Abstract
Great efforts in the field of solid organ transplantation are being devoted to identifying biomarkers that allow a transplanted patient’s immune status to be established. Recently, it has been well documented that epigenetic mechanisms like DNA methylation and histone modifications regulate the expression of immune system-related genes, modifying the development of the innate and adaptive immune responses. An in-depth knowledge of these epigenetic mechanisms could modulate the immune response after transplantation and to develop new therapeutic strategies. Epigenetic modifiers, such as histone deacetylase (HDAC) inhibitors have considerable potential as anti-inflammatory and immunosuppressive agents, but their effect on transplantation has not hitherto been known. Moreover, the detection of epigenetic marks in key immune genes could be useful as biomarkers of rejection and progression among transplanted patients. Here, we describe recent discoveries concerning the epigenetic regulation of the immune system, and how this knowledge could be translated to the field of transplantation.
Introduction
The immune response to an allograft is one of the biggest barriers to successful transplantation. The type and strength of response is determined by the cell type and cellular conditions under which it occurs. External (infections, chemical agents and drugs) and internal (cytokines and hormones) environmental stimuli can modify the epigenetic profile of a gene, directly influencing its expression and, ultimately, the cell type and immune response. Our knowledge of the epigenetic mechanisms involved in the development and differentiation of the immune system, and consequently that of related pathologies (autoimmune diseases and hematological disorders), has advanced considerably in recent years.Citation1,Citation2 Transplantation was the last component to be added to the list, showing that monitoring these epigenetic changes or modifying the immune response through epigenetic modifiers could help follow up transplanted patients and reduce the adverse effects of current immunosuppressive therapies.Citation3-Citation5
The epigenome connects the genome with the cellular environment and determines cellular identity and functionality. DNA methylation and histone post-translational modifications act in concert to establish an open or closed chromatin structure and ultimately define active or inactive gene transcription (). In general, DNA hypermethylation is associated with repressive chromatin while histone marks are known to work in a synergistic way to activate or silence genes.Citation6,Citation7 These epigenetic modifications are established in specific residues by the action of several enzymes, including DNA methyltransferases (DNMTs), histone acetyltransferases (HATs), histone deacetylases (HDACs), histone methyltransferases (HMTs) and phosphatases, among others. Aberrant changes in these proteins or their targets enable gene expression to be modified and disrupt the cellular machinery contributing to alterations of the immune cell homeostasis. Thus, defining and modifying the “normal” epigenetic landscape of cells of the immune system could have important clinical consequences for the diagnosis and treatment of transplanted patients.
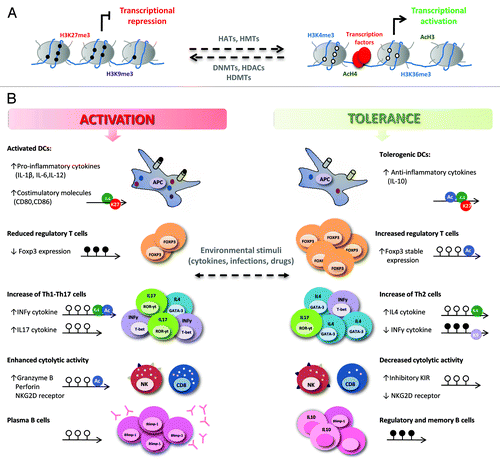
Figure 1. Epigenetic regulation of the immune response in transplantation. (A) DNA methylation and histone modifications allow changes in chromatin structure that influence gene transcription. DNA methylation (black circles) and repressive histone marks (H3K27me3, H3K9me3, etc.) are associated with closed chromatin and gene repression while DNA demethylation (white circles) and active histone modifications (AcH3, AcH4, H3K4me3, H3K36me3, etc.) are correlated with an open chromatin structure, facilitating transcription factor binding and transcription. Enzymes such as histone acetyltransferases (HATs), histone deacetylases (HDACs), histone methyltransferases (HMTs), histone demethylases (HDMTs) and DNA methyltransferases (DNMTs), establish and maintain the balance between these modifications. (B) Changes in the chromatin structure of key genes in the immune responses that take place in organ transplantation can contribute to alloimmunity or tolerance. Epigenetic modifications (DNA methylation and histone modifications) regulate gene expression in different cell types in several ways: (1) the expression of pro-inflammatory (IL12, IL-1β, IL-6, TNF-α), regulatory (IL-10) cytokines, and co-stimulatory molecules (CD80, CD86, CD40) is modified in antigen-presenting cells (APCs); (2) DNA demethylation and histone deacetylation increase the stable expression of the FoxP3 transcription factor, strengthening the suppressive function of Treg cells; (3) in CD4+ T cells, the differentiation of naive CD4+ T cells into “helper” T cells (Th1, Th2, Th17) and the plasticity among them is modulated; (4) the transcription of cytotoxic molecules (granzyme B and perforin) and activating receptors (NKG2D) is inhibited, and the expression of inhibitory KIR is enhanced in memory CD8+ T cells and NK cells, promoting allograft acceptance; (5) mature B cell differentiation into antibody-producing cells is controlled, decreasing allorecognition by donor-specific antibodies and preventing graft rejection. DNA methylation and demethylation are represented by black and white lollipops, respectively; histone modifications are shown as circles: green, H3K4me3; red, H3K27me3; purple, H3K9me3; blue, acetylation of H3 or H4.
Against this background, several international projects such as the ENCODE project (Encyclopedia of DNA Elements, www.encodeproject.org/ENCODE) and the NIH Roadmap Epigenomics Program (www.roadmapepigenomics.org) have been undertaken to create a resource of epigenomic maps in a variety of primary human cells and tissues in order to determine how these epigenetic mechanisms contribute to disease.Citation8,Citation9 Additionally, the BLUEPRINT (www.blueprint-epigenome.eu) and DEEP (www.deutsches-epigenom-programm.de) consortia are focused on creating reference epigenomes of hematopoietic cells from healthy donors and from patients with malignant diseases (leukemias and autoimmune diseases) or cells and tissues from metabolic and inflammatory diseases, respectively.Citation10 All these projects, now coordinated by the International Human Epigenome Consortium (IHEC; www.ihec-epigenomes.org), seem likely to provide a deep understanding of the epigenome of the immune system in the near future.
Herein, we review the recent advances in the knowledge of the epigenetic mechanisms involved in establishing cell-fate programs in the immune system and the way they are modified in response to certain environmental conditions (infection and inflammation). Special emphasis is placed on the plasticity between effector T helper (Th) subsets (Th1, Th2 and Th17) and regulatory T (Treg) CD4+ T cells—key players in the post-transplant immune response—and how it could be modulated by epigenetic modifiers. We also discuss possible clinical applications of DNA methylation and histone modification patterns as biomarkers for monitoring the immune status of transplanted patients. In-depth knowledge of these epigenetic mechanisms would help us develop novel strategies to modulate the immune responses to transplantation and identify new diagnostic biomarkers.
