the kinase activity of PKMζ, leading to suppression of GluR2 recycling. (C) In endosomal trafficking of TfnR, KIBRA does not affect internalization, but affects trafficking from the endosome sorting compartment to the endosomal recycling compartment. (D) In directional migration, KIBRA can affect vesicular trafficking through interaction with the exocyst complex and aPKC.
Potential Importance of KIBRA-aPKC Pathway in Membrane Trafficking
How can these phenotypes in membrane trafficking be explained at the molecular level? The subcellular localization of endogenously expressed KIBRA seems to depend on the cell type; however, it matches with the localization of cell polarity regulators. In cultured hippocampal neurons, KIBRA colocalizes with MAP2 at shaft regions and also with postsynaptic marker proteins.Citation17 Biochemically, KIBRA was enriched in the synaptosome and postsynaptic density fraction,Citation14,Citation17 which supports the localization pattern obtained by immunofluorescence. KIBRA localizes in the apical domain and at cell-cell junctions in cultured epithelial cells (MDCK cells).Citation15 This localization is conserved in vivo, such as in the distal tubular epithelial cells in mouse kidneyCitation15 and in the follicular epithelial cells in Drosophila.Citation5-Citation7 In migrating cells, KIBRA accumulates in the leading edge.Citation2,Citation4 The PAR complex localizes in the synaptic region of cultured hippocampal neurons and in the apical domain of epithelial cells.Citation21-Citation23 Furthermore, aPKC, PAR3 and PATJ localize in the leading edge of migrating cells.Citation24 These findings suggest that KIBRA functions together with polarity proteins in many biological contexts.
KIBRA can be phosphorylated by aPKC,Citation25 but can also directly inhibit the kinase activity of aPKC in a competitive manner.Citation15 Because the kinase activity of aPKC is required for the exocytosis of VACs,Citation26 the inhibitory function of KIBRA can explain the phenotype of KIBRA-depleted MDCK cells in which the knockdown of KIBRA increases the kinase activity of aPKC, resulting in enhanced apical exocytosis (). Consistent with these results, the treatment with an aPKC inhibitor rescues the formation of VACs in KIBRA-knockdown cells.Citation15 Furthermore, overexpression of the aPKC-binding region, which possesses an inhibitory activity against aPKC in vitro, induced the formation of VACs in wild-type MDCK cells.Citation15 These results indicate that the interaction between KIBRA and aPKC as well as the KIBRA-mediated inhibition of the kinase activity of aPKC are important for the regulation of membrane trafficking to the cell surface.
Taken together with the recent discovery by Makuch et al.,Citation14 the inhibitory function of KIBRA against aPKC in membrane trafficking can explain how KIBRA influences memory formation at the molecular level (). Protein kinase M (PKM) ζ is a brain-specific, constitutively active form of aPKC that is transcribed from an alternative promoterCitation27,Citation28 and is necessary and sufficient for preserving late-long-term potentiation, presenting a cellular model of synaptic plasticity.Citation29 Expression of PKMζ increases the surface localization of AMPA receptors.Citation30 This process is involved in N-ethylmaleimide-sensitive factor-mediated dissociation of PICK1 from the GluR2 subunit.Citation31 KIBRA binds to and inhibits the activity of not only aPKC, but also of PKMζ (unpublished results). Importantly, as mentioned above, KIBRA suppresses recycling of the AMPA receptors to the cell surface, possibly through the interaction with PICK1.Citation14 Based on these results, we propose a model that KIBRA, through an interaction with PICK1, inhibits the kinase activity of constitutively active PKMζ (). KIBRA may be phosphorylated by PKMζ in the regulation of AMPA receptor trafficking, as suggested by Makuch et al.Citation14 However, the knockdown of KIBRA accelerates the surface expression of AMPA receptors,Citation14 which is similar to the overexpression of PKMζ.Citation30 Thus, it is highly possible that KIBRA inhibits the kinase activity of PKMζ during AMPA receptor trafficking similar to that in apical membrane trafficking in epithelial cells. Interestingly, it has been reported that the mRNA level of PKMζ decreases, whereas that of KIBRA increases in several Alzheimer patients.Citation13,Citation32,Citation33 Considering the antagonistic relationship between PKMζ and KIBRA, it is tempting to speculate that the kinase activity of PKMζ is dramatically decreased by the overexpression of KIBRA leading to severe defects in AMPA receptor surface trafficking. It will be interesting to test whether the interaction between PKMζ and KIBRA is regulated by neuronal activity.
