Abstract
Genomic imprinting characterizes genes with a monoallelic expression, which is dependent on the parental origin of each allele. Approximately 150 imprinted genes are known to date, in humans and mice but, though computational searches have tried to extract intrinsic characteristics of these genes to identify new ones, the existing list is probably far from being comprehensive. We used a high-throughput strategy by diverting the classical use of genotyping microarrays to compare the genotypes of mRNA/cDNA vs. genomic DNA to identify new genes presenting monoallelic expression, starting from human placental material. After filtering of data, we obtained a list of 1,082 putative candidate monoallelic SNPs located in more than one hundred candidate genes. Among these, we found known imprinted genes, such as IPW, GRB10, INPP5F and ZNF597, which contribute to validate the approach. We also explored some likely candidates of our list and identified seven new imprinted genes, including ZFAT, ZFAT-AS1, GLIS3, NTM, MAGI2, ZC3H12Cand LIN28B, four of which encode zinc finger transcription factors. They are, however, not imprinted in the mouse placenta, except for Magi2. We analyzed in more details the ZFAT gene, which is paternally expressed in the placenta (as ZFAT-AS1, a non-coding antisense RNA) but biallelic in other tissues. The ZFAT protein is expressed in endothelial cells, as well as in syncytiotrophoblasts. The expression of this gene is, moreover, downregulated in placentas from complicated pregnancies. With this work we increase by about 10% the number of known imprinted genes in humans.
Introduction
In mammalian cells, both alleles of autosomal genes are assumed to contribute equally to the overall expression level. However, genes submitted to genomic imprinting do not follow this rule of thumb. These genes are characterized by a monoallelic expression that is dependent on the parental origin of the expressed allele, meaning that some genes are expressed only from the maternally inherited copy whereas others are expressed only from the allele derived from the father. Imprinted genes present a series of common, though not essential, features: localization within clusters, complex epigenetic regulation, preferential expression in placenta and brain, non-coding antisense RNAs, etc.Citation1 Up to now, about 150 imprinted genes are known in mice and humans (http://igc.otago.ac.nz; www.geneimprint.com; www.mousebook.org/catalog.php?catalog=imprinting).
As mentioned above, most of the known imprinted genes are highly expressed in the placenta, and they often play important roles in placental and fetal development. They are therefore interesting candidates for understanding mother/fetus exchanges and placental pathologies. Imprinted genes participate to an intertwined regulation network where they contribute to control fetal growth.Citation2 This network structure is quite important to regulate the system leading to nutrient exchanges. Furthermore, it has been postulated that deregulations or mutations of a so far unknown imprinted gene could be responsible for some cases of preeclampsia, a pregnancy specific disease caused by a dysfunctional placenta.Citation3 In these perspectives, other preeclampsia candidate genes, STOX1 and CDKN1C (p57kip2), have been intensively studied on the basis of their imprinted status.Citation4-Citation8
Attempts to identify new imprinted genes in humans and mice by computational meansCitation9-Citation15 or using parthenogenetic tissuesCitation16-Citation19 have been reported, using putative genomic specificities such as gene organization or presence of repeated elements, to provide with lists of candidate genes. However, validation approaches to confirm the imprinted status of these computed genes have globally failed to identify numerous new imprinted genes.Citation20 Nevertheless it appears crucial to obtain an exhaustive list of human imprinted genes.
In order to identify new genes that could be imprinted or monoallelic in the human placenta, we designed a strategy using high throughput genotyping arrays. This strategy is decomposed in a first high-throughput genomic search for monoallelic expression followed by a second step of validation where (1) the monoallelic expression is confirmed and (2) the maternal or paternal transmission of the expression profile is explored. We describe here the identification of seven new imprinted genes expressed in the human placenta.
Results
Strategy for a global search for monoallelic expression in the human placenta
In order to identify new genes exhibiting a monoallelic expression profile (and, therefore, potentially imprinted) in the human placenta, we designed a genome-wide strategy using and “diverting” high-throughput genotyping microarrays, depicted in Figure S1. We hybridized in parallel two 250K genotyping Affymetrix arrays with either cDNA (cDNA) or genomic DNA (gDNA), both extracted from a single human placenta. We performed this approach for five unrelated placentas obtained from normal pregnancies. We called the genotypes for all SNPs of the arrays in order to compare the results obtained between cDNA and gDNA from each placenta. As expected, the quality of the genotypes obtained from cDNAs was lower than from genomic DNA (70% interpretable vs. 96%, respectively). We therefore performed a filtration step on the quality of the genotypes to keep only the most secure results (detailed in the Supplemental Material). Quality filtration removed about 88% of the genotypes. To demonstrate a monoallelic pattern, an informative SNP should present the following characteristics: (1) a necessary heterozygosity on gDNA and (2) an apparent homozygosity on the corresponding cDNA. We selected all SNPs meeting these criteria (15,125 SNPs). We made another selection to concentrate on SNPs where at least two placentas out of the five tested showed this potential monoallelic profile. This left us with 1,088 candidate SNPs. The next step was to associate genes to the selected SNPs, leading to 1,082 SNPs effectively connected to at least one gene (provided as Table S2) among which some known imprinted genes could be identified, such as IPW, GRB10, INPP5F and ZNF597. In particular, ZNF597 is a transcription factor reported as imprinted in human white blood cells.Citation21 Using rs12737 and/or rs37824, we confirmed that ZNF597 is expressed following a monoallelic pattern in all human placentas tested (n = 7) and that the active allele is the one inherited from the mother (n = 3 informative mothers) (Fig. S2). We explored this gene (Zfp597) in the murine placenta using rs4152238 but discovered a biallelic expression in E14.5 and E17.5 total placentas as well as in E17.5 labyrinth and spongiotrophoblast.
The list was concentrated to a total of 171 genes that were called by at least two SNPs or by an exonic SNP.
Validation of candidate genes
A list of 20 genes was chosen for further validation, with a priority given to genes with monoallelic SNPs according to the following criteria: i) redundancy (n > 2 SNP in the gene), ii) location in exonic sequences, or iii) located in introns. For each gene, one or two exonic (coding or UTR) polymorphisms were chosen from dbSNP (NCBI database), likely to be frequent in Caucasian populations. Specific primers were designed, in order to genotype these SNPs, with specific amplifications, different for genomic DNA and cDNA, when possible (see Table S1). More than 30 placental genomic DNAs were genotyped to identify heterozygous individuals. We then sequenced the cDNA obtained from RNA extracted from the same placental tissue. For 12 genes, a biallelic signal was observed on cDNA despite the microarray result (Table S3), demonstrating false positives. However, for the other candidate genes, sequencing of placental cDNAs from heterozygous individuals led to observe a consistent monoallelic expression. summarizes the list of monoallelic genes identified (as exemplified for the ZFAT gene in ). For all SNPs and genes, either one or the other known alleles could be observed as monoallelic among the studied samples, rejecting the hypothesis that the nucleotidic variations per se or an associated haplotype could be responsible for the extinction of one allele. None of these genes localizes in close proximity to known imprinted genes or cluster of imprinted genes.
Table 1. Summary of newly identified imprinted genes
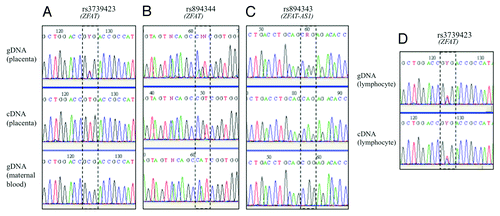
Figure 1. Sequencing results of SNPs in the human ZFAT and ZFAT-AS1 genes. Imprinted expression was deduced by comparing genotypes of gDNA, cDNA and maternal gDNA, in placenta using SNPs rs rs3739423 (A), rs894344 (B) and rs894343 (C) and in lymphocytes (D).
The new genes identified as monoallelic seem to be involved in various functions, including diabetes, growth and cancer. The transcription factor LIN28B (Lin28 C. elegans homolog B) together with its paralogous gene LIN28A, has been the subject of many publications in relation to miRNA regulation and cell transformation. However, it is worth noting that four are transcription factors harboring zinc finger domains: (1) LIN28B, (2) ZFAT (zinc finger gene in Autoimmune Thyroid disease or zinc finger and AT hook domain containing or ZNF406), (3) GLIS3 (GLIS family zinc finger protein 3 or ZNF515) and (4) ZC3H12C. Two other genes are involved in neuronal cell adhesion [NTM (Neurotrimin) and MAGI2 (Membrane-associated Guanylate Kinase, WW and PDZ domain containing 2 or S-SCAM for Synaptic Scaffolding molecule)].