Epigenetic Regulation of the Immune Response and Allotolerance
The immune response produced after solid organ transplantation (SOT) is an ongoing dialog between the innate and adaptive immune systems. The development of these responses is orchestrated by a diverse group of functionally specialized cells and is highly influenced by the cellular microenvironment in which they are triggered. These cells lineages are established during hematopoietic differentiation through the interplay of multiple transcription factors and epigenetic modifications. However, these epigenetic patterns can be altered under certain environmental conditions (inflammation, viral infections and chemical reagents), changing chromatin structure and modifying gene transcription, contributing to cell plasticity and, ultimately, determining the strength of post-transplant immune responses ().
Dendritic Cells: Activation or Tolerance
Dendritic cells (DCs) are essential mediators of the innate and adaptive immune response, and can induce immunity or tolerance.Citation11,Citation12 In the initial stages after transplantation, DCs orchestrate inflammation subsequent to ischemia-reperfusion injury or indirectly lead to the activation of effector T lymphocytes and natural killer cells. However, in the absence of inflammatory signals and because of their phenotypic plasticity, DCs may also induce tolerance. Epigenetic mechanisms are involved during the differentiation of human monocyte-derived DCs (moDCs) and the critical “decision” to become activated or tolerogenic cells. Exposure to appropriate stimuli promotes differentiation of monocytes into DCs, leading to CD14 downregulation and increased expression of DC-SIGN (CD209), an essential molecule for trafficking DC and establishing DC-T cell contact. This process is regulated by the acquisition of an active histone mark (H3K9Ac) in CD209 gene along with the loss of repressive marks (H3K9me3, H4K20me3) and DNA methylation.Citation13 Additionally, the balance between the active H3K4me3 and repressive H3K27me3 histone marks in specific genes is associated with changes in gene expression, allowing the differentiation and function of activated or tolerized moDCs.Citation14
Cell therapy with tolerogenic DCs (TolDCs) is currently an attractive approach to minimizing the use of immunosuppressive drugs in transplantation.Citation15 Administration of TolDCs in several animal models of transplantation has shown improved graft survival and function although the current goal is to transfer this knowledge to humans. Epigenetic modifiers, such as HDAC inhibitors, can affect DC functions, revealing broader implications for immunotherapeutic strategies.Citation16 Treatment of DCs with Valproate and Butyrate reduce the expression of co-stimulatory molecules (CD40, CD80 and CD86) and secretion of pro-inflammatory cytokines (TNFα, IL-1β, IL-6 and IL12), inhibiting the activation of Th1 and Th17 responses and the expression of granzyme B in stimulated CD8+ T cells.Citation17,Citation18 Disruption of HDAC11 in antigen presenting cells (APCs) upregulates the expression of the IL10 gene and impairs the antigen-specific T cell responses, making this a promising target for inducing immune tolerance.Citation19
Plasticity of the CD4 T cell subsets: Effector and Regulatory Balance
After antigen recognition, naive T cells differentiate into effector T “helper” cells (Th1, Th2, and Th17) or regulatory T (Treg) cells with a suppressive functionCitation20 (). The appropriate balance between effector and regulatory T cell subsets determines the rejection or tolerance of the transplanted graft. Differentiation of naive CD4+ T cells toward one or other lineage is determined by the strength of the encounter with foreign antigens presented by the APCs and the presence of cytokines in the microenvironment where they occur. These signals enable changes in the expression of transcription factors that are correlated with epigenetic changes at specific loci, thereby establishing stable differentiation lineages.Citation21,Citation22
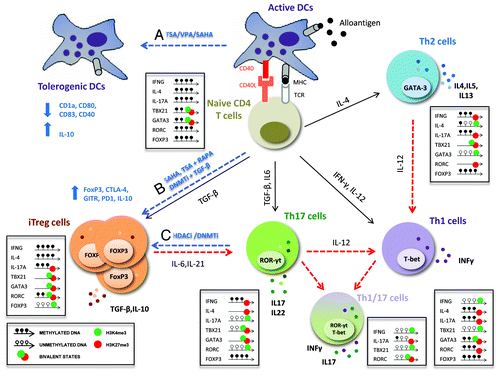
Figure 2. Targeting the activation and plasticity of CD4 T cells by HDAC inhibitors. After activation, CD4 T cells are directed toward different subsets of effector T cells (Th1, Th2 or Th17) or regulatory T cells (Treg) with specialized functions. These processes are regulated by epigenetic modifications that allow stable and heritable lineages but at the same time maintain the capacity to respond to environmental changes and switch from one lineage to another (plasticity). Dashed red lines indicate the plasticity and flexibility among CD4+ T cell subsets regulated by epigenetic mechanisms; dashed blue lines show the epigenetic treatments proposed for providing tolerance after transplantation. Epigenetic status of key transcription factors and cytokines essential for plasticity are shown for each CD4 T cell subset. This molecular mechanism may be related to poised, bivalent epigenetic stages (i.e., permissive H3K4me3 plus repressive H3K27me3 marks) in opposing lineages. HDAC inhibitors (HDACi) are believed to modulate the balance between immunity and tolerance: (A) TSA, VPA and SAHA diminish the expression of MHC class II and co-stimulatory molecules (CD1a, CD40, CD80, CD83), and disruption of HDAC11 increases IL-10 expression in DCs, favoring immune tolerance; (B) TSA and SAHA increase mRNA levels of FoxP3, CTLA4, GITR, PD-1 and IL-10, promoting the peripheral conversion of T cells into iTreg cells and enhancing suppressive function in vitro and in vivo; (C) an interesting approach is the use of epigenetic inhibitors to block the conversion of iTreg into Th17/Th1 cells in an inflammatory environment or the differentiation of effector T cells (Th1, Th17) into regulatory T cells with suppressive functions.
