Abstract
The HepA-related protein (HARP/SMARCAL1) is an ATP-dependent annealing helicase that is capable of rewinding DNA structures that are stably unwound due to binding of the single-stranded DNA (ssDNA)-binding protein Replication Protein A (RPA). HARP has been implicated in maintaining genome integrity through its role in DNA replication and repair, two processes that generate RPA-coated ssDNA. In addition, mutations in HARP cause a rare disease known as Schimke immuno-osseous dysplasia. In this study, we purified HARP containing complexes with the goal of identifying the predominant factors that stably associate with HARP. We found that HARP preferentially interacts with RPA molecules that are bound to the DNA-dependent protein kinase (DNA-PK). We also found that RPA is phosphorylated by DNA-PK in vitro, while the RPA-HARP complexes are not. Our results suggest that, in addition to its annealing helicase activity, which eliminates the natural binding substrate for RPA, HARP blocks the phosphorylation of RPA by DNA-PK.
Keywords: :
Introduction
The annealing helicase HepA-related protein (HARP; also known as SMARCAL1) is an ATP-dependent enzyme capable of rewinding DNA that is bound by the single-stranded DNA (ssDNA)-binding protein replication protein A (RPA).Citation1 RPA binds with high-affinity to ssDNA and can prevent cDNA from annealing.Citation2 Such RPA-bound structures can arise during normal processes that involve unwinding of the DNA duplex, such as DNA replication, DNA repair and transcription. HARP has been shown to play an important role during both DNA replication and repair, and cells lacking HARP exhibit replication defects and persistent RPA binding.Citation3-Citation7 The importance of HARP has been further highlighted by the finding that mutations in human HARP cause a rare disorder known as Schimke immuno-osseous dysplasia (SIOD).Citation8 While the molecular basis of SIOD is not fully understood, a potential link between an increase in chromosome breakage and mutations in HARP is consistent with a role of HARP in DNA repair.Citation9
HARP binds preferentially and with high affinity to forked DNA structures (i.e., split ends), which are formed at the ends of ssDNA regions.Citation1 HARP interacts directly with RPA through a conserved motif near the N terminus of HARP.Citation3-Citation7 While this interaction is dispensable for the annealing helicase activity of HARP, it helps localize HARP to sites of DNA replication and repair.Citation3-Citation7 In addition to the annealing helicase activity of HARP, which regulates the amount of RPA-bound ssDNA in the cell, RPA can be regulated through phosphorylation.Citation10-Citation15 RPA, a heterotrimer consisting of RPA1, RPA2 and RPA3, is preferentially phosphorylated on the N terminus of its RPA2 subunit. Several kinases phosphorylate RPA including cyclin-dependent kinase (CDK), ataxia-telangiectasia mutated (ATM), ATM and Rad3-related (ATR), and the DNA-dependent protein kinase (DNA-PK).Citation10-Citation15 Changes in RPA phosphorylation have been observed during cell-cycle progression and in response to DNA damage, which ultimately leads to hyperphoshorylation of RPA. The downstream effects of RPA phosphorylation depend on the residues that are phosphorylated and include checkpoint activation, the regulation of DNA repair, and the modulation of RPA-DNA interactions.Citation16-Citation19
Like RPA, HARP is also phosphorylated in response to replication stress and DNA damage,Citation6,Citation7,Citation20,Citation21 although the significance of HARP phosphorylation is less clear. ATM, ATR and DNA-PK are all capable of phosphorylating HARP.Citation6,Citation7,Citation20,Citation21 Despite the knowledge that HARP associates with RPA, and that both factors are substrates for multiple kinases, little is known about which factors form the preferred HARP complex. In this study, we found that, after RPA, DNA-PK is the most abundant factor in the HARP complex. A stable interaction of DNA-PK with HARP is mediated by RPA and suggests a tendency for HARP to associate with RPA bound by DNA-PK. Using DNA-PK purified from HeLa cells by a rapid DNA-affinity protocol, we found that while DNA-PK is able to phosphorylate RPA in vitro, it is unable to phosphorylate RPA in the presence of HARP. DNA-PK also phosphorylates HARP in vitro and the phosphorylation of HARP by DNA-PK does not significantly affect its annealing helicase activity. Together, our data suggest that HARP inhibits the function or activity of RPA by both eliminating ssDNA via the annealing helicase activity of HARP and by preventing RPA phosphorylation by DNA-PK.