Genome-wide screening in C.elegans showed that the PAR complex seems to be involved in endosomal trafficking.Citation34 This study showed that the suppression of PAR complex by dominant-negative Cdc42 or dominant-negative PAR6 inhibited internalization of TfnR in HeLa cells, but did not affect its recycling. Trafficking of TfnR depends on clathrin, but interestingly, in the case of clathrin-independent pathways such as the major histocompatibility complex (MHC) class I endocytosis and recycling, inhibition of the PAR complex does not affect MHC class I internalization, but rather suppresses its recycling to the cell surface. Considering that KIBRA does not affect the internalization of TfnR,Citation16 we cannot explain currently how KIBRA and the PAR complex cooperate to regulate TfnR trafficking. We speculate that KIBRA and the PAR complex can independently regulate TfnR trafficking. It would be interesting to investigate whether KIBRA cooperates with the PAR complex to regulate the clathrin-independent pathway of exocytosis. If a cooperation between KIBRA and the PAR complex exists, then the knockdown of KIBRA may enhance the recycling of MHC class I molecules to the cell surface due to the increased activity of the PAR complex.
The importance of the KIBRA-aPKC pathway seems to be conserved in directional migration (). Duning et al.Citation2 demonstrated that the knockdown of KIBRA in cultured podocytes increased the migrating speed, but caused a frequent change in the direction of migration, resulting in inefficient directional migration. Cell migration requires vesicular transport, endocytosis of the plasma membrane at the lagging edge, and exocytosis at the leading edge. The knockdown of KIBRA may enhance the exocytosis at the leading edge, resulting in hastened dysregulation of cell migration. Indeed, Rosse and colleaguesCitation4 showed that KIBRA binds not only to aPKC, but also to a component of the exocyst complex, Sec3, which is required for vesicle tethering and targeting in exocytosis, and the knockdown of KIBRA in normal rat kidney epithelial cells results in mislocalization of aPKC, leading to suppression of cell migration in a wound healing assay. These findings raise the possibility that KIBRA plays a critical role in cell migration by regulating vesicular trafficking through its interaction with aPKC.
Conclusions and Future Directions
As reviewed above, KIBRA seems to play important roles in plasma membrane homeostasis through the regulation of exocytosis from the endosome to the cell surface. However, we do not know the precise step in membrane trafficking in which KIBRA participates, such as vesicle fusion and membrane budding. Furthermore, we know very little about how KIBRA, in conjunction with aPKC, affects membrane regulatory machineries, such as the SNARE complex. Importantly, KIBRA binds to a component of the exocyst complex, Sec3, which regulates vesicular targeting and tethering during exocytosis. During apical membrane exocytosis, the components of the exocyst complex, Sec8, Sec10 and Sec15a, mediate the activity of Rab11a and Rab8 to target apical-containing vesicles to the PAR3-localizing cell-cell contact site ().Citation35 Furthermore, PKC phosphorylates the exocyst complex and regulates vesicular exocytosis.Citation36 Taken together, the exocyst complex seems to be the key in clarifying the nature of the KIBRA-aPKC connection. Furthermore, identification of unknown aPKC targets, whose phosphorylation is regulated by KIBRA, will give us novel insights in this area. Finally, how the cellular functions of KIBRA described here are related to other functions of KIBRA, including the regulation of the Hippo pathway, remains an intriguing question to be answered in the future.