The first studies in humans showed that Th1 and Th2 cells are true lineages regulated by epigenetic modifications in IFNG, IL4 and IL13 genes allowing differentiation into a lineage with the extinction of the opposite fate. The INFG promoter is hypermethylated in human naive T cells and is only demethylated during the differentiation to Th1 cells.Citation23 By contrast, IL4 promoter is highly methylated in naive and Th1 cells, and only partial demethylation, specific to Th2 cells, is observed in intron 2 of IL4 and the IL13 promoter region.Citation24 Moreover, epigenetic histone marks are also essential for the Th1/Th2 cell fate decisions. Key transcription factors for the Th1 (STAT4 and T-bet) or Th2 (STAT6 and GATA-3) lineage choice are critical to the establishment of histone marks across the IFNG locus. This gene displays acetylation of H4 (AcH4) and H3K4-trimethylation marks in Th1 cells in combination with H3K27 di- and tri-methylation in Th2 cells.Citation25 The histone methylase SUV39H1, which is involved in the trimethylation of H3K9 (H3K9me3), has recently been implicated in the silencing of the Th1 locus and the subsequent promotion of stability of Th2 cells.Citation26
The discovery of two new, more flexible lineages of CD4+ T cells, Th17 and induced regulatory T (iTreg) cells, has greatly complicated the landscape of CD4+ T cell differentiation.Citation27,Citation28 Th17 cells are defined by the specific expression of the RORγt (also known as RORC) transcription factor and IL17 production and potentially contribute to allograft rejection.Citation29,Citation30 By contrast, Treg cells are fundamental for maintaining immunological tolerance and immune homeostasis.Citation31 In vivo-isolated human Th17 cells are characterized by DNA demethylation in IL17A and RORC loci correlated with specific expression in these cells, and the presence of bivalent H3K4me3/H3K27me3 domains at the TBX1 (T-box 21) promoter, suggesting that this gene is poised to be rapidly expressed under particular conditions.Citation32 Treg cells can originate as a stable subset from the thymus or be induced at the periphery by exposure to TGF-β, in both cases being characterized by the continuous expression of the transcription factor FoxP3.Citation33 DNA demethylation in a conserved noncoding region of the FOXP3 gene, known as TSDR (Treg cell-specific demethylation region), and hyperacetylation of histones are responsible for maintaining the stable expression of FOXP3 and the correct function of Treg cells.Citation34-Citation36
As mentioned above, epigenetic mechanisms are intricately coupled to the differentiation of CD4+T cells, facilitating the heritability and lineage stability of these subsets. However, simultaneously, these mechanisms may be altered at any point, favoring plasticity and producing a more effective immune response.Citation37-Citation40 This elevated plasticity among CD4+ T cell types has revealed the immunological circumstances of rejection to be more complicated than previously thought. During chronic inflammation, Th17 cells can express IFN-γ and ILFoxp317 concomitantly, giving rise to an intermediate Th1/17 phenotype. These cells have an epigenetic profile similar to that of Th17 cells, but retain the demethylated IFNG gene and lose the repressive H3K27me3 mark of the bivalent domain in TBX21 gene, inducing IFN-γ expression and contributing to the damage of the transplanted graft.Citation32 Moreover, Th17 cells have the capacity to differentiate into FoxP3+ cells with a suppressive function after continuous in vitro TCR stimulation by partial demethylation of the FOXP3 gene, although this matter is still controversial.Citation41
A challenge for the treatment of allograft rejection and induction of tolerance is to focus on cell therapy with Treg cells.Citation42 A major inconvenience, however, is the loss of FoxP3 expression and reprogramming into effector T cells secreting IL-17 and IFN-ɣ in a pro-inflammatory microenvironment. Memory Tregs can become IL-17+ cells through DNA demethylation of the RORC locus.Citation43 However, although Treg cells can lose FoxP3 expression, they maintain the demethylated status of TSDR and retain the ability to reactivate FoxP3 expression and the suppressive function upon activation.Citation44 Thus, the DNA methylation status of TSRD could be useful for specifically identifying genuine suppressor Treg cells during clinical follow-up of transplanted patients.Citation45
Additionally, the use of epigenetic treatments could be exploited to generate functionally stable Treg cells. Inhibition of DNA methylation by DNMT inhibitors (5AzaC or DAC), in the presence or absence of TGF-β, promotes stable induction of FoxP3 expression). The inhibition of the histone deacetylases HDAC6, HDAC9 or Sirtuin-1 (Sirt1) is known to enhance the suppressive function of Treg cells additively.Citation46,Citation47 Targeting these enzymes with specific pharmacological inhibitors is an exciting therapeutic option for increasing Treg function in transplanted patients.
Establishing T-Cell Memory and NK-Cell Identity
The immunological history of a transplant recipient influences the graft outcome. Transplanted patients with a higher frequency of donor-specific memory T cells have a higher risk of alloreactive memory T cell-mediated rejection and of being a barrier to tolerance.Citation48 During memory T cell formation, epigenetic changes in transcription factors (T-bet and eomesodermin), cytokines (IL-2 and IFN-γ) and other molecules (CD70, CD40L, ITGAL, PRF and CCR6) are essential for controlling the transcriptional profiles and function of these cells.Citation49 For example, in memory CD4+ T cells, demethylation of the CCR6 gene enables the stable expression of this chemokine, facilitating migration of these cells toward the renal proximal tubular epithelial cells, where they carry out their function.Citation50 In memory CD8+ T cells, hyperacetylation of histone 3 lysine 9 (H3K9Ac) in effector molecules (Eomes, PRF and GRZ) permits a stronger and quicker cytotoxic response to subsequent antigen re-encounters.Citation51 It is thought that these chromatin changes are established during the effector phase and maintained in the memory phase in addition to new modifications in gene loci that are differentially expressed in memory T cells. Genome-wide analysis of histone methylation (H3K4me3 and H3K427me3) in CD8+ memory T cells identifies four epigenetic states (active, poised, bivalent and repressed), reflecting the function of various genes in these cells.Citation52
During chronic viral infections (CMV, HCV and HIV), demethylation in the PDCD1 locus (encodes PD-1) in exhausted memory CD8+ T cells enables its expression, damaging the resolution of the infection.Citation53,Citation54 Understanding how the epigenetic modifications are established in memory T cells will allow us to develop strategies for targeting these cells and avoiding unwanted immune responses to transplanted grafts, and for improving long-term protection against infections.
NK cells play a fundamental role in the alloimmune and anti-infectious responses, since they are associated with inflammatory and regulatory functions.Citation55 Depending on the early activation of NK cells after SOT and the type of NK cell receptors expressed, these cells can promote rejection or tolerance. This is particularly relevant in the early stages of transplantation when it is essential to avoid elimination of the graft by NK cells while maintaining its anti-viral immune response function that commonly develops in transplanted patients. Several studies have demonstrated that epigenetic modifications modulate the cytolytic activity of NK cells regulating the expression of some NK cell receptors (KIR, NCRs and NKG2D) and cytotoxic molecules (GRZ and PRF).Citation56-Citation58 Treatments with demethylating agents (5azaC and DAC) increase the expression of inhibitory KIRs, while HDAC inhibitors decrease the expression of the activating NKp46 and NKp30 receptors. Our group has recently demonstrated that the NKG2D-activating receptor is regulated by histone acetylation (H3K9Ac) and that inhibition of histone acetyltransferase (HAT) activity by curcumin not only downregulates its expression at the cell surface but also leads to a marked reduction in the lytic capacity of NK cells.Citation59 Further studies are needed to clarify the epigenetic mechanisms involved in regulating all the inhibitory and activator receptors and their contribution to NK cell function.