Results and Discussion
As previously reported, we purified HARP from HeLa cells that constitutively express a FLAG-tagged copy of HARP by tandem-affinity purification.Citation3 Following the purification, we observed that in addition to RPA, three other factors stably co-purify with HARP. These proteins were identified by mass spectrometry as the subunits of the DNA-PK heterotrimer (i.e., DNA-PKcs, Ku80, and Ku70). Other reports have identified DNA-PK subunits as part of a group of potential cofactors for HARP,Citation4,Citation6,Citation22 but our analysis reveals that RPA and DNA-PK are the two most abundant factors that stably co-purify with HARP (). Under our purification conditions, we did not detect WRN helicase, a factor previously reported to associate with HARP,Citation22 suggesting that WRN may be part of a less abundant or less stable HARP complex.
Figure 1. DNA-PK copurifies with HARP and RPA. (A) The HARP-containing complex was purified from HeLa cells by tandem anti-FLAG/anti-HA purification and resolved by SDS-PAGE and silver staining. DNA-PK was identified in the complex by mass spectrometry. (B) HARP, RPA, and DNA-PK co-fractionate on glycerol gradients. The purified HARP complex was resolved on a 10–30% glycerol gradient and the migration of the proteins determined by SDS-PAGE and silver staining.
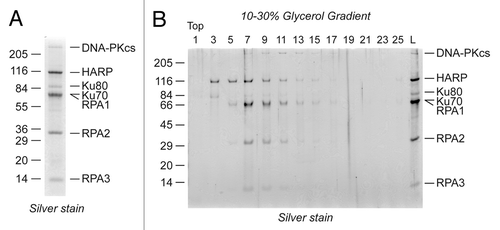
To gain insight into the stability of the HARP-RPA-DNA-PK interactions, we resolved the complex by glycerol gradient sedimentation. We collected fractions off the top of the gradient and analyzed them by SDS-PAGE and silver staining. We found that the DNA-PK heterotrimer is present in the same fractions that contain both HARP and RPA (, fractions 7 to 11), suggesting that HARP, RPA, and DNA-PK associate in a single, stable complex.
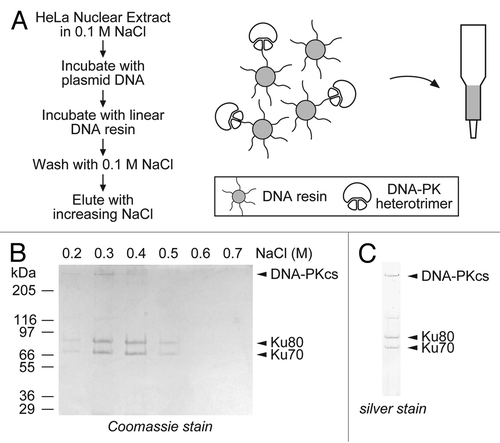
To examine how DNA-PK affects the activity of HARP, we purified the DNA-PK heterotrimer from HeLa cells using a one-step DNA-affinity protocol (). We first crosslinked sonicated poly-[dI:dC] DNA fragments to Sepharose beads and then used the free DNA ends to purify DNA-PK from HeLa nuclear extracts. We initially prepared nuclear extracts from 4 L of HeLa cells and pre-incubated the extracts with plasmid DNA (which has no free DNA ends) to remove non-specific DNA-binding proteins. We then incubated the pre-cleared extracts with the DNA-resin that contains free DNA ends. The beads were then washed to remove the unbound proteins, and the bound proteins were eluted using a step gradient of increasing NaCl. Under these conditions, the collected fractions consisted almost entirely of the DNA-PK heterotrimer, which eluted from the resin in the fractions containing between 0.3 to 0.4 M of NaCl (). The identity of the eluted proteins was confirmed by mass spectrometry. A scaled-down version of the protocol was used to extract DNA-PK from HeLa cells from a single 15 cm dish, showing that small-scale preparations of DNA-PK can be rapidly made ().
