Abstract
Dosage compensation of X-linked genes is a phenomenon of concerted, chromosome-wide regulation of gene expression underpinned by sustained and tightly regulated histone modifications and chromatin remodeling, coupled with constrains of nuclear architecture. This elaborate process allows the accomplishment of regulated expression of genes on the single male X chromosome to levels comparable to those expressed from the two X chromosomes in females. The ribonucleoprotein Male Specific Lethal (MSL) complex is enriched on the male X chromosome and is intricately involved in this process in Drosophila melanogaster. In this review we discuss the recent advances that highlight the complexity lying behind regulation of gene expression by just two-fold.
The need to compensate for the hemizygosity of the X-linked genes in heterogametic males and to equalize their expression levels with that of the two X chromosomes in females has led to the development of dosage compensation mechanisms. These mechanisms have evolved through the combination of pre-existing proteins acquiring new functions in the context of de novo complexes and gene products specifically dedicated to the new dosage compensation activity. The fascinating insights into the sex-specific dosage compensation phenomena are mainly derived from three groups of organisms: fruit flies (Drosophila melanogaster), round worms (Caenorhabditis elegans) and mammals (mice, Mus musculus and humans, Homo sapiens) (). In these model organisms, males are the heteromorphic sex (XY in Drosophila and mammals and XO in C. elegans), whereas the homomorphic animals (XX) are either females (Drosophila and mammals) or hermaphrodites (C. elegans).
In flies, dosage compensation of the X-linked genes leads to a two-fold upregulation in males in comparison to females. This is mediated by the dosage compensation complex (DCC) also known as Male Specific Lethal complex (MSL), due to the male specific lethality phenotype upon loss-of-function of its major components. The Drosophila MSL is a ribonucleoprotein complex and is composed of at least five proteins, namely MSL-1 (male-specific lethal 1, scaffolding protein), MSL-2 (male-specific lethal 2, RING finger protein), MSL-3 (male-specific lethal 3, chromodomain protein), MOF (males absent on the first, histone acetyltransferase) and MLE (maleless, RNA helicase), and two functionally redundant long non-coding RNAs: roX1 (RNA on the X 1) and roX2 (RNA on the X 2) ( and ). The MSL complex decorates the male X chromosome which is also hyperacetylated at histone H4 lysine 16 (reviewed in refs. Citation1 and Citation2).
In worms, dosage compensation is achieved by downregulation by half of the transcriptional output from both X chromosomes in hermaphrodites (). This process is mediated by a multiprotein complex, also known as the DCC, which consists of a number of condensin-like proteins. Similar to flies, the worm DCC is also enriched on the dosage compensated X chromosome (reviewed in ref. Citation3). However, in contrast to Drosophila or mammals (see below), non-coding RNAs contributing to its function have not been identified so far.Citation3 The exact mechanism by which the C. elegans DCC is targeted to the X chromosomes is unknown, but discrete recruitment elements, namely rexCitation4,Citation5 and doxCitation6,Citation7 sites on the X chromosome have recently been identified.
In mammals, dosage compensation is achieved by inactivation of one of the two female X chromosomes. Interestingly, X chromosome inactivation requires the concerted action of a number of non-coding RNAs (reviewed in ref. Citation8). A recent new addition to the list of non-coding RNAs involved in X inactivation is Jpx.Citation9 Work over the last two decades has shown that X-inactive-specific transcript (Xist), a non-coding RNA expressed from the X chromosome inactivation center (Xic), plays a major role in X inactivation. It acts in cis by coating the entire chromosome from which it is produced and triggers a cascade of chromatin modification events (). The search for protein components mediating X chromosome inactivation is currently ongoing. Nevertheless, recruitment of the Polycomb repressive complexes and histone H3 lysine 27 methylation has been implicated.Citation10–Citation12 Furthermore, the E3 ubiquitin ligase RNF12 has recently been shown to also play an important role in X inactivation.Citation13,Citation14
In this review, we focus on recent advances in our understanding of the dosage compensation process in Drosophila.