While most identified genes show a strict monoallelic expression in all cDNA samples tested (from 8 to 11 independent samples), two genes have a less stringent profile. For MAGI2 and NTM, about half of the heterozygous samples show a monoallelic expression while other samples maintain the expression of both alleles.
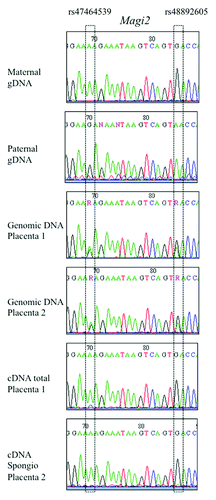
Genotyping of genomic DNAs from the respective mothers, when available, was also performed in order to determine the allelic parental transmission. For a given gene, all informative samples (a heterozygous child with an homozygous mother) gave the same parental inheritance. Though this strict parental origin cannot be formally demonstrated, the scores calculated in tend to validate this specific inheritance. Therefore, six genes (ZFAT, ZFAT-AS1, GLIS3, ZC3H12C, MAGI2 and LIN28B) were expressed only from the paternal copy while for two genes (NTM and ZNF597), the maternally inherited allele was the only active one ().
Therefore, we report the identification of seven new imprinted genes, matching both criteria of monoallelic expression and parental dependent expression.
New imprinted genes are mainly specific to humans
We explored the expression of the newly identified imprinted genes in placentas from F1 mice resulting from the mating of two distinct murine sub-species, Mus musculus domesticus and Mus musculus molossinus. Exonic SNP whose allelic versions are different between the parental strains were chosen for each gene from the dbSNP and MGI databases. Heterozygosity was first confirmed for all SNPs on genomic DNA extracted from each fetus whose placenta was studied. Sequencing cDNAs from the corresponding E17.5 total placentas (n = 2) produced a biallelic profile in the cases of Zfat, Glis3, Zc3h12c and Lin28b, contrary to human placentas (as exemplified for Zfat in ).
Figure 2. Sequencing results of SNPs in the murine (A) and bovine (B) Zfat genes. Imprinted expression was deduced by comparing genotypes of gDNA, cDNA and maternal gDNA, in placenta and other tissues.
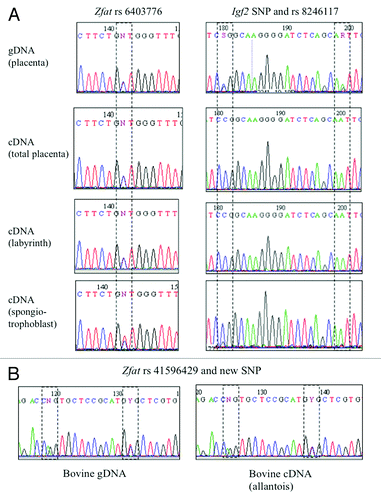
For the Zfat gene, we also explored cDNAs extracted from E14.5 total placentas, and from separated labyrinth and spongiotrophoblast layers of murine placentas (n = 2). Whereas the amplification always gives perfectly equilibrated allelic peaks for total placentas and labyrinth, the spongiotrophoblast amplification (n = 2) exhibited a slight excess of the maternal allele. We hypothesized that this might be due either to a contamination of this thin sample by maternal endometrial tissue or to an incomplete imprinting in this tissue. Amplification of the imprinted and paternally expressed Igf2 gene around both SNP rs8246117 and an unreported nearby SNP showed a strict paternal expression in the labyrinth and in total placenta, whereas a small maternal peak was also observed in cDNAs from the spongiotrophoblast layer, in favor of a contamination of this material by maternal tissue (). Therefore, the Zfat gene does not seem to be imprinted in the murine placenta.
We had access to 26 samples of bovine placentas from early and mid-gestation. We genotyped DNAs extracted from these placentas for SNPs rs41596429 located in the 3′UTR of the bovine Zfat gene. Three samples showed heterozygosity for this SNP, and one of them was, in addition, heterozygous for two new polymorphisms located within the same amplicon. Sequencing of the Zfat cDNA from various fetal tissues (cotyledon, allantois, amnios, liver, brain and heart) demonstrated a biallelic signal in all cases () and therefore an absence of imprinting in this species, as in mice.
Only the Magi2 gene seemed to be also imprinted in mice and cattle. F1 heterozygous placentas exhibited a monoallelic signal for cDNA for rs2956343, a SNP located within the 5′UTR region of the longest isoform. When studying two other SNPs located within a central exon common to all 3 alternative isoforms (rs47464539 and rs48892605), a similar monoallelic signal was obtained, in total placental cDNA, as well as in labyrinth and spongiotrophoblast extracts. The expressed allele was the one inherited from the mother (), which is different from the paternal expression of the human MAGI2 gene. In the bovine species, genomic annotation describes two MAGI2 genes in close neighborhood. For the most upstream one (XM_595287), we could find a polymorphic SNP (rs42342806). In embryonic and extra-embryonic tissues of 18 d (cotyledon, brain) and 60 d (allantoid, amnios, muscle, brain, heart, intestine) heterozygous animals, a monoallelic signal was observed in Magi2 cDNAs. The expressed allele was the one inherited from the mother, as in mice.
The ZFAT gene presents characteristics of classic imprinted genes
We demonstrated that the ZFAT gene presents both basic characteristics of an imprinted gene: a monoallelic expression pattern in placental tissue and a strict parental (paternal) origin of the expressed allele. ZFAT codes for a widely expressed transcription factor, mainly studied in lymphocyte signaling but strongly expressed in the placenta and was recently shown to be involved in capillary formation and organization of the endothelium.Citation22 We analyzed more deeply other traits that are shared by reported imprinted genes.
A non-coding antisense RNA overlaps the ZFAT locus
A non-coding antisense RNA, overlapping the ZFAT genomic region and transcribed in the opposite direction to ZFAT has been reported in the literature and named ZFAT-AS1 (map in Fig. S3).Citation23 The second exon of ZFAT-AS1 partially overlaps with ZFAT exon 7 and both ZFAT-AS1 exons overlap with the alternative extended version of ZFAT exon 7. We identified SNP rs894343 as present on both ZFAT and ZFAT-AS1 coding sequences and genotyped it in placental gDNAs. We then amplified in placental cDNAs a specific ZFAT-AS1 product containing this polymorphism. Sequencing of cDNAs from heterozygous carriers showed a perfect monoallelic expression of the ZFAT-AS1 transcript in 10 of the 10 cases available (). For three placentas, access to the informative genotype of the mother allowed us to deduce that the only ZFAT-AS1 expressed allele was also the one transmitted by the father. This indicates that ZFAT and ZFAT-AS1 are both imprinted in the human placenta, with an expression of the paternal alleles.
Gene and CpG island localization, methylation levels
ZFAT is located on chromosome 8q24. The nearest known human imprinted genes are located distally at about 5 Mb (KCNK9), 8 Mb (LY6D) and 10 Mb (GPT). The murine Zfat gene is carried by chromosome 15 in a region syntenic to its human equivalent and is also relatively distant from validated imprinted genes (Kcnk9, Peg 13 and Slc38a4). It is interesting to note the presence of two miRNA coding genes, MIR30B and MIR30D, near ZFAT (Fig. S3).
We searched the ZFAT genomic sequence for CpG-rich regions using the newcpgreport, cpgplot and newcpgseek software. They were consistently located in three regions: (1) near the miRNA genes about 120 kb upstream of ZFAT, (2) at the beginning of the gene in the transcription start region and (3) at the end of the gene. These three CpG rich regions are approximately 1.5 kb (162 CpGs), 1.5 kb (143 CpGs) and 3.5 kb (215 CpGs) long, respectively, and are conserved in mammals. We designed primers to amplify these regions from bisulfite-treated placental genomic DNAs. After direct sequencing, we could read 24 CpGs from the first region, 8 from the second and 23 from the third one. All non-CpG cytosines in the first two regions had been fully transformed by the bisulfite treatment and no trace of resistance due to methylation could be observed at any position and in any of the ten placental genomic DNAs tested, reflecting an unmethylated status. CpGs in the third region were however completely resistant to bisulfite treatment, showing a complete methylated status (data not shown). Therefore, we could not observe the specific differentially methylated profile of many genuine imprinted genes. This suggests that the imprinted status of the locus could be related to a differential methylation profile of other more distant CpG islands, or to other epigenetic mechanisms of regulation of monoallelic expression.