| Abbreviations: | ||
| AMPA | = | α-amino-3-hydroxyl-5-methyl-4-isoxazole-propionate |
| aPKC | = | atypical protein kinase C |
| DDR1 | = | discoidin domain receptor 1 |
| FERM | = | Band 4.1, Ezrin, Radixin and Moesin |
| KIBRA | = | kidney and brain protein |
| MAPK | = | mitogen-activated protein kinase |
| MAP2 | = | microtubule-associated protein 2 |
| MDCK | = | Madin-Darby canine kidney |
| MHC | = | major histocompatibility complex |
| NF2 | = | neurofibromin 2 |
| PAR | = | partitioning defective |
| PATJ | = | protein associated with tight junction |
| PDZ | = | postsynaptic density 95 (PSD95)/discs large (Dlg)/zonula occludens-1 (ZO-1) |
| PICK1 | = | protein interacting with C kinase 1 |
| PKMζ | = | protein kinase Mζ |
| SNARE | = | soluble NSF attachment protein receptor |
| SNP | = | single nucleotide polymorphism |
| TfnR | = | transferrin receptor |
| VAC | = | vacuolar apical compartment |
| YAP | = | yes-associated protein |
References
- Kremerskothen J, Plaas C, Büther K, Finger I, Veltel S, Matanis T, et al. Characterization of KIBRA, a novel WW domain-containing protein. Biochem Biophys Res Commun 2003; 300:862 - 7; http://dx.doi.org/10.1016/S0006-291X(02)02945-5; PMID: 12559952
- Duning K, Schurek EM, Schlüter M, Bayer M, Reinhardt HC, Schwab A, et al. KIBRA modulates directional migration of podocytes. J Am Soc Nephrol 2008; 19:1891 - 903; http://dx.doi.org/10.1681/ASN.2007080916; PMID: 18596123
- Hilton HN, Stanford PM, Harris J, Oakes SR, Kaplan W, Daly RJ, et al. KIBRA interacts with discoidin domain receptor 1 to modulate collagen-induced signalling. Biochim Biophys Acta 2008; 1783:383-93.
- Rosse C, Formstecher E, Boeckeler K, Zhao Y, Kremerskothen J, White MD, et al. An aPKC-exocyst complex controls paxillin phosphorylation and migration through localised JNK1 activation. PLoS Biol 2009; 7:e1000235; http://dx.doi.org/10.1371/journal.pbio.1000235; PMID: 19885391
- Genevet A, Wehr MC, Brain R, Thompson BJ, Tapon N. Kibra is a regulator of the Salvador/Warts/Hippo signaling network. Dev Cell 2010; 18:300 - 8; http://dx.doi.org/10.1016/j.devcel.2009.12.011; PMID: 20159599
- Yu J, Zheng Y, Dong J, Klusza S, Deng WM, Pan D. Kibra functions as a tumor suppressor protein that regulates Hippo signaling in conjunction with Merlin and Expanded. Dev Cell 2010; 18:288 - 99; http://dx.doi.org/10.1016/j.devcel.2009.12.012; PMID: 20159598
- Baumgartner R, Poernbacher I, Buser N, Hafen E, Stocker H. The WW domain protein Kibra acts upstream of Hippo in Drosophila. Dev Cell 2010; 18:309 - 16; http://dx.doi.org/10.1016/j.devcel.2009.12.013; PMID: 20159600
- Grusche FA, Richardson HE, Harvey KF. Upstream regulation of the hippo size control pathway. Curr Biol 2010; 20:R574 - 82; http://dx.doi.org/10.1016/j.cub.2010.05.023; PMID: 20619814
- Genevet A, Tapon N. The Hippo pathway and apico-basal cell polarity. Biochem J 2011; 436:213 - 24; http://dx.doi.org/10.1042/BJ20110217; PMID: 21568941
- Xiao L, Chen Y, Ji M, Volle DJ, Lewis RE, Tsai MY, et al. KIBRA protein phosphorylation is regulated by mitotic kinase aurora and protein phosphatase 1. J Biol Chem 2011; 286:36304 - 15; http://dx.doi.org/10.1074/jbc.M111.246850; PMID: 21878642