Targeting of Plasma Cell Differentiation
There is clear evidence of the role played by B cells, plasma cells and their secreted antibodies in the acute cellular rejection and in acute and chronic antibody-mediated rejection.Citation60 Studies using “omics” assays showed a functional balance between different B-cell subsets, some aimed at inducing organ injury (plasmatic B cells) and others involved in tolerance (transitional and regulatory B cells).Citation61
Differentiation of hematopoietic stem cells into antibody-producing B cells requires a complex epigenetic regulatory mechanism that produces changes in chromatin structure throughout B cell development.Citation62-Citation64 Altered DNA demethylation patterns within gene bodies outside of CpG islands are essential for the correct differentiation and function of B cells.Citation65 At early stages, demethylation of transcription factors (Pax5 and PU1) and their target genes [CD79A (mb1), CD19 and IRF4] is essential to establish a functional expression pattern in B cells and to initiate immunoglobulin V(D)J recombination.Citation66-Citation70 In later stages, naive B cells migrate to germinal centers, where they are activated by antigen encounter and become antigen-experienced plasma cells producing antibodies or memory B cells.Citation71 The activation-induced by cytidine deaminase (AID) enzyme, which plays a critical role in the processes of somatic hypermutation and class switch recombination, resulting in diversification and high-affinity antibody production, is involved in the active DNA demethylation process.Citation72,Citation73 Moreover, the transcriptional repressor Blimp-1, which suppresses the mature B-cell gene expression favoring formation of antibody-secreting cells, functions by interacting with the LSD1 histone lysine demethylase.Citation74 Further studies of these mechanisms are needed to determine whether targeting these enzymes could help attenuate antibody production.
Epigenetic Biomarkers and New Therapeutic Targets
Identification of tolerant patients is the cornerstone of transplantation medicine. There is currently an absence of adequate robust biomarkers, which are necessary for monitoring the immune response. These are needed to reduce the level of immunosuppressive therapy and to improve long-term transplant outcomes definitively.Citation75,Citation76 Therefore, it is necessary to develop non-invasive biomarkers that allow the early detection of patients who have developed immunological unresponsiveness (total or partial) toward their graft, either spontaneously or through active tolerance induction. In recent years, European and other international networks (IOT, ITN and RISET) have integrated various “omics” technologies (genomics, proteomics and transcriptomics) and platforms to identify biomarkers reflecting the immune status of transplanted patients.Citation77-Citation81
The use of epigenetic biomarkers, mainly based on DNA methylation, is well established for the diagnosis and prognosis of tumors and has begun to be understood in other pathologies such as autoimmune diseases.Citation82 The identification of DNA methylation and histone modification marks in immune function-related genes could encourage the development of new epigenetic-based post-transplant biomarkers that have direct applications in clinical practice (). Recently, our group has reported that methylation changes in blood from patients with hematopoietic cell transplant are linked to physiological changes during transplant evolution.Citation5 Identification of the DNA methylation level for IFNG, FASL and IL-10 genes could be used as potential biomarkers for the development of graft-versus-host diseases. Moreover, demethylation of FoxP3 gene in biopsies from kidney transplant patients with subclinical rejection has been associated with a better long-term allograft outcome.Citation45 Novel epigenetic biomarkers will help to establish an “immunological fingerprint” of rejection risk or tolerance during follow-up of transplanted patients.
Table 1. Epigenetic modifications as potential biomarkers of tolerance after solid organ transplantation
From a clinical perspective, one of the most important characteristics of epigenetic modifications is that they can be reverted using enzymatic treatments (such as DNMT and HDAC inhibitors). Some of these epigenetic drugs (5-Azacytidine, Decitabine, Vorinostat and Romidepsin) have been approved by the US Food and Drug administration (FDA) for the treatment of hematologic diseases,Citation83 and many phase II and III clinical trials are under development. Although it has not been extensively studied, preliminary data suggest that treatment with epigenetic modifiers in combination with current immunosuppressant therapies prolongs allograft survival in animal models of cardiac and renal transplant.Citation84-Citation86 In humans, curcumin (a HAT inhibitor) in combination with cyclosporine A can suppress the induction of Th1 cytokines in ex vivo peripheral blood lymphocytes from kidney transplant patients.Citation87
Current approaches for clinical use are based on the in vitro generation or ex-vivo expansion of regulatory immune cells, with the aims of creating a suppressive environment and improving long-term allograft survival or inducing transplantation tolerance.Citation88-Citation90 As we previously reported, epigenetic drugs have the ability to modify the expression of key genes during the generation of diverse types of regulatory cell (). Chromatin modifications are critical for the differentiation and maintenance of the suppressive functions of Treg cells, regulating the balance between effector and regulatory T cells, differentiating tolerogenic dendritic cells, or during the polarization toward regulatory macrophages, suggesting the potential utility of epigenetic treatments for producing immunoregulatory cells. Moreover, in recent years, cell therapies with mesenchymal stem cells (MSCs) have come to be recognized as an attractive strategy for tolerance induction in organ transplantation. The first clinical trials are underway.Citation91 MSCs have immunomodulatory properties, damping the effector T-cell response and promoting the increase of regulatory T cells. However, during long-term cultures for therapies, these properties and the self-renewing capacity can be lost by chromatin alterations via epigenetic mechanisms. HDAC inhibitors are believed to maintain the biological characteristics of MSCs, thereby improving their usefulness in therapy.Citation92
One of the main inconveniences of the use of epigenetic treatments is that they are unspecific and may induce chromosomal instability that predisposes to cancer and makes it difficult to use for treating other pathologies. New therapeutic strategies aim to selectively target proteins that recognize modified histones, such as BET (bromodomain and extra terminal domain) family proteins, or alter DNA methylation in a specific way through artificial transcription factors or the thymine DNA glycosylase (TGD) enzyme.Citation93-Citation96 These new epigenetic approaches will allow the stable and heritable manipulation of the gene expression of target genes with significant functional relevance in immune system diseases and related conditions.
Concluding Remarks
Changes in chromatin structure arising from DNA methylation and histone modifications are involved in critical immune function pathways, such as generating monocyte-derived DC, differentiation and plasticity of “helper” CD4+ T cells and regulatory T cells, establishing a memory phenotype, and producing antibodies by plasma cells. Our growing knowledge of the epigenetic mechanisms involved in regulating the immune response will give us a more thorough understanding of the immunological processes involved in rejection and allograft tolerance. Therefore, we may be confident that in the coming years the identification of epigenetic alterations that occur during immune system deregulation after transplantation will lead to the development of new biomarkers. The advantage of these is that it will be possible to use non-invasive and stable DNA samples, combined with rapid, specific and quantitative methods such as pyrosequencing. These biomarkers are essential for identifying not only tolerant patients whose immunosuppressive therapy could be minimized or withdrawn, but also the early occurrence of graft rejection.
Additional studies will determine whether epigenetic treatments may one day become a valid therapeutic strategy for modulating the balance between immunity and tolerance. The potential usefulness of these epigenetic modifiers in combination with the immunosuppressant drugs currently used in clinical practice and their role in the ex vivo differentiation and maintenance of immunoregulatory cells needs to be established.