Imprinted expression of ZFAT is not observed in other human tissues
We then wanted to check if the monoallelic pattern of ZFAT was also present in other human tissues. We explored lymphocytes of individuals genotyped as heterozygous for rs3739423 and/or rs894344. In six independent cases (five simple heterozygotes and a double heterozygote), ZFAT cDNAs showed a biallelic pattern (). Therefore, we consider that the ZFAT gene is not imprinted in lymphocytes.
Endometrial tissues were also explored. In two samples heterozygous for either SNP of the ZFAT gene, a biallelic expression could be observed (data not shown) and, therefore, suggested that ZFAT is not imprinted in this cell type. We genotyped four patients affected with thyroid tumors for ZFAT and ZFAT-AS1 SNPs. Three of them were found concordantly heterozygous on both lymphocyte and tumor genomic DNAs, in order to exclude loss of heterozygosity in the tumor due to genic rearrangements. ZFAT and ZFAT-AS1 expression remained biallelic in the tumor. In conclusion, ZFAT monoallelic expression is not ubiquitously distributed in the body.
ZFAT expression in pathological placentas
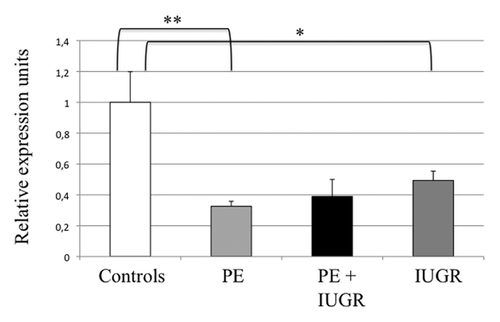
The expression of ZFAT was estimated by real-time RT-PCR in placental samples from pregnancies complicated by either preeclampsia, preeclampsia associated with IUGR, isolated IUGR or vascular IUGR (n = 14, n = 4, n = 9 and n = 7, respectively), and compared with placentas from uncomplicated pregnancies (n = 16). Interestingly, ZFAT appears to be under-expressed in all pathological placentas, particularly in preeclampsia where the average level is at least 3 times lower than in controls (p = 0.002) ().
Figure 4. Real-time RT-PCR of ZFAT in human placentas. Expression of the ZFAT gene was compared between Controls (n = 17), IUGR (pregnancies with intrauterine growth restriction) (n = 16), PE (pregnancies complicated with preeclampsia) (n = 15) and PE + IUGR (n = 5), after normalization by the SDHA housekeeping gene. *p = 0,02, ** p = 0,002 in a group vs. the control group.
The expression of ZFAT-AS1 was also challenged, but this antisense gene seems to be expressed at a much lower level (about 1,000 times less) than the sense ZFAT gene and was therefore too close to the detection threshold to conclude.
ZFAT protein expression in placentas
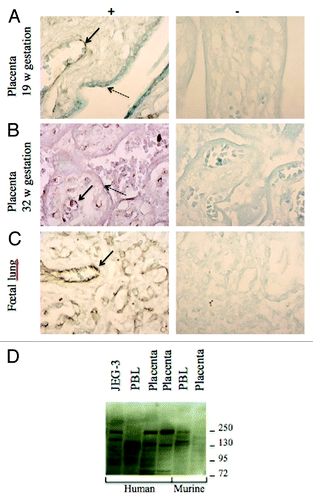
We used an anti-ZFAT antibody to reveal ZFAT expression profile on human placenta sections. Labeling was strong in endothelial cells, in both 19 and 32 weeks of amenorrhea placentas, with a cytoplasmic localization of the protein (). A fainter labeling was sometimes present in the syncytiotrophoblast layer, as a thin border. As a positive control, fetal lung also harbored an endothelial expression pattern (). A western blot using various human and murine tissues exhibited multiple bands compatible with the theoretical molecular masses of the main isoforms of ZFAT (139, 137 and 94 kDa for proteins Q9P243–1, -2 and -3, respectively, and 128 kDa for another isoform), although some slight differences in isoform expression can be observed between tissues and species ().
Figure 5. Immunohistochemistry to detect ZFAT on sections of human placentas. (A and B) (19 and 32 weeks of amenorrhea respectively) and (C) (fetal lung). (+) shows the detection with the primary antibody sc-87510 and (-) the negative control of detection without this antibody. Plain arrows show the strong labeling of endothelial cells while dotted arrows point to the syncytiotrophoblast labeling. (D) A western blot of human and mouse tissues revealed by an anti-ZFAT antibody. PBL stands for peripheral blood leukocytes whereas JEG-3 is a human choriocarcinoma cell line commonly used as a model of trophoblast.
LIN28B
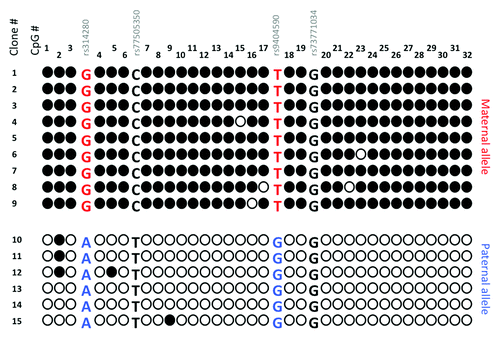
The LIN28B gene, a regulator of miRNA expression, was also more deeply analyzed. LIN28B is surrounded by CpG islands, and we chose to analyze the one co-localizing with its promoter. Direct sequencing of bisulfite treated-genomic DNA from 10 normal human placentas revealed double peaks at the 19 CpG positions available (data not shown). This suggested the presence of a differentially methylated region (DMR) characteristic of imprinted genes. To further characterize it, we cloned and sequenced this fragment for four individuals. shows the analysis of this region for one representative placenta, heterozygous for two SNPs and whose mother was moreover informative. We observed that CpGs in this region are completely methylated on the maternal allele, whereas the paternal allele shows a global unmethylated state. This is coherent with the extinction of the maternal copy of LIN28B and the expression of its paternal allele. This region therefore behaves as a differentially methylated region (DMR).
Figure 6. Cloning and sequencing of PCR fragments obtained from bisulfite-treated placental gDNA in the promoter CpG island of the LIN28B gene. Black and white circles represent methylated and unmethylated cytosines respectively over the 32 CpG dinucleotides under study. SNPs are present within this fragment and allow the study of the allelic segregation and parental transmission.
In agreement with an expression profile mainly embryonic, we could not amplify the LIN28B cDNA in lymphocytes or endometrial tissue and therefore could not investigate a putative imprinting status in other cell types than placenta.
Expression analysis of the LIN28B gene in pathological placentas did not show any significant variation in comparison to samples from normal pregnancies (data not shown).
Discussion
By diverting high-throughput genotyping arrays, we explored the putative imprinted status of candidate human genes.
Historically, the first maps of imprinted genes have been established thanks to the use of mice harboring uniparental disomies.Citation24 These large and complex chromosomal rearrangements together with ad hoc crosses allowed the production of mice carrying a large genomic region in the expected double copies but inherited from only one or the other parent. The resulting phenotypes were severe and allowed the identification of large clusters of imprinted genes, controlled by an imprinting control region (ICR). Putative imprinted genes, disseminated across the genome in opposition to those located in clusters, might have been missed by this screen as they are unlikely to participate to such large phenotypic syndromes and as they are probably not under the control of a long range ICR. In addition, imprinted genes having a restricted expression profile, either spatially or temporally, or with an imprinted pattern limited to a precise developmental stage, organ, or isoform, are probably very difficult to identify, particularly in humans. For example, in spite of controversial reports suggesting or excluding an imprinted status for the well-studied retinoblastoma gene RB1, it is only very recently that it was shown to undergo genomic imprinting.Citation25
Attempts to exhaustively identify imprinted genes have been developed, either using computational approaches or with high throughput strategies.Citation9,Citation10,Citation13-Citation15 If bioinformatical screenings have suggested that imprinted genes might be much more numerous than the actual known list, up to 600 in miceCitation12 and 150 in humans,Citation11 validation analyses to confirm the imprinted status of the candidate genes have not been very successful and the list has not increased much. A previous approach using genotyping arrays failed to reveal new imprinted genes.Citation26 However, a similar screening led to the identification of the random monoallelic pattern of lymphocytes.Citation27 Difficulties to identify new imprinted genes partially rely on the lack of strict characteristics of sequence, gene organization, etc., common to all imprinted genes and that could be used as criterion to identify new candidates.