- Xiao L, Chen Y, Ji M, Dong J. KIBRA regulates Hippo signaling activity via interactions with large tumor suppressor kinases. J Biol Chem 2011; 286:7788 - 96; http://dx.doi.org/10.1074/jbc.M110.173468; PMID: 21233212
- Papassotiropoulos A, Stephan DA, Huentelman MJ, Hoerndli FJ, Craig DW, Pearson JV, et al. Common Kibra alleles are associated with human memory performance. Science 2006; 314:475 - 8; http://dx.doi.org/10.1126/science.1129837; PMID: 17053149
- Schneider A, Huentelman MJ, Kremerskothen J, Duning K, Spoelgen R, Nikolich K. KIBRA: A New Gateway to Learning and Memory?. Front Aging Neurosci 2010; 2:4; PMID: 20552044
- Makuch L, Volk L, Anggono V, Johnson RC, Yu Y, Duning K, et al. Regulation of AMPA receptor function by the human memory-associated gene KIBRA. Neuron 2011; 71:1022 - 9; http://dx.doi.org/10.1016/j.neuron.2011.08.017; PMID: 21943600
- Yoshihama Y, Sasaki K, Horikoshi Y, Suzuki A, Ohtsuka T, Hakuno F, et al. KIBRA suppresses apical exocytosis through inhibition of aPKC kinase activity in epithelial cells. Curr Biol 2011; 21:705 - 11; http://dx.doi.org/10.1016/j.cub.2011.03.029; PMID: 21497093
- Traer CJ, Rutherford AC, Palmer KJ, Wassmer T, Oakley J, Attar N, et al. SNX4 coordinates endosomal sorting of TfnR with dynein-mediated transport into the endocytic recycling compartment. Nat Cell Biol 2007; 9:1370 - 80; http://dx.doi.org/10.1038/ncb1656; PMID: 17994011
- Johannsen S, Duning K, Pavenstädt H, Kremerskothen J, Boeckers TM. Temporal-spatial expression and novel biochemical properties of the memory-related protein KIBRA. Neuroscience 2008; 155:1165 - 73; http://dx.doi.org/10.1016/j.neuroscience.2008.06.054; PMID: 18672031
- Hung AY, Sheng M. PDZ domains: structural modules for protein complex assembly. J Biol Chem 2002; 277:5699 - 702; http://dx.doi.org/10.1074/jbc.R100065200; PMID: 11741967
- Weisz OA, Rodriguez-Boulan E. Apical trafficking in epithelial cells: signals, clusters and motors. J Cell Sci 2009; 122:4253 - 66; http://dx.doi.org/10.1242/jcs.032615; PMID: 19923269
- Vega-Salas DE, Salas PJ, Rodriguez-Boulan E. Exocytosis of vacuolar apical compartment (VAC): a cell-cell contact controlled mechanism for the establishment of the apical plasma membrane domain in epithelial cells. J Cell Biol 1988; 107:1717 - 28; http://dx.doi.org/10.1083/jcb.107.5.1717; PMID: 3053735
- Zhang H, Macara IG. The PAR-6 polarity protein regulates dendritic spine morphogenesis through p190 RhoGAP and the Rho GTPase. Dev Cell 2008; 14:216 - 26; http://dx.doi.org/10.1016/j.devcel.2007.11.020; PMID: 18267090
- Zhang H, Macara IG. The polarity protein PAR-3 and TIAM1 cooperate in dendritic spine morphogenesis. Nat Cell Biol 2006; 8:227 - 37; http://dx.doi.org/10.1038/ncb1368; PMID: 16474385
- Suzuki A, Ohno S. The PAR-aPKC system: lessons in polarity. J Cell Sci 2006; 119:979 - 87; http://dx.doi.org/10.1242/jcs.02898; PMID: 16525119