In conclusion, epigenomics is a growing area of transplant immunology research that could not only inspire new ways of immunologically monitoring transplanted patients but also lead to the development of new cell-based immunosuppressive strategies.
| Abbreviations: | ||
| IL | = | interleukin |
| IFNγ | = | interferon gamma |
| CD40L | = | CD40 ligand |
| MHC | = | major histocompatibility complex |
| TCR | = | T cell receptor |
| TGFβ | = | tumor growth factor beta |
| TSA | = | trichostatin A |
| SAHA | = | suberoylanilide hydroxamic acid |
| VPA | = | valproic acid |
| DNMTi | = | DNMT inhibitors |
| RAPA | = | rapamycin |
Acknowledgments
This work was supported by Spanish grants from Fondo de Investigaciones Sanitaria-Fondos FEDER European Union (FIS PI08/0566 and PI12/02587) and the Red de Investigación Renal (REDinREN RD12/0021) from the Institute Carlos III.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- Rodriguez-Cortez VC, Hernando H, de la Rica L, Vento R, Ballestar E. Epigenomic deregulation in the immune system. Epigenomics 2011; 3:697 - 713; http://dx.doi.org/10.2217/epi.11.99; PMID: 22126290
- Suarez-Alvarez B, Rodriguez RM, Fraga MF, López-Larrea C. DNA methylation: a promising landscape for immune system-related diseases. Trends Genet 2012; 28:506 - 14; http://dx.doi.org/10.1016/j.tig.2012.06.005; PMID: 22824525
- Schildberg FA, Hagmann CA, Böhnert V, Tolba RH. Improved transplantation outcome by epigenetic changes. Transpl Immunol 2010; 23:104 - 10; http://dx.doi.org/10.1016/j.trim.2010.05.001; PMID: 20471480
- McCaughan JA, McKnight AJ, Courtney AE, Maxwell AP. Epigenetics: time to translate into transplantation. Transplantation 2012; 94:1 - 7; http://dx.doi.org/10.1097/TP.0b013e31824db9bd; PMID: 22437848
- Rodriguez RM, Suarez-Alvarez B, Salvanés R, Muro M, Martínez-Camblor P, Colado E, et al. DNA methylation dynamics in blood after hematopoietic cell transplant. PLoS One 2013; 8:e56931; http://dx.doi.org/10.1371/journal.pone.0056931; PMID: 23451113
- Kulis M, Esteller M. DNA methylation and cancer. Adv Genet 2010; 70:27 - 56; http://dx.doi.org/10.1016/B978-0-12-380866-0.60002-2; PMID: 20920744
- Bannister AJ, Kouzarides T. Regulation of chromatin by histone modifications. Cell Res 2011; 21:381 - 95; http://dx.doi.org/10.1038/cr.2011.22; PMID: 21321607
- Dunham I, Kundaje A, Aldred SF, Collins PJ, Davis CA, Doyle F, et al, ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature 2012; 489:57 - 74; http://dx.doi.org/10.1038/nature11247; PMID: 22955616
- Chadwick LH. The NIH Roadmap Epigenomics Program data resource. Epigenomics 2012; 4:317 - 24; http://dx.doi.org/10.2217/epi.12.18; PMID: 22690667
- Adams D, Altucci L, Antonarakis SE, Ballesteros J, Beck S, Bird A, et al. BLUEPRINT to decode the epigenetic signature written in blood. Nat Biotechnol 2012; 30:224 - 6; http://dx.doi.org/10.1038/nbt.2153; PMID: 22398613
- Merad M, Collin M, Bromberg J. Dendritic cell homeostasis and trafficking in transplantation. Trends Immunol 2007; 28:353 - 9; http://dx.doi.org/10.1016/j.it.2007.06.003; PMID: 17618832
- Steinman RM, Banchereau J. Taking dendritic cells into medicine. Nature 2007; 449:419 - 26; http://dx.doi.org/10.1038/nature06175; PMID: 17898760
- Bullwinkel J, Lüdemann A, Debarry J, Singh PB. Epigenotype switching at the CD14 and CD209 genes during differentiation of human monocytes to dendritic cells. Epigenetics 2011; 6:45 - 51; http://dx.doi.org/10.4161/epi.6.1.13314; PMID: 20818162
- Huang Y, Min S, Lui Y, Sun J, Su X, Liu Y, et al. Global mapping of H3K4me3 and H3K27me3 reveals chromatin state-based regulation of human monocyte-derived dendritic cells in different environments. Genes Immun 2012; 13:311 - 20; http://dx.doi.org/10.1038/gene.2011.87; PMID: 22278394
- Moreau A, Varey E, Bouchet-Delbos L, Cuturi MC. Cell therapy using tolerogenic dendritic cells in transplantation. Transplant Res 2012; 1:13; http://dx.doi.org/10.1186/2047-1440-1-13; PMID: 23369513
- Frikeche J, Peric Z, Brissot E, Grégoire M, Gaugler B, Mohty M. Impact of HDAC inhibitors on dendritic cell functions. Exp Hematol 2012; 40:783 - 91; http://dx.doi.org/10.1016/j.exphem.2012.06.008; PMID: 22728031
- Nencioni A, Beck J, Werth D, Grünebach F, Patrone F, Ballestrero A, et al. Histone deacetylase inhibitors affect dendritic cell differentiation and immunogenicity. Clin Cancer Res 2007; 13:3933 - 41; http://dx.doi.org/10.1158/1078-0432.CCR-06-2903; PMID: 17606727
- Wang B, Morinobu A, Horiuchi M, Liu J, Kumagai S. Butyrate inhibits functional differentiation of human monocyte-derived dendritic cells. Cell Immunol 2008; 253:54 - 8; http://dx.doi.org/10.1016/j.cellimm.2008.04.016; PMID: 18522857
- Villagra A, Cheng F, Wang HW, Suarez I, Glozak M, Maurin M, et al. The histone deacetylase HDAC11 regulates the expression of interleukin 10 and immune tolerance. Nat Immunol 2009; 10:92 - 100; http://dx.doi.org/10.1038/ni.1673; PMID: 19011628
- Luckheeram RV, Zhou R, Verma AD, Xia B. CD4⁺T cells: differentiation and functions. Clin Dev Immunol 2012; 2012:925135; http://dx.doi.org/10.1155/2012/925135; PMID: 22474485
- Wilson CB, Rowell E, Sekimata M. Epigenetic control of T-helper-cell differentiation. Nat Rev Immunol 2009; 9:91 - 105; http://dx.doi.org/10.1038/nri2487; PMID: 19151746
- Wei G, Wei L, Zhu J, Zang C, Hu-Li J, Yao Z, et al. Global mapping of H3K4me3 and H3K27me3 reveals specificity and plasticity in lineage fate determination of differentiating CD4+ T cells. Immunity 2009; 30:155 - 67; http://dx.doi.org/10.1016/j.immuni.2008.12.009; PMID: 19144320
- Schoenborn JR, Dorschner MO, Sekimata M, Santer DM, Shnyreva M, Fitzpatrick DR, et al. Comprehensive epigenetic profiling identifies multiple distal regulatory elements directing transcription of the gene encoding interferon-gamma. Nat Immunol 2007; 8:732 - 42; http://dx.doi.org/10.1038/ni1474; PMID: 17546033