High-throughput techniques as in this work allow the screening of a huge number of genes simultaneously, though the location of these SNPs, not gene-focused, is not ideal. Therefore, the background is high, particularly for the hybridization of cDNA and needs a careful selection of pertinent data. We used filtering steps to concentrate on the more robust set of data in terms of quality and reproducibility of the results and performed a complete validation step.
By sequencing the cDNA of placentas previously genotyped as heterozygous for SNPs located within exons, we could distinguish true or false candidates. Our validation step allowed the exclusion of some false positive genes. These might be due to a lack of sensibility of the technique; indeed, the software is designed to provide the genetic composition of genomic DNA (heterozygosity/homozygosity) whereas cDNA signals might deviate from this clear-cut situation and lead to a misinterpretation. In addition, our validation approach concentrates on the analysis of the major isoform or all isoforms of a given gene and might therefore miss some imprinting effects that could be specific of a particular alternative isoform of the gene (potentially targeted by the array’s probe), as already observed for GRB10, for example.Citation28
In our screen, ZFAT, the best candidate from the arrays, was targeted by six monoallelic SNPs, in at least two of the five placentas under study. It is noteworthy that these six SNPs are located in intronic regions of the ZFAT gene. Re-examination of the complete data showed that six additional SNPs, also located in intronic sequences, presented a monoallelic pattern in the only informative of the five placentas. It is therefore likely that, as we hypothesized prior to undertaking this study and as already mentioned,Citation27 a significant portion of pre-messengers is present in the RNA pool. This contributes efficiently to the hybridization process and produces consistent and robust results leading to the identification of monoallelic genes. This result strengthens the choice of the Affymetrix genotyping arrays that unfortunately harbor only 5% of coding SNPs, but that contain a large proportion of intronic SNPs.
This approach led to the confirmation of known imprinted genes such as INPP5F, IPW, GRB10 and ZNF597. This latter gene, a zinc finger transcription factor, had been cited as imprinted in lymphocytesCitation21 and was recently identified by screens for differentially methylated regions in uniparental disomiesCitation29 and hydatidiform moles.Citation16 We confirmed that this gene was also imprinted in the human placenta and that the expressed allele was the maternally inherited one, as recently reported.Citation29 In the murine placenta, the Zpf597 gene displayed a biallelic expression. Homozygous mice invalidated for this gene (also known as HIT-4) are early embryonic lethal whereas heterozygous mice exhibit abnormal behaviors and abnormal structures in the brain.Citation30 However, the precise role of this gene in both placental and brain function remains to be analyzed.
Besides, seven novel human imprinted genes were found, which represents an increase of about 10% relative to the today list of human imprinted genes. Little is known about the genes we identified and their precise role in the placenta generally remains to be characterized. Neurotrimin (NTM) is a cell adhesion molecule that seems to participate in synaptogenesis.Citation31 MAGI2 is involved in infantile spasm.Citation32 ZFAT is a widely expressed transcription factor from the zinc finger family that appears to play a role in immune functions and apoptosis.Citation33,Citation34 GLIS3 is implicated in neonatal diabetes and pancreatic development.Citation35,Citation36 This pathway has a considerable importance in placental physiology, since glucose transport and insulin regulation are considerably affected in growth restricted fetuses, as reviewed in.Citation37,Citation38 In addition, GLIS3 is strongly expressed in endothelial cells, an issue of relevance for the normal placental physiology, also encountered for ZFAT. LIN28B controls the expression of the let-7 miRNA and is implicated in cancer progression and glucose metabolism.Citation39,Citation40 LIN28B was found associated with age at menarche in various populations.Citation41 LIN28, the paralogous gene of LIN28B, has been shown to control IGF2 expression level at the post-transcriptional level.Citation42,Citation43 The expression of LIN28B is in addition correlated with IGF2 expression, at the mRNA and peptidic level in ovarian cancers, an interesting connection between imprinted genes.Citation44 The ZC3H12C gene has been shown to regulate inflammatory pathways in mice.Citation45 The balance between pro- and anti-inflammatory pathways is an issue of extreme importance in human placental diseases, where pro-inflammatory protein encoding genes are abnormally upregulated in pathological pregnancies, especially in preeclampsia.Citation46,Citation47 The imprinted status of inflammatory factors may thus be relevant to adjust the amount of protein synthesized. Some of these seven genes have a very strong expression in the placenta (ZFAT, LIN28B) while others are barely expressed in this tissue (NTM, MAGI2, ZNF597) (data obtained using Unigene and Nextbio). A cerebral expression profile is also common. Population studies have highlighted SNPs in some of them in association with growth related phenotypes.Citation41,Citation48-Citation50
Among several independent placenta samples, a perfectly monoallelic expression could be seen for five of the seven candidate genes, showing a strict imprinting. In these cases, either one or the other allele could be observed in the cDNAs, rejecting the hypothesis that one of the nucleotidic variations could be responsible for a functional effect resulting in the decrease of the expression level of one copy. For two genes (NTM and MAGI2), however, variability among individuals was observed, as some exhibited a strict monoallelic expression, while others maintained a biallelic profile of the same gene, tested under the same conditions. This phenomenon of polymorphic imprinting had already been observed for the IGF2R gene that is imprinted in mice but shows a variable pattern in humans.Citation51 The mechanisms underlying this variability are so far unknown.
Genes identified by this screen are located away from known imprinted genes or clusters. As no other signals in their neighborhood could be deduced from the arrays, they could therefore a priori belong to the class of isolated imprinted genes. It is striking to find four out of seven new imprinted genes coding for transcription factors with zinc finger domains. Analysis of 3′ exons of genes from this family had highlighted the coexistence of different and contradictory histone marks that could be comparable to those of imprinted genes.Citation52 The expression of a selection of ZF genes was however found biallelic by these authors and the presence of these marks was rather correlated with the repeats of ZF domains. In addition to the genes identified in this screen (ZFAT, GLIS3, ZC3H12C and LIN28B), other ZF genes are known to be imprinted: ZNF264, ZIM2, PLAGL1, PEG3, among others.
Conservation of the imprinted status is not strict between mammalian species. We explored murine placentas and observed a biallelic profile for most of the newly identified imprinted genes. Therefore, the placental imprinted profile of these genes seems to be specific to the human species. The L3MBTL1 gene, coding for a zinc finger transcription factor and potential tumor suppressor, is the only other case of an imprinted gene (also expressed from the paternal copy in human tissues) but escaping imprinting in mice.Citation53 However, it is difficult to refute or affirm categorically the imprinted status of a gene, particularly in the human species, as this phenomenon can be very restricted in its temporal and/or spatial profile.
The Magi2/S-Scam gene, however, seems to be also imprinted in the murine and bovine species. This gene produces three alternative isoforms, all of which seem to be imprinted from our results. A partial knockout mouse was performed that lacks the expression of the longest isoform (S-SCAMα) but maintains the two others (S-SCAMβ and γ).Citation54 No reproduction failure was reported but these mice die shortly after birth and seem to have abnormal signaling responses in dendrites. It is likely that this isoform, whose function is necessary in the brain, is not crucial in the murine placenta and that the presence of the two other isoforms is sufficient for the placental function of the Magi2 gene, whatever it is. In humans, deletion of this gene was associated with infantile spasms.Citation32 It was a surprise to see that the human MAGI2 gene expressed the paternal copy while in murine placenta and bovine tissues, the active copy was the maternal one. This situation has already been described for the ZIM2 geneCitation55 and some imprinted genes also show variable imprinting effects according to different isoforms and tissues.