Figure 2. Isolation of native DNA-PK by a single-step DNA-affinity purification method. (A) DNA was crosslinked to Sepharose beads and used to extract DNA-PK, which preferentially binds to free DNA ends. (B) Nuclear extracts from 4 L of HeLa cells were pre-incubated with circular plasmid DNA to bind non-specific DNA binding proteins. The extract was then passed through the crosslinked beads, the unbound proteins were washed off, and the bound proteins were eluted by increasing salt step gradient. The eluted proteins were analyzed by SDS-PAGE followed by Coomassie staining. (C) A scaled-down purification was performed with nuclear extracts prepared from a single 15 cm dish. The peak fraction from the salt gradient was analyzed by SDS-PAGE followed by silver staining.
Using the native DNA-PK purified from HeLa cell extracts, we first asked whether DNA-PK interacts directly with HARP or associates with the complex through an interaction with RPA. Following several independent purifications of the HARP complex, we noticed that the DNA-PK is often less abundant than HARP and RPA. We considered the possibility that DNA-PK, which is known to associate with RPA, is present in the HARP complex through a direct interaction with RPA. To determine whether RPA is required for DNA-PK to stably associate with the HARP complex, we tested whether a mutant version of HARP, which contains a 10 amino acid substitution in the N-terminal RPA-interaction motif,Citation3 can interact with DNA-PK. We incubated purified RPA, DNA-PK with either wild-type or mutant HARP, immunoprecipitated the HARP proteins using anti-FLAG antibodies, and analyzed the co-precipitating factors by western blotting (). As previously reported, we found that wild-type but not mutant HARP interacts with RPA.Citation3 Moreover, DNA-PK readily co-precipitates with wild-type HARP but not the mutant HARP, suggesting that a direct interaction between HARP and DNA-PK is not stable. A similar finding was observed using a truncated version of HARP.Citation22 We cannot rule out the possibility that RPA and DNA-PK compete for the same binding site on HARP and that the mutant HARP fails to interact with both RPA and DNA-PK. However, we think this is unlikely as our gradient analysis suggests that HARP, RPA, and DNA-PK all appear to stably interact in one single complex. Our findings suggest that HARP exhibits a preference, or tendency, to interact with RPA bound to DNA-PK.
Figure 3. Interaction and kinase activity of DNA-PK with HARP and RPA. (A) Purified wild-type (WT) or mutant (Mut) HARP proteins were purified by a baculovirus system and incubated with bacterially expressed RPA and native DNA-PK. (B) The HARP proteins were then immunoprecipitated using anti-FLAG antibodies, and the immunoprecipitated material was analyzed by western blotting. (C) In vitro kinase assays were performed by incubating purified DNA-PK with HARP, RPA, [32P]-labeled ATP and plasmid DNA. The proteins were then resolved by SDS-PAGE and analyzed by phosphorimaging analysis. (D) The in vitro kinase assays were performed in the presence of equal amounts of WT or Mut HARP proteins.
![Figure 3. Interaction and kinase activity of DNA-PK with HARP and RPA. (A) Purified wild-type (WT) or mutant (Mut) HARP proteins were purified by a baculovirus system and incubated with bacterially expressed RPA and native DNA-PK. (B) The HARP proteins were then immunoprecipitated using anti-FLAG antibodies, and the immunoprecipitated material was analyzed by western blotting. (C) In vitro kinase assays were performed by incubating purified DNA-PK with HARP, RPA, [32P]-labeled ATP and plasmid DNA. The proteins were then resolved by SDS-PAGE and analyzed by phosphorimaging analysis. (D) The in vitro kinase assays were performed in the presence of equal amounts of WT or Mut HARP proteins.](/cms/asset/0bf83337-3377-4bb3-8b5f-b4b1ece2c5c4/kepi_a_10928310_f0003.gif)