- Du D, Xu F, Yu L, Zhang C, Lu X, Yuan H, et al. The tight junction protein, occludin, regulates the directional migration of epithelial cells. Dev Cell 2010; 18:52 - 63; http://dx.doi.org/10.1016/j.devcel.2009.12.008; PMID: 20152177
- Büther K, Plaas C, Barnekow A, Kremerskothen J. KIBRA is a novel substrate for protein kinase Czeta. Biochem Biophys Res Commun 2004; 317:703 - 7; http://dx.doi.org/10.1016/j.bbrc.2004.03.107; PMID: 15081397
- Horikoshi Y, Suzuki A, Yamanaka T, Sasaki K, Mizuno K, Sawada H, et al. Interaction between PAR-3 and the aPKC-PAR-6 complex is indispensable for apical domain development of epithelial cells. J Cell Sci 2009; 122:1595 - 606; http://dx.doi.org/10.1242/jcs.043174; PMID: 19401335
- Hirai T, Niino YS, Chida K. PKC ζ II, a small molecule of protein kinase C ζ, specifically expressed in the mouse brain. Neurosci Lett 2003; 348:151 - 4; http://dx.doi.org/10.1016/S0304-3940(03)00780-8; PMID: 12932816
- Hernandez AI, Blace N, Crary JF, Serrano PA, Leitges M, Libien JM, et al. Protein kinase M ζ synthesis from a brain mRNA encoding an independent protein kinase C ζ catalytic domain. Implications for the molecular mechanism of memory. J Biol Chem 2003; 278:40305 - 16; http://dx.doi.org/10.1074/jbc.M307065200; PMID: 12857744
- Ling DS, Benardo LS, Serrano PA, Blace N, Kelly MT, Crary JF, et al. Protein kinase Mzeta is necessary and sufficient for LTP maintenance. Nat Neurosci 2002; 5:295 - 6; http://dx.doi.org/10.1038/nn829; PMID: 11914719
- Ling DS, Benardo LS, Sacktor TC. Protein kinase Mzeta enhances excitatory synaptic transmission by increasing the number of active postsynaptic AMPA receptors. Hippocampus 2006; 16:443 - 52; http://dx.doi.org/10.1002/hipo.20171; PMID: 16463388
- Yao Y, Kelly MT, Sajikumar S, Serrano P, Tian D, Bergold PJ, et al. PKM ζ maintains late long-term potentiation by N-ethylmaleimide-sensitive factor/GluR2-dependent trafficking of postsynaptic AMPA receptors. J Neurosci 2008; 28:7820 - 7; http://dx.doi.org/10.1523/JNEUROSCI.0223-08.2008; PMID: 18667614
- Burgess JD, Pedraza O, Graff-Radford NR, Hirpa M, Zou F, Miles R, et al. Association of common KIBRA variants with episodic memory and AD risk. Neurobiol Aging 2011; 32:557 e1-9.
- Corneveaux JJ, Liang WS, Reiman EM, Webster JA, Myers AJ, Zismann VL, et al. Evidence for an association between KIBRA and late-onset Alzheimer’s disease. Neurobiol Aging 2010; 31:901 - 9; http://dx.doi.org/10.1016/j.neurobiolaging.2008.07.014; PMID: 18789830
- Balklava Z, Pant S, Fares H, Grant BD. Genome-wide analysis identifies a general requirement for polarity proteins in endocytic traffic. Nat Cell Biol 2007; 9:1066 - 73; http://dx.doi.org/10.1038/ncb1627; PMID: 17704769
- Bryant DM, Datta A, Rodríguez-Fraticelli AE, Peränen J, Martín-Belmonte F, Mostov KE. A molecular network for de novo generation of the apical surface and lumen. Nat Cell Biol 2010; 12:1035 - 45; http://dx.doi.org/10.1038/ncb2106; PMID: 20890297
- Chen XW, Leto D, Xiao J, Goss J, Wang Q, Shavit JA, et al. Exocyst function is regulated by effector phosphorylation. Nat Cell Biol 2011; 13:580 - 8; http://dx.doi.org/10.1038/ncb2226; PMID: 21516108