Imprinted genes have in common a complex epigenetic regulation involving different mechanisms including antisense, miRNAs, differential methylation and histone modifications. Though not a strict requirement, the presence of antisense RNA is a hallmark of imprinted genes. ZFAT-AS1 is a 2-exon gene transcribed from the opposite strand and partially overlapping some ZFAT exons. We could show that this antisense gene also presents a monoallelic expression in the placenta and that the expressed copy is also of paternal origin. According to the ncRNAimprint database, all antisense RNAs described in the context of imprinted genes present this paternal specificity.Citation56 The antisense ZFAT-AS1 RNA together with the described antisenses for GLIS3 and MAGI2 seem to be human specificities absent in other mammalian species, from available sequence data of these genomic regions. The nearest genes distal to ZFAT, located about 80 kb upstream, are two miRNAs. The ZFAT gene has been hypothesized to be a potential target of these miR30B and miR30D genes (http://services.bio.ifi.lmu.de/mirsel). These miR30 RNAs, however, putatively target other genes, including the known imprinted genes RB1, CSF2 and CDKN1C. The small size and the paucity of frequent SNPs within these sequences make it difficult to analyze the expression profile and the potential imprinted status of these miRNAs, whose interactions with ZFAT might be of great interest.
The LIN28B and ZFAT genes were further studied in order to characterize their potential role in placenta function, as an imprinted gene.
We then explored the methylation status of the regions surrounding ZFAT and LIN28B. If the epigenetic control of known imprinted genes, generally located within clusters under the control of an ICR, is difficult to fully characterize, it appears even more difficult to anticipate how an isolated imprinted gene is regulated and at which distance are the crucial elements responsible for the differential imprints affixed during gametogenesis. In spite of this apparent lack of differentially methylated regions in ZFAT CpG islands, binding sites for transcription factors known to regulate imprinted genes can be detected in the ZFAT promoter, such as CTCF and YY1. In addition, other mechanisms than DNA methylation could be involved in the regulation of the ZFAT expression profile. For LIN28B, the CpG island located around the promoter showed, on the contrary, a differentially methylated profile that is likely to be an important regulatory region, a DMR as observed in most imprinted genes.
We then explored the imprinted status of ZFAT and LIN28B in other human tissues but failed to observe a monoallelic expression in other tissues than the placenta: in lymphocytes and endometrial tissue, ZFAT is strictly biallelic while LIN28B is not expressed. Therefore, the imprinted profile of ZFAT and LIN28B seems to be rather specific and may be restricted to the placenta. However, ZFAT could be successfully amplified from different tissues and also seems expressed in many tissues including spleen, thymus and brain.Citation57 Other genes are also known to have a restricted imprinted profile whereas others maintain their imprint within the complete bodyCitation58; even IGF2 escapes imprinting in the brain.Citation59
Null mice for Zfat present an embryonic lethality and do not survive after E8.5.Citation60 The phenotype is not observed in heterozygous mice, in agreement with our finding of a biallelic expression of Zfat in mouse. A placental phenotype was observed, as the spongiotrophoblast does not develop correctly. At E8, Zfat is normally expressed in the endothelial and hematopoietic progenitors of blood islands but the authors show in null mice an impaired differentiation of hematopoietic progenitors in blood islands of the yolk sac. ZFAT is also suspected to play a role in angiogenesis and hematopoiesis.Citation22 By immunohistochemistry we could find ZFAT expression in endothelial cells and syncytiotrophoblast. In addition, the gene was consistently downregulated in placentas from preeclampsia and/or IUGR, whereas this gene was described as stable along the human gestation period.Citation61 As a loss of imprinting would rather be synonym of increased expression, this deregulation is likely due to a transcriptional mechanism in the pathological context. Very interestingly, a recent study revealed that SNP variants within the ZFAT gene are associated with hypertension.Citation62 Our data, together with the literature coincide to propose ZFAT as a regulator of the differentiation of endothelial cells within and outside the placenta. This is consistent with the role of this gene in the assembly of endothelial cells during the angiogenesis process, and the formation of capillary networks, particularly in HUVECs.Citation22 Therefore, ZFAT appears as an interesting candidate to have a role in placental development that needs to be further characterized. Labeling of slides from pathological placentas might bring additional information on this abnormal expression profile.
In conclusion, in this work we could validate and identify new human imprinted genes whose function in the placenta remains to be explored. Increasing the number of identified imprinted genes and completing exhaustively their list will help to extract the specificities of this class of particular genes in terms of epigenetic regulation, physiological function and pathological alterations. These data are of particular interest in the present context, where approaches using new technologies such a RNA-seq to identify imprinted genes in mice tend to question some previously known imprinted genes and failed to identify many new imprinted genes.Citation63-Citation65 Indeed, some genes exhibiting a maternal expression could have been misinterpreted as imprinted after contamination of the murine placental tissue by highly expressing maternal uterine cells. The exploration of human placental tissue circumvents this problem, as its size and volume make it easier to avoid maternal contaminations.
Material and Methods
Tissue samples
Human placentas were collected from caesarean sections of normal and pathological pregnancies. Protocols were approved by the local ethics’ committee and all patients signed an informed consent. Patients’ description was previously reported.Citation66 After removal of the maternal membranes, a small piece of villosities was collected as previously described.Citation67 Human lymphocytes were purified from blood samples of mothers, patients or anonymous donors. Endometrial tissues were collected as described.Citation68 Thyroid tumors were also collected.
Mouse placentas were collected at embryonic day E14.5 and E17.5 after mating of a Mus musculus domesticus C57B6 female and a Mus musculus molossinus male. At E17.5, placentas were either kept complete or dissected to separate the labyrinth from the spongiotrophoblast layer, whereas E14.5 placentas were kept complete. Fetal material was also collected for DNA extraction and genotyping.
Bovine placental and fetal (trophoblast, cotyledon, allantois, liver, brain and heart) samples were collected from crosses between a female Holstein and a male Charolais, at 18 or 60 d post-coitum.
Whatever the source, all collected samples were divided in two aliquots and stored frozen either dry or in Trizol, for further DNA and RNA extraction respectively.
DNA and RNA extraction
Extractions were performed under standard procedures.Citation67 Briefly for DNA extraction, samples were first digested with proteinase K and SDS. Phenol-chloroform extractions were followed by alcohol precipitation of genomic DNA. Gel electrophoresis and OD measurement (Nanodrop) were performed to quantify the genomic DNA and check for its integrity. For RNA extraction, samples were lysed using beads in the Qiagen TissueLyser II apparatus. A Trizol extraction was then performed according to the manufacturer’s recommendations (Invitrogen, Cergy, France).Citation69
Genotyping arrays
After validation of the RNA quality with the Agilent Bioanalyzer 2100 (using Agilent RNA6000 nano chip kit) controlling for the absence of genomic contamination, 4 μg of total RNA were reverse transcribed using the One Cycle Target Labeling Kit (Affymetrix): briefly, a first strand cDNA synthesis was performed using Superscript II and T7oligodT followed by a second strand cDNA synthesis using E.coli DNA ligase, E.coli DNA polymerase and RNase H. The double strand cDNA was then purified using cDNA clean up spin column, eluted and quantified with the Nanodrop ND1000 UV-Vis Spectrophotometer (Nanodrop Technologies, Inc.). All cDNA samples were independently processed and final concentrations were adjusted to 100 ng/μl. The processing of cDNA samples was then identical to the processing of genomic DNA samples. Genotyping analysis with Affymetrix NspI chip was performed for both sample types following manufacturer’s guidelines. Briefly either 250 ng of double stranded cDNA or 250 ng of genomic DNA were restricted with NspI. NspI adaptors were then ligated to restricted fragments and subjected to PCR using the universal primer PCR002 provided by the kit. PCR fragments were then purified and 90 μg used for fragmentation and end-labeling with biotin using Terminal Transferase. Labeled targets were then hybridized overnight to Genechip® human 250K NspI array (Affymetrix) at 49°C. Chips were washed on the fluidic station FS450 following specific protocols (Affymetrix) and scanned using the GCS3000 7G. The image was then analyzed with the GCOS software to obtain raw data (cel files). Genotypes were called by the Affymetrix GType software using the Dynamic Model (DM) Mapping algorithms.
PCR, RT-PCR, sequencing
cDNA synthesis was performed using the MMLV cDNA or the SuperScript II synthesis kit (Invitrogen) from 2 μg of RNA, previously treated with DNase I to eliminate putative DNA contaminations. Primers were designed using the PRIMER3 software (http://frodo.wi.mit.edu). The list, sequences and conditions of use of the primers are in Table S1. Classically, PCR was performed on 1/10 of the RT product or on 200 ng of genomic DNA, for 35 cycles on a GeneAmp 750 thermocycler (Applied Biosystems) with Platinum Taq polymerase. Sequencing was performed on an Applied Biosystems 3130XL sequencer by the local platform.