We next used the purified DNA-PK to investigate the phosphorylation of HARP and RPA. In order to examine whether HARP is a substrate for DNA-PK and whether HARP affects the phosphorylation of RPA by DNA-PK, we performed in vitro kinase assays. We incubated DNA-PK with HARP, the RPA heterotrimer, or both in the presence of double-stranded plasmid DNA and [32P]-ATP. We then resolved the proteins by SDS-PAGE, dried and scanned the gel using a phosphorimager. We found that both HARP and RPA can be independently phosphorylated in vitro by DNA-PK (). As previously reported, DNA-PK preferentially phosphorylates the RPA2 subunit of RPA. When both HARP and RPA are present in the reactions, the phosphorylation of HARP and RPA2 by DNA-PK is substantially reduced (). In fact, we were unable to detect any RPA phosphorylation in the presence of HARP. One potential mechanism by which HARP could block the phosphorylation of RPA is by binding and masking the phosphorylation residues of RPA2. This idea is supported by the fact that the N terminus of HARP contains an RPA2-interacting motif.Citation3-Citation7 On the other hand, it is also possible that HARP interferes with RPA2 phosphorylation through a more subtle mechanism, such as by interfering with the kinase activity of DNA-PK or by weakening DNA-PK-RPA2 interactions. In order to address these potential models, we compared the levels of phosphorylation of RPA2 by DNA-PK in the presence of wild-type and mutant HARP proteins. The mutant HARP does not stably interact with RPA and should be less likely to mask RPA2 phosphorylation sites. We found that both wild-type and mutant HARP proteins are able to block RPA2 phosphorylation by DNA-PK (). In addition, while the levels of phosphorylation of wild-type HARP are reduced in the presence of RPA, the levels of phosphorylation of mutant HARP remain high in the presence of RPA. These findings suggest that HARP does not simply mask the phosphorylation sites of RPA through the stable interaction of the two factors. It is likely that a more complex mechanism is behind the ability of HARP to block the phosphorylation of RPA2. For example, in addition to the stable interaction between HARP and RPA that is mediated by the conserved N terminus of HARP, multiple weaker interactions may exist between other regions of HARP and RPA. These interactions may be sufficient for both wild-type and mutant HARP to block the phosphorylation of RPA2. Furthermore, interactions between HARP and DNA-PK may exist that are not strong enough for the mutant HARP to co-purify with DNA-PK, but are strong enough for HARP to affect the ability of DNA-PK to phosphorylate RPA. In contrast, the observed reduction in the phosphorylation of HARP by DNA-PK and in the presence of RPA may be explained by the masking of phosphorylation sites on HARP by RPA. This would explain why mutant HARP, which fails to stably interact with RPA, does not show the reduced levels of phosphorylation seen with wild-type HARP.
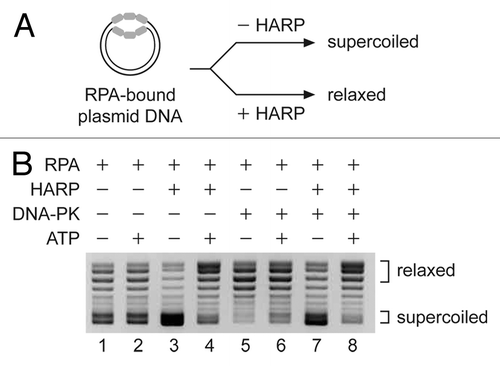
The role that phosphorylation of HARP plays in the cell is poorly understood although it has been suggested that phosphorylation of some residues may affect the activity of HARP.Citation20,Citation21 To test whether DNA-PK affects the ATP-dependent activity of HARP, we performed annealing helicase assays in the presence or absence of purified DNA-PK. We generated RPA-bound DNA substrates using purified RPA and plasmid DNA (). These plasmid substrates contain ssDNA bubbles that are bound by RPA. Once deproteinized and precipitated, the unwound plasmid DNA becomes supercoiled. In contrast, if the RPA-bound substrate is rewound, the resulting double-stranded plasmid DNA will be in a relaxed form following precipitation. Supercoiled and relax DNA are resolved by agarose gel electrophoresis. As previously reported, we observed that HARP, in the absence of DNA-PK, rewinds the unwound regions of the DNA in an ATP-dependent manner (, lanes 3 and 4). We also found that addition of DNA-PK to the RPA-bound DNA in the absence of HARP resulted in a modest decrease in the stability of the RPA-ssDNA complex in an ATP-independent manner (, lanes 5 and 6). This is indicated by the presence of more relaxed DNA. Reactions in which both HARP and DNA-PK were added to the RPA-bound substrate did not show any change in the annealing helicase activity of HARP (, lanes 7 and 8). Under the conditions we tested, HARP is able to fully rewind the RPA-bound DNA in the presence of DNA-PK and ATP. It should be noted that in the reactions that contained both HARP and DNA-PK, we pre-incubated HARP with DNA-PK in the presence or absence of ATP to allow for HARP phosphorylation before the addition of the RPA-bound DNA. Based on this finding, and the fact that DNA-PK is present in the HARP complex through its binding to RPA, it appears likely that RPA is the main target of the kinase activity of DNA-PK.