- Santangelo S, Cousins DJ, Winkelmann N, Triantaphyllopoulos K, Staynov DZ. Chromatin structure and DNA methylation of the IL-4 gene in human T(H)2 cells. Chromosome Res 2009; 17:485 - 96; http://dx.doi.org/10.1007/s10577-009-9040-3; PMID: 19521787
- Aune TM, Collins PL, Chang S. Epigenetics and T helper 1 differentiation. Immunology 2009; 126:299 - 305; http://dx.doi.org/10.1111/j.1365-2567.2008.03026.x; PMID: 19178593
- Allan RS, Zueva E, Cammas F, Schreiber HA, Masson V, Belz GT, et al. An epigenetic silencing pathway controlling T helper 2 cell lineage commitment. Nature 2012; 487:249 - 53; http://dx.doi.org/10.1038/nature11173; PMID: 22763435
- Muranski P, Restifo NP. Essentials of Th17 cell commitment and plasticity. Blood 2013; 121:2402 - 14; http://dx.doi.org/10.1182/blood-2012-09-378653; PMID: 23325835
- Ohkura N, Hamaguchi M, Morikawa H, Sugimura K, Tanaka A, Ito Y, et al. T cell receptor stimulation-induced epigenetic changes and Foxp3 expression are independent and complementary events required for Treg cell development. Immunity 2012; 37:785 - 99; http://dx.doi.org/10.1016/j.immuni.2012.09.010; PMID: 23123060
- Yuan X, Paez-Cortez J, Schmitt-Knosalla I, D’Addio F, Mfarrej B, Donnarumma M, et al. A novel role of CD4 Th17 cells in mediating cardiac allograft rejection and vasculopathy. J Exp Med 2008; 205:3133 - 44; http://dx.doi.org/10.1084/jem.20081937; PMID: 19047438
- Hanidziar D, Koulmanda M. Inflammation and the balance of Treg and Th17 cells in transplant rejection and tolerance. Curr Opin Organ Transplant 2010; 15:411 - 5; http://dx.doi.org/10.1097/MOT.0b013e32833b7929; PMID: 20613526
- Issa F, Chandrasekharan D, Wood KJ. Regulatory T cells as modulators of chronic allograft dysfunction. Curr Opin Immunol 2011; 23:648 - 54; http://dx.doi.org/10.1016/j.coi.2011.06.005; PMID: 21752619
- Cohen CJ, Crome SQ, MacDonald KG, Dai EL, Mager DL, Levings MK. Human Th1 and Th17 cells exhibit epigenetic stability at signature cytokine and transcription factor loci. J Immunol 2011; 187:5615 - 26; http://dx.doi.org/10.4049/jimmunol.1101058; PMID: 22048764
- Josefowicz SZ, Rudensky A. Control of regulatory T cell lineage commitment and maintenance. Immunity 2009; 30:616 - 25; http://dx.doi.org/10.1016/j.immuni.2009.04.009; PMID: 19464984
- Baron U, Floess S, Wieczorek G, Baumann K, Grützkau A, Dong J, et al. DNA demethylation in the human FOXP3 locus discriminates regulatory T cells from activated FOXP3(+) conventional T cells. Eur J Immunol 2007; 37:2378 - 89; http://dx.doi.org/10.1002/eji.200737594; PMID: 17694575
- Lal G, Zhang N, van der Touw W, Ding Y, Ju W, Bottinger EP, et al. Epigenetic regulation of Foxp3 expression in regulatory T cells by DNA methylation. J Immunol 2009; 182:259 - 73; PMID: 19109157
- van Loosdregt J, Vercoulen Y, Guichelaar T, Gent YY, Beekman JM, van Beekum O, et al. Regulation of Treg functionality by acetylation-mediated Foxp3 protein stabilization. Blood 2010; 115:965 - 74; http://dx.doi.org/10.1182/blood-2009-02-207118; PMID: 19996091
- Nakayamada S, Takahashi H, Kanno Y, O’Shea JJ. Helper T cell diversity and plasticity. Curr Opin Immunol 2012; 24:297 - 302; http://dx.doi.org/10.1016/j.coi.2012.01.014; PMID: 22341735
- Hirahara K, Vahedi G, Ghoreschi K, Yang XP, Nakayamada S, Kanno Y, et al. Helper T-cell differentiation and plasticity: insights from epigenetics. Immunology 2011; 134:235 - 45; http://dx.doi.org/10.1111/j.1365-2567.2011.03483.x; PMID: 21977994
- Murphy KM, Stockinger B. Effector T cell plasticity: flexibility in the face of changing circumstances. Nat Immunol 2010; 11:674 - 80; http://dx.doi.org/10.1038/ni.1899; PMID: 20644573
- Zhou L, Chong MM, Littman DR. Plasticity of CD4+ T cell lineage differentiation. Immunity 2009; 30:646 - 55; http://dx.doi.org/10.1016/j.immuni.2009.05.001; PMID: 19464987
- Ye J, Su X, Hsueh EC, Zhang Y, Koenig JM, Hoft DF, et al. Human tumor-infiltrating Th17 cells have the capacity to differentiate into IFN-γ+ and FOXP3+ T cells with potent suppressive function. Eur J Immunol 2011; 41:936 - 51; http://dx.doi.org/10.1002/eji.201040682; PMID: 21381020
- Issa F, Robb RJ, Wood KJ. The where and when of T cell regulation in transplantation. Trends Immunol 2013; 34:107 - 13; http://dx.doi.org/10.1016/j.it.2012.11.003; PMID: 23228885
- Schmidl C, Hansmann L, Andreesen R, Edinger M, Hoffmann P, Rehli M. Epigenetic reprogramming of the RORC locus during in vitro expansion is a distinctive feature of human memory but not naïve Treg. Eur J Immunol 2011; 41:1491 - 8; http://dx.doi.org/10.1002/eji.201041067; PMID: 21469109
- Miyao T, Floess S, Setoguchi R, Luche H, Fehling HJ, Waldmann H, et al. Plasticity of Foxp3(+) T cells reflects promiscuous Foxp3 expression in conventional T cells but not reprogramming of regulatory T cells. Immunity 2012; 36:262 - 75; http://dx.doi.org/10.1016/j.immuni.2011.12.012; PMID: 22326580
- Bestard O, Cuñetti L, Cruzado JM, Lucia M, Valdez R, Olek S, et al. Intragraft regulatory T cells in protocol biopsies retain foxp3 demethylation and are protective biomarkers for kidney graft outcome. Am J Transplant 2011; 11:2162 - 72; http://dx.doi.org/10.1111/j.1600-6143.2011.03633.x; PMID: 21749644
- Beier UH, Wang L, Han R, Akimova T, Liu Y, Hancock WW. Histone deacetylases 6 and 9 and sirtuin-1 control Foxp3+ regulatory T cell function through shared and isoform-specific mechanisms. Sci Signal 2012; 5:ra45; http://dx.doi.org/10.1126/scisignal.2002873; PMID: 22715468
- Beier UH, Wang L, Hancock WW. Combination of isoform-selective histone/protein deacetylase inhibitors improves Foxp3+ T-regulatory cell function. Cell Cycle 2012; 11:3351 - 2; http://dx.doi.org/10.4161/cc.21876; PMID: 22918251