Methylation analysis
Various softwares were used to locate CpG rich regions (newcpgreport, Cpgblot Cpgseek, CpGprod) at http://mobile.pasteur.fr. Aliquots of genomic DNA were treated with sodium bisulphite using the EZ DNA Methylation-Gold kit (Zymo Research). These DNAs were used as templates for amplification and direct sequencing of specific CpG islands and subsequent cloning into the pJET vector (Fermentas). Positive clones were checked by PCR and sequenced.
Real-time PCR
Real time PCR was performed using the LightCycler 480, the corresponding LC480 SYBRGreen Master kit and 96-well plates (Roche). Conditions were as follows: 95°C for 5 min, and 40 cycles of 3 temperature steps (95°C for 10 sec, 55°C for 10 sec and 72°C for 10 sec). Finally, samples were submitted to a progressive temperature ramping resulting in a melting curve, validating the specificity of the amplification. Amplification products were also checked by agarose gel electrophoresis and sequencing. The threshold cycle numbers (Ct) were obtained using the LightCycler 480 software (Roche) and the second derivate maximum method. Data from target genes were normalized using the succinate dehydrogenase subunit A (SDHA) used as reference gene and shown previously to be stable and highly expressed in the human placenta.Citation70 Data were analyzed using the delta Ct method.Citation71 Experiments were conducted in quadruplicates.
Immunoblotting and immunohistochemistry
Placenta, peripheral blood lymphocytes (PBL) or human choriocarcinoma JEG-3 cells were solubilized in RIPA buffer for 1h at +4°C. Fifty µg of each sample were loaded per well and separated on 4–12% NuPage Bis-Tris precast gels (Invitrogen) in MOPS SDS running buffer and then electro-transferred to PVDF membrane under standard conditions. The ZFAT protein was detected with a goat anti-ZFAT IgG (sc-87510, obtained from Santa Cruz Biotechnology) at 1:750 and donkey anti-goat peroxydase conjugated IgG at 1:7500 revealed by the enhanced Chemiluminescence (ECL) detection system (Millipore).
Human placental and fetal lung slides were hybridized with the same antibody (1:100) and revealed using the Universal LSAB + HRP Detection Kit (Dako). A counterstaining of slides was performed with Methyl Green.
| Abbreviations: | ||
| gDNA | = | genomic DNA |
| cDNA | = | complementary DNA |
| SNP | = | single nucleotide polymorphism |
| IUGR | = | intra uterine growth restriction |
Additional material
Download Zip (338.4 KB)Acknowledgments
We are grateful to all participating patients, as well as to the Centre d’Investigation Clinique en Périnatalogie of the Port Royal maternity for their expertise and help. We would like to thank Pr Francis Jaubert (Hôpital Necker) for his advice, Dr Houria Sahli (Hôpital Saint Vincent de Paul), Astrid Doutreluigne for her contribution and Céline Méhats for her critical reading of the manuscript.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Financial Disclosures
G.G.L. was supported by a fellowship from the Fondation pour la Recherche Médicale and by the Association de Néonatalogie de Port-Royal.
Supplemental Material
Supplemental materials may be found here: www.landesbioscience.com/journals/epigenetics/article/21495
References
- Reik W, Walter J. Genomic imprinting: parental influence on the genome. Nat Rev Genet 2001; 2:21 - 32; http://dx.doi.org/10.1038/35047554; PMID: 11253064
- Varrault A, Gueydan C, Delalbre A, Bellmann A, Houssami S, Aknin C, et al. Zac1 regulates an imprinted gene network critically involved in the control of embryonic growth. Dev Cell 2006; 11:711 - 22; http://dx.doi.org/10.1016/j.devcel.2006.09.003; PMID: 17084362
- Graves JA. Genomic imprinting, development and disease--is pre-eclampsia caused by a maternally imprinted gene?. Reprod Fertil Dev 1998; 10:23 - 9; http://dx.doi.org/10.1071/R98014; PMID: 9727590
- Fenstad MH, Johnson MP, Løset M, Mundal SB, Roten LT, Eide IP, et al. STOX2 but not STOX1 is differentially expressed in decidua from pre-eclamptic women: data from the Second Nord-Trondelag Health Study. Mol Hum Reprod 2010; 16:960 - 8; http://dx.doi.org/10.1093/molehr/gaq064; PMID: 20643876
- Kanayama N, Takahashi K, Matsuura T, Sugimura M, Kobayashi T, Moniwa N, et al. Deficiency in p57Kip2 expression induces preeclampsia-like symptoms in mice. Mol Hum Reprod 2002; 8:1129 - 35; http://dx.doi.org/10.1093/molehr/8.12.1129; PMID: 12468647
- Rigourd V, Chauvet C, Chelbi ST, Rebourcet R, Mondon F, Letourneur F, et al. STOX1 overexpression in choriocarcinoma cells mimics transcriptional alterations observed in preeclamptic placentas. PLoS One 2008; 3:e3905; http://dx.doi.org/10.1371/journal.pone.0003905; PMID: 19079545
- van Dijk M, Mulders J, Poutsma A, Könst AA, Lachmeijer AM, Dekker GA, et al. Maternal segregation of the Dutch preeclampsia locus at 10q22 with a new member of the winged helix gene family. Nat Genet 2005; 37:514 - 9; http://dx.doi.org/10.1038/ng1541; PMID: 15806103
- van Dijk M, van Bezu J, Poutsma A, Veerhuis R, Rozemuller AJ, Scheper W, et al. The pre-eclampsia gene STOX1 controls a conserved pathway in placenta and brain upregulated in late-onset Alzheimer’s disease. J Alzheimers Dis 2010; 19:673 - 9; PMID: 20110611
- Brideau CM, Eilertson KE, Hagarman JA, Bustamante CD, Soloway PD. Successful computational prediction of novel imprinted genes from epigenomic features. Mol Cell Biol 2010; 30:3357 - 70; http://dx.doi.org/10.1128/MCB.01355-09; PMID: 20421412
- Hutter B, Bieg M, Helms V, Paulsen M. Imprinted genes show unique patterns of sequence conservation. BMC Genomics 2010; 11:649; http://dx.doi.org/10.1186/1471-2164-11-649; PMID: 21092170
- Luedi PP, Dietrich FS, Weidman JR, Bosko JM, Jirtle RL, Hartemink AJ. Computational and experimental identification of novel human imprinted genes. Genome Res 2007; 17:1723 - 30; http://dx.doi.org/10.1101/gr.6584707; PMID: 18055845
- Luedi PP, Hartemink AJ, Jirtle RL. Genome-wide prediction of imprinted murine genes. Genome Res 2005; 15:875 - 84; http://dx.doi.org/10.1101/gr.3303505; PMID: 15930497
- Pollard KS, Serre D, Wang X, Tao H, Grundberg E, Hudson TJ, et al. A genome-wide approach to identifying novel-imprinted genes. Hum Genet 2008; 122:625 - 34; http://dx.doi.org/10.1007/s00439-007-0440-1; PMID: 17955261
- Wang Z, Fan H, Yang HH, Hu Y, Buetow KH, Lee MP. Comparative sequence analysis of imprinted genes between human and mouse to reveal imprinting signatures. Genomics 2004; 83:395 - 401; http://dx.doi.org/10.1016/j.ygeno.2003.09.007; PMID: 14962665
- Yang HH, Hu Y, Edmonson M, Buetow K, Lee MP. Computation method to identify differential allelic gene expression and novel imprinted genes. Bioinformatics 2003; 19:952 - 5; http://dx.doi.org/10.1093/bioinformatics/btg127; PMID: 12761057
- Choufani S, Shapiro JS, Susiarjo M, Butcher DT, Grafodatskaya D, Lou Y, et al. A novel approach identifies new differentially methylated regions (DMRs) associated with imprinted genes. Genome Res 2011; 21:465 - 76; http://dx.doi.org/10.1101/gr.111922.110; PMID: 21324877