Figure 4. The presence of DNA-PK does not significantly affect the annealing helicase activity of HARP. Annealing helicase assays were performed in the presence or absence of DNA-PK using a plasmid partially loaded with RPA. For the reactions that contain both DNA-PK and HARP, the two factors were preincubated in the presence or absence of ATP prior to addition of the RPA-bound DNA substrate. Following the reactions, the purified DNA was analyzed by agarose gel electrophoresis and visualized by ethidium bromide (EtBr) staining. The migration positions of supercoiled and relaxed DNA are indicated.
The role of phosphorylation in the regulation of the activities of RPA is not entirely understood. Some studies have reported that DNA-PK preferentially phosphorylates RPA that is bound to ssDNA near junctions with dsDNA,Citation13,Citation23 such as at fork DNA, which are also the preferred binding sites for HARP. This suggests that HARP, RPA, and DNA-PK may function together at these unique DNA sites. Our in vitro pull-down experiments were performed in the absence of DNA and demonstrate that HARP, RPA, and DNA-PK do not require DNA for their interaction (). Whether significant amounts of HARP, RPA, and DNA-PK are interacting without DNA in the nucleus is unclear. The purification of the HARP complex may result in extraction of the three factors off of the DNA. Our study also found that HARP interferes with RPA phosphorylation both by its annealing helicase activity, which limits the amount of ssDNA in the cell, and by preventing phosphorylation of RPA by protein:protein interactions. Thus, it appears that HARP plays a multifaceted role in regulating the function of RPA. The connection between mutations in HARP and SIOD, a disease with a pleiotropic phenotype, is not fully understood. The diverse roles of HARP and RPA in modulating the levels of ssDNA in the cell likely impact many processes such as DNA replication, repair, and transcription, ultimately contributing to the complex nature of SIOD.
Materials and Methods
Native HARP complex
The native HARP-containing complex was purified from HeLa cell nuclear extracts by tandem-affinity purification as previously reported.Citation3 The glycerol gradients were performed by loading the purified HARP complex onto a 4 ml, 10 to 30% linear gradient in HARP buffer (20 mM HEPES, pH 7.6, 1.5 mM MgCl2, 10% glycerol, 0.05% NP40, 1 mM DTT, 0.2 mM PMSF) + 0.1 M KCl. The gradients were centrifuged at 170 000 × g for 3 h at 4 °C.
Single-step purification of DNA-PK
The native DNA-PK heterotrimer was purified by a single step DNA-affinity chromatography method. Poly-[dI:dC] (Amersham) was resuspended in H2O and briefly sonicated (three times for 2 s each) to fragment the polymers into smaller fragments. The fragments were then crosslinked to CNBr-activated Sepharose (GE) according to the manufacturer protocol. Nuclear extracts were prepared from HeLa cells using 0.42 M KCl extraction as previously describedCitation3,Citation24 and dialyzed to 0.1 M KCl. The extracts were then incubated with plasmid DNA (at a concentration of 130 µg/ml) on ice for 10 min. The extract was then centrifuged at 16 000 × g for 10 min to remove precipitations. The clarified extract was then incubated with the crosslinked Sepharose beads for 10 min at 4 °C on a rotator. The mix was poured into a 10 ml disposable column and the resin was allowed to settle. The column was drained and the resin washed four times with 5 column volumes of HARP Buffer + 0.1 M NaCl. The bound proteins were eluted by a step gradient of increasing NaCl.
Pull-down and in vitro kinase assays
Recombinant human HARP was purified by baculovirus expression as previously described.Citation1 The human RPA used in all of the assays was expressed and purified from bacteria as previously described.Citation1,Citation15 The pull-down experiments were performed essentially as described.Citation3 HARP (0.5 µg), RPA (0.5 µg), DNA-PK (0.1 µg) were incubated in 20 µl of Binding buffer (20 mM Hepes [K+], pH 7.6, 0.1 M KCl, 5 mM MgCl2, 3% [v/v] glycerol, 0.25 mg/ml BSA, 0.05 mM EDTA, 0.5 mM DTT, 0.01% (v/v) NP-40) for 20 min at 30 °C. The reactions were then diluted to 200 µl with Binding buffer and incubated with 10 µl of anti-FLAG resin for 3 h at 4 °C. The resin was washed three times with Binding buffer and the proteins eluted with SDS-sample buffer. For the in vitro kinase assays, HARP (0.4 µg), RPA (0.5 µg), DNA-PK (0.3 µg) were incubated in 20 µl of Binding buffer with plasmid DNA (60 ng) and [32P]-ATP for 15 min at 30 °C. The reactions were stopped by the addition of SDS sample buffer and resolved by SDS-PAGE. The gel was dried and the labeled proteins were detected by phosphorimaging.