- Page AJ, Ford ML, Kirk AD. Memory T-cell-specific therapeutics in organ transplantation. Curr Opin Organ Transplant 2009; 14:643 - 9; http://dx.doi.org/10.1097/MOT.0b013e328332bd4a; PMID: 19779342
- Weng NP, Araki Y, Subedi K. The molecular basis of the memory T cell response: differential gene expression and its epigenetic regulation. Nat Rev Immunol 2012; 12:306 - 15; http://dx.doi.org/10.1038/nri3173; PMID: 22421787
- Steinfelder S, Floess S, Engelbert D, Haeringer B, Baron U, Rivino L, et al. Epigenetic modification of the human CCR6 gene is associated with stable CCR6 expression in T cells. Blood 2011; 117:2839 - 46; http://dx.doi.org/10.1182/blood-2010-06-293027; PMID: 21228329
- Araki Y, Fann M, Wersto R, Weng NP. Histone acetylation facilitates rapid and robust memory CD8 T cell response through differential expression of effector molecules (eomesodermin and its targets: perforin and granzyme B). J Immunol 2008; 180:8102 - 8; PMID: 18523274
- Araki Y, Wang Z, Zang C, Wood WH 3rd, Schones D, Cui K, et al. Genome-wide analysis of histone methylation reveals chromatin state-based regulation of gene transcription and function of memory CD8+ T cells. Immunity 2009; 30:912 - 25; http://dx.doi.org/10.1016/j.immuni.2009.05.006; PMID: 19523850
- Youngblood B, Oestreich KJ, Ha SJ, Duraiswamy J, Akondy RS, West EE, et al. Chronic virus infection enforces demethylation of the locus that encodes PD-1 in antigen-specific CD8(+) T cells. Immunity 2011; 35:400 - 12; http://dx.doi.org/10.1016/j.immuni.2011.06.015; PMID: 21943489
- Youngblood B, Wherry EJ, Ahmed R. Acquired transcriptional programming in functional and exhausted virus-specific CD8 T cells. Curr Opin HIV AIDS 2012; 7:50 - 7; http://dx.doi.org/10.1097/COH.0b013e32834ddcf2; PMID: 22134341
- Villard J. The role of natural killer cells in human solid organ and tissue transplantation. J Innate Immun 2011; 3:395 - 402; http://dx.doi.org/10.1159/000324400; PMID: 21411978
- Ogbomo H, Michaelis M, Kreuter J, Doerr HW, Cinatl J Jr.. Histone deacetylase inhibitors suppress natural killer cell cytolytic activity. FEBS Lett 2007; 581:1317 - 22; http://dx.doi.org/10.1016/j.febslet.2007.02.045; PMID: 17349632
- Schmiedel BJ, Arélin V, Gruenebach F, Krusch M, Schmidt SM, Salih HR. Azacytidine impairs NK cell reactivity while decitabine augments NK cell responsiveness toward stimulation. Int J Cancer 2011; 128:2911 - 22; http://dx.doi.org/10.1002/ijc.25635; PMID: 20960460
- Li G, Yu M, Weyand CM, Goronzy JJ. Epigenetic regulation of killer immunoglobulin-like receptor expression in T cells. Blood 2009; 114:3422 - 30; http://dx.doi.org/10.1182/blood-2009-01-200170; PMID: 19628706
- Fernández-Sánchez A, Baragaño Raneros A, Carvajal Palao R, Sanz AB, Ortiz A, Ortega F, et al. DNA demethylation and histone H3K9 acetylation determine the active transcription of the NKG2D gene in human CD8+ T and NK cells. Epigenetics 2013; 8:66 - 78; http://dx.doi.org/10.4161/epi.23115; PMID: 23235109
- Zarkhin V, Sarwal MM. The coin toss of B cells in rejection and tolerance: danger versus defense. Semin Immunol 2012; 24:86 - 91; http://dx.doi.org/10.1016/j.smim.2011.09.003; PMID: 22035649
- Newell KA, Asare A, Kirk AD, Gisler TD, Bourcier K, Suthanthiran M, et al, Immune Tolerance Network ST507 Study Group. Identification of a B cell signature associated with renal transplant tolerance in humans. J Clin Invest 2010; 120:1836 - 47; http://dx.doi.org/10.1172/JCI39933; PMID: 20501946
- Ramírez J, Lukin K, Hagman J. From hematopoietic progenitors to B cells: mechanisms of lineage restriction and commitment. Curr Opin Immunol 2010; 22:177 - 84; http://dx.doi.org/10.1016/j.coi.2010.02.003; PMID: 20207529
- Santos P, Arumemi F, Park KS, Borghesi L, Milcarek C. Transcriptional and epigenetic regulation of B cell development. Immunol Res 2011; 50:105 - 12; http://dx.doi.org/10.1007/s12026-011-8225-y; PMID: 21717070
- Parra M. Epigenetic events during B lymphocyte development. Epigenetics 2009; 4:462 - 8; http://dx.doi.org/10.4161/epi.4.7.10052; PMID: 19875937
- Lee ST, Xiao Y, Muench MO, Xiao J, Fomin ME, Wiencke JK, et al. A global DNA methylation and gene expression analysis of early human B-cell development reveals a demethylation signature and transcription factor network. Nucleic Acids Res 2012; 40:11339 - 51; http://dx.doi.org/10.1093/nar/gks957; PMID: 23074194
- McManus S, Ebert A, Salvagiotto G, Medvedovic J, Sun Q, Tamir I, et al. The transcription factor Pax5 regulates its target genes by recruiting chromatin-modifying proteins in committed B cells. EMBO J 2011; 30:2388 - 404; http://dx.doi.org/10.1038/emboj.2011.140; PMID: 21552207
- Amaravadi L, Klemsz MJ. DNA methylation and chromatin structure regulate PU.1 expression. DNA Cell Biol 1999; 18:875 - 84; http://dx.doi.org/10.1089/104454999314737; PMID: 10619599
- Maier H, Ostraat R, Gao H, Fields S, Shinton SA, Medina KL, et al. Early B cell factor cooperates with Runx1 and mediates epigenetic changes associated with mb-1 transcription. Nat Immunol 2004; 5:1069 - 77; http://dx.doi.org/10.1038/ni1119; PMID: 15361869
- Walter K, Bonifer C, Tagoh H. Stem cell-specific epigenetic priming and B cell-specific transcriptional activation at the mouse Cd19 locus. Blood 2008; 112:1673 - 82; http://dx.doi.org/10.1182/blood-2008-02-142786; PMID: 18552207
- Matheson LS, Corcoran AE. Local and global epigenetic regulation of V(D)J recombination. Curr Top Microbiol Immunol 2012; 356:65 - 89; http://dx.doi.org/10.1007/82_2011_137; PMID: 21695632
- Shaknovich R, Cerchietti L, Tsikitas L, Kormaksson M, De S, Figueroa ME, et al. DNA methyltransferase 1 and DNA methylation patterning contribute to germinal center B-cell differentiation. Blood 2011; 118:3559 - 69; http://dx.doi.org/10.1182/blood-2011-06-357996; PMID: 21828137