- Ruf N, Dünzinger U, Brinckmann A, Haaf T, Nürnberg P, Zechner U. Expression profiling of uniparental mouse embryos is inefficient in identifying novel imprinted genes. Genomics 2006; 87:509 - 19; http://dx.doi.org/10.1016/j.ygeno.2005.12.007; PMID: 16455231
- Smith RJ, Dean W, Konfortova G, Kelsey G. Identification of novel imprinted genes in a genome-wide screen for maternal methylation. Genome Res 2003; 13:558 - 69; http://dx.doi.org/10.1101/gr.781503; PMID: 12670997
- Sritanaudomchai H, Ma H, Clepper L, Gokhale S, Bogan R, Hennebold J, et al. Discovery of a novel imprinted gene by transcriptional analysis of parthenogenetic embryonic stem cells. Hum Reprod 2010; 25:1927 - 41; http://dx.doi.org/10.1093/humrep/deq144; PMID: 20522441
- Ruf N, Bähring S, Galetzka D, Pliushch G, Luft FC, Nürnberg P, et al. Sequence-based bioinformatic prediction and QUASEP identify genomic imprinting of the KCNK9 potassium channel gene in mouse and human. Hum Mol Genet 2007; 16:2591 - 9; http://dx.doi.org/10.1093/hmg/ddm216; PMID: 17704508
- Pant PV, Tao H, Beilharz EJ, Ballinger DG, Cox DR, Frazer KA. Analysis of allelic differential expression in human white blood cells. Genome Res 2006; 16:331 - 9; http://dx.doi.org/10.1101/gr.4559106; PMID: 16467561
- Yoshida Y, Tsunoda T, Takashima Y, Fujimoto T, Doi K, Sasazuki T, et al. ZFAT is essential for endothelial cell assembly and the branch point formation of capillary-like structures in an angiogenesis model. Cell Mol Biol Lett 2010; 15:541 - 50; http://dx.doi.org/10.2478/s11658-010-0028-y; PMID: 20645017
- Shirasawa S, Harada H, Furugaki K, Akamizu T, Ishikawa N, Ito K, et al. SNPs in the promoter of a B cell-specific antisense transcript, SAS-ZFAT, determine susceptibility to autoimmune thyroid disease. Hum Mol Genet 2004; 13:2221 - 31; http://dx.doi.org/10.1093/hmg/ddh245; PMID: 15294872
- Cattanach BM, Kirk M. Differential activity of maternally and paternally derived chromosome regions in mice. Nature 1985; 315:496 - 8; http://dx.doi.org/10.1038/315496a0; PMID: 4000278
- Kanber D, Berulava T, Ammerpohl O, Mitter D, Richter J, Siebert R, et al. The human retinoblastoma gene is imprinted. PLoS Genet 2009; 5:e1000790; http://dx.doi.org/10.1371/journal.pgen.1000790; PMID: 20041224
- Daelemans C, Ritchie ME, Smits G, Abu-Amero S, Sudbery IM, Forrest MS, et al. High-throughput analysis of candidate imprinted genes and allele-specific gene expression in the human term placenta. BMC Genet 2010; 11:25; http://dx.doi.org/10.1186/1471-2156-11-25; PMID: 20403199
- Gimelbrant A, Hutchinson JN, Thompson BR, Chess A. Widespread monoallelic expression on human autosomes. Science 2007; 318:1136 - 40; http://dx.doi.org/10.1126/science.1148910; PMID: 18006746
- Blagitko N, Mergenthaler S, Schulz U, Wollmann HA, Craigen W, Eggermann T, et al. Human GRB10 is imprinted and expressed from the paternal and maternal allele in a highly tissue- and isoform-specific fashion. Hum Mol Genet 2000; 9:1587 - 95; http://dx.doi.org/10.1093/hmg/9.11.1587; PMID: 10861285
- Nakabayashi K, Trujillo AM, Tayama C, Camprubi C, Yoshida W, Lapunzina P, et al. Methylation screening of reciprocal genome-wide UPDs identifies novel human-specific imprinted genes. Hum Mol Genet 2011; 20:3188 - 97; http://dx.doi.org/10.1093/hmg/ddr224; PMID: 21593219
- Tanabe Y, Hirano A, Iwasato T, Itohara S, Araki K, Yamaguchi T, et al. Molecular characterization and gene disruption of a novel zinc-finger protein, HIT-4, expressed in rodent brain. J Neurochem 2010; 112:1035 - 44; http://dx.doi.org/10.1111/j.1471-4159.2009.06525.x; PMID: 19968752
- Gil OD, Zanazzi G, Struyk AF, Salzer JL. Neurotrimin mediates bifunctional effects on neurite outgrowth via homophilic and heterophilic interactions. J Neurosci 1998; 18:9312 - 25; PMID: 9801370
- Marshall CR, Young EJ, Pani AM, Freckmann ML, Lacassie Y, Howald C, et al. Infantile spasms is associated with deletion of the MAGI2 gene on chromosome 7q11.23-q21.11. Am J Hum Genet 2008; 83:106 - 11; http://dx.doi.org/10.1016/j.ajhg.2008.06.001; PMID: 18565486
- Doi K, Fujimoto T, Koyanagi M, Tsunoda T, Tanaka Y, Yoshida Y, et al. ZFAT is a critical molecule for cell survival in mouse embryonic fibroblasts. Cell Mol Biol Lett 2011; 16:89 - 100; http://dx.doi.org/10.2478/s11658-010-0041-1; PMID: 21225468
- Fujimoto T, Doi K, Koyanagi M, Tsunoda T, Takashima Y, Yoshida Y, et al. ZFAT is an antiapoptotic molecule and critical for cell survival in MOLT-4 cells. FEBS Lett 2009; 583:568 - 72; http://dx.doi.org/10.1016/j.febslet.2008.12.063; PMID: 19162026
- Kang HS, Kim YS, ZeRuth G, Beak JY, Gerrish K, Kilic G, et al. Transcription factor Glis3, a novel critical player in the regulation of pancreatic beta-cell development and insulin gene expression. Mol Cell Biol 2009; 29:6366 - 79; http://dx.doi.org/10.1128/MCB.01259-09; PMID: 19805515
- Yang Y, Chang BH, Yechoor V, Chen W, Li L, Tsai MJ, et al. The Krüppel-like zinc finger protein GLIS3 transactivates neurogenin 3 for proper fetal pancreatic islet differentiation in mice. Diabetologia 2011; 54:2595 - 605; http://dx.doi.org/10.1007/s00125-011-2255-9; PMID: 21786021
- Cetin I, Antonazzo P. The role of the placenta in intrauterine growth restriction (IUGR). Z Geburtshilfe Neonatol 2009; 213:84 - 8; http://dx.doi.org/10.1055/s-0029-1224143; PMID: 19536707
- Jansson T, Myatt L, Powell TL. The role of trophoblast nutrient and ion transporters in the development of pregnancy complications and adult disease. Curr Vasc Pharmacol 2009; 7:521 - 33; http://dx.doi.org/10.2174/157016109789043982; PMID: 19485888
- King CE, Wang L, Winograd R, Madison BB, Mongroo PS, Johnstone CN, et al. LIN28B fosters colon cancer migration, invasion and transformation through let-7-dependent and -independent mechanisms. Oncogene 2011; 30:4185 - 93; http://dx.doi.org/10.1038/onc.2011.131; PMID: 21625210
- Zhu H, Shyh-Chang N, Segrè AV, Shinoda G, Shah SP, Einhorn WS, et al, DIAGRAM Consortium, MAGIC Investigators. The Lin28/let-7 axis regulates glucose metabolism. Cell 2011; 147:81 - 94; http://dx.doi.org/10.1016/j.cell.2011.08.033; PMID: 21962509
- Elks CE, Ong KK. Whole genome associated studies for age at menarche. Brief Funct Genomics 2011; 10:91 - 7; http://dx.doi.org/10.1093/bfgp/elq030; PMID: 21436305
- Balzer E, Heine C, Jiang Q, Lee VM, Moss EG. LIN28 alters cell fate succession and acts independently of the let-7 microRNA during neurogliogenesis in vitro. Development 2010; 137:891 - 900; http://dx.doi.org/10.1242/dev.042895; PMID: 20179095
- Polesskaya A, Cuvellier S, Naguibneva I, Duquet A, Moss EG, Harel-Bellan A. Lin-28 binds IGF-2 mRNA and participates in skeletal myogenesis by increasing translation efficiency. Genes Dev 2007; 21:1125 - 38; http://dx.doi.org/10.1101/gad.415007; PMID: 17473174