Annealing helicase assays
The annealing helicase assay was performed as previously described.Citation1 In brief, plasmid DNA was incubated with RPA in 1× TE in the presence of topoisomerase I. This results in the loading of RPA onto the plasmid due to transient opening of the DNA. The RPA bound substrate is then incubated with HARP in Annealing Helicase buffer (10 mM TRIS-HCl, pH 7.9, 20 mM KCl, 2 mM MgCl2, 0.5 mM DTT, 0.1 mM EDTA), and in the presence or absence of ATP (UTP was included in the “minus” ATP reactions). The reactions are stopped and deproteinized with SDS and phenol extraction. The DNA is then precipitated and resolved by agarose gel electrophoresis. Where indicated, 0.5 µg of DNA-PK was included and in reactions that contained both DNA-PK and HARP, the DNA-PK and HARP were pre-incubated with or without ATP for 15 min before combining with the RPA-bound DNA substrate.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank James T Kadonaga and Patricia Szajner for critical reading of this manuscript. This work was supported by the Dana-Farber Cancer Institute and the Ellison Medical Foundation. Initial studies were also supported with the NIH grant RO1GM058272 to James T Kadonaga.
References
- Yusufzai T, Kadonaga JT. HARP is an ATP-driven annealing helicase. Science 2008; 322:748 - 50; http://dx.doi.org/10.1126/science.1161233; PMID: 18974355
- Lao Y, Lee CG, Wold MS. Replication protein A interactions with DNA. 2. Characterization of double-stranded DNA-binding/helix-destabilization activities and the role of the zinc-finger domain in DNA interactions. Biochemistry 1999; 38:3974 - 84; http://dx.doi.org/10.1021/bi982371m; PMID: 10194309
- Yusufzai T, Kong X, Yokomori K, Kadonaga JT. The annealing helicase HARP is recruited to DNA repair sites via an interaction with RPA. Genes Dev 2009; 23:2400 - 4; http://dx.doi.org/10.1101/gad.1831509; PMID: 19793863
- Yuan J, Ghosal G, Chen J. The annealing helicase HARP protects stalled replication forks. Genes Dev 2009; 23:2394 - 9; http://dx.doi.org/10.1101/gad.1836409; PMID: 19793864
- Ciccia A, Bredemeyer AL, Sowa ME, Terret ME, Jallepalli PV, Harper JW, Elledge SJ. The SIOD disorder protein SMARCAL1 is an RPA-interacting protein involved in replication fork restart. Genes Dev 2009; 23:2415 - 25; http://dx.doi.org/10.1101/gad.1832309; PMID: 19793862
- Bansbach CE, Bétous R, Lovejoy CA, Glick GG, Cortez D. The annealing helicase SMARCAL1 maintains genome integrity at stalled replication forks. Genes Dev 2009; 23:2405 - 14; http://dx.doi.org/10.1101/gad.1839909; PMID: 19793861
- Postow L, Woo EM, Chait BT, Funabiki H. Identification of SMARCAL1 as a component of the DNA damage response. J Biol Chem 2009; 284:35951 - 61; http://dx.doi.org/10.1074/jbc.M109.048330; PMID: 19841479
- Boerkoel CF, Takashima H, John J, Yan J, Stankiewicz P, Rosenbarker L, André JL, Bogdanovic R, Burguet A, Cockfield S, et al. Mutant chromatin remodeling protein SMARCAL1 causes Schimke immuno-osseous dysplasia. Nat Genet 2002; 30:215 - 20; http://dx.doi.org/10.1038/ng821; PMID: 11799392
- Simon AJ, Lev A, Jeison M, Borochowitz ZU, Korn D, Lerenthal Y, Somech R. Novel SMARCAL1 Bi-allelic Mutations Associated with a Chromosomal Breakage Phenotype in a Severe SIOD Patient. J Clin Immunol 2013; Forthcoming PMID: 24197801