- Teperek-Tkacz M, Pasque V, Gentsch G, Ferguson-Smith AC. Epigenetic reprogramming: is deamination key to active DNA demethylation?. Reproduction 2011; 142:621 - 32; http://dx.doi.org/10.1530/REP-11-0148; PMID: 21911441
- Kuraoka M, Holl TM, Liao D, Womble M, Cain DW, Reynolds AE, et al. Activation-induced cytidine deaminase mediates central tolerance in B cells. Proc Natl Acad Sci U S A 2011; 108:11560 - 5; http://dx.doi.org/10.1073/pnas.1102571108; PMID: 21700885
- Su ST, Ying HY, Chiu YK, Lin FR, Chen MY, Lin KI. Involvement of histone demethylase LSD1 in Blimp-1-mediated gene repression during plasma cell differentiation. Mol Cell Biol 2009; 29:1421 - 31; http://dx.doi.org/10.1128/MCB.01158-08; PMID: 19124609
- Roedder S, Vitalone M, Khatri P, Sarwal MM. Biomarkers in solid organ transplantation: establishing personalized transplantation medicine. Genome Med 2011; 3:37; http://dx.doi.org/10.1186/gm253; PMID: 21658299
- Naesens M, Sarwal MM. Molecular diagnostics in transplantation. Nat Rev Nephrol 2010; 6:614 - 28; http://dx.doi.org/10.1038/nrneph.2010.113; PMID: 20736923
- Viklicky O, Krystufkova E, Brabcova I, Sekerkova A, Wohlfahrt P, Hribova P, et al. B-cell-related biomarkers of tolerance are up-regulated in rejection-free kidney transplant recipients. Transplantation 2013; 95:148 - 54; http://dx.doi.org/10.1097/TP.0b013e3182789a24; PMID: 23222918
- Chowdhury P, Hernandez-Fuentes MP. Non-invasive biomarkers to guide management following renal transplantation: the need for a multiplatform approach. Curr Opin Organ Transplant 2013; 18:1 - 5; http://dx.doi.org/10.1097/MOT.0b013e32835c8025; PMID: 23283248
- Li L, Khatri P, Sigdel TK, Tran T, Ying L, Vitalone MJ, et al. A peripheral blood diagnostic test for acute rejection in renal transplantation. Am J Transplant 2012; 12:2710 - 8; http://dx.doi.org/10.1111/j.1600-6143.2012.04253.x; PMID: 23009139
- Londoño MC, Danger R, Giral M, Soulillou JP, Sánchez-Fueyo A, Brouard S. A need for biomarkers of operational tolerance in liver and kidney transplantation. Am J Transplant 2012; 12:1370 - 7; http://dx.doi.org/10.1111/j.1600-6143.2012.04035.x; PMID: 22486792
- Lozano JJ, Pallier A, Martinez-Llordella M, Danger R, López M, Giral M, et al. Comparison of transcriptional and blood cell-phenotypic markers between operationally tolerant liver and kidney recipients. Am J Transplant 2011; 11:1916 - 26; http://dx.doi.org/10.1111/j.1600-6143.2011.03638.x; PMID: 21827613
- Heyn H, Esteller M. DNA methylation profiling in the clinic: applications and challenges. Nat Rev Genet 2012; 13:679 - 92; http://dx.doi.org/10.1038/nrg3270; PMID: 22945394
- Ho AS, Turcan S, Chan TA. Epigenetic therapy: use of agents targeting deacetylation and methylation in cancer management. Onco Targets Ther 2013; 6:223 - 32; PMID: 23569385
- Wu SL, Pan CE, Yu L, Meng KW. Immunosuppression by combined use of cyclosporine and resveratrol in a rat liver transplantation model. Transplant Proc 2005; 37:2354 - 9; http://dx.doi.org/10.1016/j.transproceed.2005.03.112; PMID: 15964415
- Kinugasa F, Nagatomi I, Nakanishi T, Noto T, Mori H, Matsuoka H, et al. Effect of the immunosuppressant histone deacetylase inhibitor FR276457 in a canine renal transplant model. Transpl Immunol 2009; 21:198 - 202; http://dx.doi.org/10.1016/j.trim.2009.04.006; PMID: 19409992
- Zhang X, Han S, Kang Y, Guo M, Hong S, Liu F, et al. SAHA, an HDAC inhibitor, synergizes with tacrolimus to prevent murine cardiac allograft rejection. Cell Mol Immunol 2012; 9:390 - 8; http://dx.doi.org/10.1038/cmi.2012.28; PMID: 22922441
- Bharti AC, Panigrahi A, Sharma PK, Gupta N, Kumar R, Shukla S, et al. Clinical relevance of curcumin-induced immunosuppression in living-related donor renal transplant: an in vitro analysis. Exp Clin Transplant 2010; 8:161 - 71; PMID: 20565374
- Wood KJ, Bushell A, Hester J. Regulatory immune cells in transplantation. Nat Rev Immunol 2012; 12:417 - 30; http://dx.doi.org/10.1038/nri3227; PMID: 22627860
- Sagoo P, Lombardi G, Lechler RI. Relevance of regulatory T cell promotion of donor-specific tolerance in solid organ transplantation. Front Immunol 2012; 3:184; http://dx.doi.org/10.3389/fimmu.2012.00184; PMID: 22811678
- Ohkura N, Kitagawa Y, Sakaguchi S. Development and maintenance of regulatory T cells. Immunity 2013; 38:414 - 23; http://dx.doi.org/10.1016/j.immuni.2013.03.002; PMID: 23521883
- Casiraghi F, Perico N, Remuzzi G. Mesenchymal stromal cells to promote solid organ transplantation tolerance. Curr Opin Organ Transplant 2013; 18:51 - 8; http://dx.doi.org/10.1097/MOT.0b013e32835c5016; PMID: 23254705
- Wang Y, Chen T, Yan H, Qi H, Deng C, Ye T, et al. Role of histone deacetylase inhibitors in the aging of human umbilical cord mesenchymal stem cells. [Epub ahead of print] J Cell Biochem 2013; http://dx.doi.org/10.1002/jcb.24569; PMID: 23564418
- Nicodeme E, Jeffrey KL, Schaefer U, Beinke S, Dewell S, Chung CW, et al. Suppression of inflammation by a synthetic histone mimic. Nature 2010; 468:1119 - 23; http://dx.doi.org/10.1038/nature09589; PMID: 21068722
- Gregory DJ, Mikhaylova L, Fedulov AV. Selective DNA demethylation by fusion of TDG with a sequence-specific DNA-binding domain. Epigenetics 2012; 7:344 - 9; http://dx.doi.org/10.4161/epi.19509; PMID: 22419066
- Dalton SR, Bellacosa A. DNA demethylation by TDG. Epigenomics 2012; 4:459 - 67; http://dx.doi.org/10.2217/epi.12.36; PMID: 22920184
- Rivenbark AG, Stolzenburg S, Beltran AS, Yuan X, Rots MG, Strahl BD, et al. Epigenetic reprogramming of cancer cells via targeted DNA methylation. Epigenetics 2012; 7:350 - 60; http://dx.doi.org/10.4161/epi.19507; PMID: 22419067