- Lu L, Katsaros D, Shaverdashvili K, Qian B, Wu Y, de la Longrais IA, et al. Pluripotent factor lin-28 and its homologue lin-28b in epithelial ovarian cancer and their associations with disease outcomes and expression of let-7a and IGF-II. Eur J Cancer 2009; 45:2212 - 8; http://dx.doi.org/10.1016/j.ejca.2009.05.003; PMID: 19477633
- Liang J, Wang J, Azfer A, Song W, Tromp G, Kolattukudy PE, et al. A novel CCCH-zinc finger protein family regulates proinflammatory activation of macrophages. J Biol Chem 2008; 283:6337 - 46; http://dx.doi.org/10.1074/jbc.M707861200; PMID: 18178554
- Lockwood CJ, Huang SJ, Krikun G, Caze R, Rahman M, Buchwalder LF, et al. Decidual hemostasis, inflammation, and angiogenesis in pre-eclampsia. Semin Thromb Hemost 2011; 37:158 - 64; http://dx.doi.org/10.1055/s-0030-1270344; PMID: 21370218
- Lockwood CJ, Krikun G, Caze R, Rahman M, Buchwalder LF, Schatz F. Decidual cell-expressed tissue factor in human pregnancy and its involvement in hemostasis and preeclampsia-related angiogenesis. Ann N Y Acad Sci 2008; 1127:67 - 72; http://dx.doi.org/10.1196/annals.1434.013; PMID: 18443332
- Kim HN, Lee EJ, Jung SC, Lee JY, Chung HW, Kim HL. Genetic variants that affect length/height in infancy/early childhood in Vietnamese-Korean families. J Hum Genet 2010; 55:681 - 90; http://dx.doi.org/10.1038/jhg.2010.88; PMID: 20668459
- Takeuchi F, Nabika T, Isono M, Katsuya T, Sugiyama T, Yamaguchi S, et al. Evaluation of genetic loci influencing adult height in the Japanese population. J Hum Genet 2009; 54:749 - 52; http://dx.doi.org/10.1038/jhg.2009.99; PMID: 19834501
- Widén E, Ripatti S, Cousminer DL, Surakka I, Lappalainen T, Järvelin MR, et al. Distinct variants at LIN28B influence growth in height from birth to adulthood. Am J Hum Genet 2010; 86:773 - 82; http://dx.doi.org/10.1016/j.ajhg.2010.03.010; PMID: 20398887
- Monk D, Arnaud P, Apostolidou S, Hills FA, Kelsey G, Stanier P, et al. Limited evolutionary conservation of imprinting in the human placenta. Proc Natl Acad Sci U S A 2006; 103:6623 - 8; http://dx.doi.org/10.1073/pnas.0511031103; PMID: 16614068
- Blahnik KR, Dou L, Echipare L, Iyengar S, O’Geen H, Sanchez E, et al. Characterization of the contradictory chromatin signatures at the 3′ exons of zinc finger genes. PLoS One 2011; 6:e17121; http://dx.doi.org/10.1371/journal.pone.0017121; PMID: 21347206
- Li J, Bench AJ, Vassiliou GS, Fourouclas N, Ferguson-Smith AC, Green AR. Imprinting of the human L3MBTL gene, a polycomb family member located in a region of chromosome 20 deleted in human myeloid malignancies. Proc Natl Acad Sci U S A 2004; 101:7341 - 6; http://dx.doi.org/10.1073/pnas.0308195101; PMID: 15123827
- Iida J, Ishizaki H, Okamoto-Tanaka M, Kawata A, Sumita K, Ohgake S, et al. Synaptic scaffolding molecule alpha is a scaffold to mediate N-methyl-D-aspartate receptor-dependent RhoA activation in dendrites. Mol Cell Biol 2007; 27:4388 - 405; http://dx.doi.org/10.1128/MCB.01901-06; PMID: 17438139
- Kim J, Bergmann A, Lucas S, Stone R, Stubbs L. Lineage-specific imprinting and evolution of the zinc-finger gene ZIM2. Genomics 2004; 84:47 - 58; http://dx.doi.org/10.1016/j.ygeno.2004.02.007; PMID: 15203203
- Zhang Y, Guan DG, Yang JH, Shao P, Zhou H, Qu LH. ncRNAimprint: a comprehensive database of mammalian imprinted noncoding RNAs. RNA 2010; 16:1889 - 901; http://dx.doi.org/10.1261/rna.2226910; PMID: 20801769
- Koyanagi M, Nakabayashi K, Fujimoto T, Gu N, Baba I, Takashima Y, et al. ZFAT expression in B and T lymphocytes and identification of ZFAT-regulated genes. Genomics 2008; 91:451 - 7; http://dx.doi.org/10.1016/j.ygeno.2008.01.009; PMID: 18329245
- Coan PM, Burton GJ, Ferguson-Smith AC. Imprinted genes in the placenta--a review. Placenta 2005; 26:Suppl A S10 - 20; http://dx.doi.org/10.1016/j.placenta.2004.12.009; PMID: 15837057
- Pham NV, Nguyen MT, Hu JF, Vu TH, Hoffman AR. Dissociation of IGF2 and H19 imprinting in human brain. Brain Res 1998; 810:1 - 8; http://dx.doi.org/10.1016/S0006-8993(98)00783-5; PMID: 9813220
- Tsunoda T, Takashima Y, Tanaka Y, Fujimoto T, Doi K, Hirose Y, et al. Immune-related zinc finger gene ZFAT is an essential transcriptional regulator for hematopoietic differentiation in blood islands. Proc Natl Acad Sci U S A 2010; 107:14199 - 204; http://dx.doi.org/10.1073/pnas.1002494107; PMID: 20660741
- Winn VD, Haimov-Kochman R, Paquet AC, Yang YJ, Madhusudhan MS, Gormley M, et al. Gene expression profiling of the human maternal-fetal interface reveals dramatic changes between midgestation and term. Endocrinology 2007; 148:1059 - 79; http://dx.doi.org/10.1210/en.2006-0683; PMID: 17170095
- Slavin TP, Feng T, Schnell A, Zhu X, Elston RC. Two-marker association tests yield new disease associations for coronary artery disease and hypertension. Hum Genet 2011; 130:725 - 33; http://dx.doi.org/10.1007/s00439-011-1009-6; PMID: 21626137
- Okae H, Hiura H, Nishida Y, Funayama R, Tanaka S, Chiba H, et al. Re-investigation and RNA sequencing-based identification of genes with placenta-specific imprinted expression. Hum Mol Genet 2012; 21:548 - 58; http://dx.doi.org/10.1093/hmg/ddr488; PMID: 22025075
- Proudhon C, Bourc’his D. Identification and resolution of artifacts in the interpretation of imprinted gene expression. Brief Funct Genomics 2010; 9:374 - 84; http://dx.doi.org/10.1093/bfgp/elq020; PMID: 20829207
- Wang X, Sun Q, McGrath SD, Mardis ER, Soloway PD, Clark AG. Transcriptome-wide identification of novel imprinted genes in neonatal mouse brain. PLoS One 2008; 3:e3839; http://dx.doi.org/10.1371/journal.pone.0003839; PMID: 19052635
- Gascoin-Lachambre G, Buffat C, Rebourcet R, Chelbi ST, Rigourd V, Mondon F, et al. Cullins in human intra-uterine growth restriction: expressional and epigenetic alterations. Placenta 2010; 31:151 - 7; http://dx.doi.org/10.1016/j.placenta.2009.11.008; PMID: 20005570
- Mondon F, Mignot TM, Rebourcet R, Jammes H, Danan JL, Ferré F, et al. Profiling of oxygen-modulated gene expression in early human placenta by systematic sequencing of suppressive subtractive hybridization products. Physiol Genomics 2005; 22:99 - 107; http://dx.doi.org/10.1152/physiolgenomics.00276.2004; PMID: 15797968
- Borghese B, Mondon F, Noël JC, Fayt I, Mignot TM, Vaiman D, et al. Gene expression profile for ectopic versus eutopic endometrium provides new insights into endometriosis oncogenic potential. Mol Endocrinol 2008; 22:2557 - 62; http://dx.doi.org/10.1210/me.2008-0322; PMID: 18818281
- Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem 1987; 162:156 - 9; http://dx.doi.org/10.1016/0003-2697(87)90021-2; PMID: 2440339
- Meller M, Vadachkoria S, Luthy DA, Williams MA. Evaluation of housekeeping genes in placental comparative expression studies. Placenta 2005; 26:601 - 7; http://dx.doi.org/10.1016/j.placenta.2004.09.009; PMID: 16085039
- Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 2001; 29:e45; http://dx.doi.org/10.1093/nar/29.9.e45; PMID: 11328886