- Din S, Brill SJ, Fairman MP, Stillman B. Cell-cycle-regulated phosphorylation of DNA replication factor A from human and yeast cells. Genes Dev 1990; 4:968 - 77; http://dx.doi.org/10.1101/gad.4.6.968; PMID: 2200738
- Dutta A, Stillman B. cdc2 family kinases phosphorylate a human cell DNA replication factor, RPA, and activate DNA replication. EMBO J 1992; 11:2189 - 99; PMID: 1318195
- Fotedar R, Roberts JM. Cell cycle regulated phosphorylation of RPA-32 occurs within the replication initiation complex. EMBO J 1992; 11:2177 - 87; PMID: 1318194
- Brush GS, Anderson CW, Kelly TJ. The DNA-activated protein kinase is required for the phosphorylation of replication protein A during simian virus 40 DNA replication. Proc Natl Acad Sci U S A 1994; 91:12520 - 4; http://dx.doi.org/10.1073/pnas.91.26.12520; PMID: 7809070
- Boubnov NV, Weaver DT. scid cells are deficient in Ku and replication protein A phosphorylation by the DNA-dependent protein kinase. Mol Cell Biol 1995; 15:5700 - 6; PMID: 7565721
- Henricksen LA, Carter T, Dutta A, Wold MS. Phosphorylation of human replication protein A by the DNA-dependent protein kinase is involved in the modulation of DNA replication. Nucleic Acids Res 1996; 24:3107 - 12; http://dx.doi.org/10.1093/nar/24.15.3107; PMID: 8760901
- Liaw H, Lee D, Myung K. DNA-PK-dependent RPA2 hyperphosphorylation facilitates DNA repair and suppresses sister chromatid exchange. PLoS One 2011; 6:e21424; http://dx.doi.org/10.1371/journal.pone.0021424; PMID: 21731742
- Shao RG, Cao CX, Zhang H, Kohn KW, Wold MS, Pommier Y. Replication-mediated DNA damage by camptothecin induces phosphorylation of RPA by DNA-dependent protein kinase and dissociates RPA:DNA-PK complexes. EMBO J 1999; 18:1397 - 406; http://dx.doi.org/10.1093/emboj/18.5.1397; PMID: 10064605
- Wang Y, Zhou XY, Wang H, Huq MS, Iliakis G. Roles of replication protein A and DNA-dependent protein kinase in the regulation of DNA replication following DNA damage. J Biol Chem 1999; 274:22060 - 4; http://dx.doi.org/10.1074/jbc.274.31.22060; PMID: 10419533
- Oakley GG, Patrick SM, Yao J, Carty MP, Turchi JJ, Dixon K. RPA phosphorylation in mitosis alters DNA binding and protein-protein interactions. Biochemistry 2003; 42:3255 - 64; http://dx.doi.org/10.1021/bi026377u; PMID: 12641457
- Couch FB, Bansbach CE, Driscoll R, Luzwick JW, Glick GG, Bétous R, Carroll CM, Jung SY, Qin J, Cimprich KA, et al. ATR phosphorylates SMARCAL1 to prevent replication fork collapse. Genes Dev 2013; 27:1610 - 23; http://dx.doi.org/10.1101/gad.214080.113; PMID: 23873943
- Carroll C, Bansbach CE, Zhao R, Jung SY, Qin J, Cortez D. Phosphorylation of a C-terminal auto-inhibitory domain increases SMARCAL1 activity. Nucleic Acids Res 2014; 42:918 - 25; http://dx.doi.org/10.1093/nar/gkt929; PMID: 24150942
- Bétous R, Glick GG, Zhao R, Cortez D. Identification and characterization of SMARCAL1 protein complexes. PLoS One 2013; 8:e63149; http://dx.doi.org/10.1371/journal.pone.0063149; PMID: 23671665
- Vidal-Eychenié S, Décaillet C, Basbous J, Constantinou A. DNA structure-specific priming of ATR activation by DNA-PKcs. J Cell Biol 2013; 202:421 - 9; http://dx.doi.org/10.1083/jcb.201304139; PMID: 23897887
- Nakatani Y, Ogryzko V. Immunoaffinity purification of mammalian protein complexes. Methods Enzymol 2003; 370:430 - 44; http://dx.doi.org/10.1016/S0076-6879(03)70037-8; PMID: 14712665