Solution for Keeping X Chromosomal Balance in Female Flies
An inevitable consequence of dosage compensation systems in which X-linked genes get transcriptionally upregulated is the risk of overexpression of the X-linked genes in females, where excess of the corresponding products could be as deleterious as their deficiency in males. Therefore, mechanisms have co-evolved with dosage compensation in males to prevent overexpression in females. In Drosophila, an exquisite interplay between two proteins MSL-2 (expressed in males) and Sex Lethal (SXL; expressed in females) ensures that the “holo” MSL complex is stably expressed only in males.
In females, expression of SXL is triggered from an “early” Sxl promoter.Citation15 In the presence of early SXL, female-specific splicing of Sxl pre-mRNA is set up and maintained in females from a “late” promoter and the translation-terminating exon 3 is removed after splicing.Citation16 Since little or no SXL is present in males, Sxl pre-mRNA is spliced by default, the sequence that carries a translational stop codon is retained and this results in a truncated and inactive SXL protein. Following this stage, Sxl expression is shut down in males but maintained in females by a positive autoregulatory loop determining sexual identity for the rest of the life cycle (reviewed in ref. Citation17).
Upon establishment of stable SXL expression in females, msl-2 expression is controlled at two levels. First, SXL binds multiple poly(U) sequences within msl-2 pre-mRNA and guides the alternative splicing of the first intron; out in males but retained in females. This leads to instability of the msl-2 mRNA and a substantial downregulation of the transcript in females.Citation18,Citation19 Second, MSL-2 is translationally silenced in Drosophila females by SXL, which binds to poly(U) stretches in the msl-2 mRNA 5′ and 3′ UTRs.Citation18,Citation20 An SXL 3′ UTR repressor complex blocks the 43S pre-initiation complex, independently of a separate 5′UTR bound SXL mechanism, which is able to inhibit ribosomal scanning.Citation21 The resulting absence of MSL-2 in females leads to destabilization and partial degradation of MSL-1,Citation22 and to a lower extent MSL-3 and roX RNAs. Consequently, this renders the dosage compensation complex inactive in females, which is a prerequisite for their viability. This is confirmed by the striking observation that ectopic MSL-2 expression in female flies induces 85% lethality which goes up to 100% upon simultaneous ectopic expression of MSL-1,Citation23 whereas reduction of MSL-1 by half completely relieves the MSL-2 overexpression toxicity.Citation22
Interestingly, a recent study has suggested that early during embryonic development, prior to the full Sxl activation, a transient assembly of MSL complexes and dosage compensation occurs. This is reliant on the maternal contribution and zygotic roX1 expression and also occurs in females. It is required for the sex determination signal accumulation, the upregulation of Sxl transcription and subsequent shut down of MSL-2 expression.Citation24 This effect is surprising given that neither the MSL-2 protein aloneCitation22,Citation25 nor the MSL complex have been previously detected in female embryos.Citation26,Citation27 However, maternal effect of msl-2 has been previously described.Citation28 Nevertheless, to firmly establish the mechanism of this regulation and whether there is indeed MSL-2 protein expression and functional MSL assembly concomitant with targeting to the Sxl locus, in female embryos, further investigation is required.
Recognizing and Targeting the Male X Chromosome
Regardless of the sex and the need to upregulate or downregulate X-linked gene expression, the recognition and targeting to the X chromosome is a major problem faced by the dosage compensation complex.
The recent genome-wide techniques such as chromatin immunoprecipitation coupled with microarray technology (ChIP-on-chip) or deep sequencing (ChIP-seq) have allowed high-resolution mapping of MSL proteins.Citation29–Citation34 One major finding was that on a genome-wide scale, the MSL targets are enriched in the gene body with a peak at their 3′endsCitation29–Citation33 (). Furthermore, not all MSL proteins behave similarly as the MOF protein also binds to promoter proximal regions on the X chromosome as well as to a large number of autosomal promoters, independently of the other MSL complex members.Citation32 Accordingly, MOF has recently been reported to be a component of a second complex named nonspecific lethal (NSL) complex which binds to more than 4,000 genes at their promoter regions and acts as a major transcriptional regulator in Drosophila.Citation35–Citation37 Fascinatingly, the existence of NSL complexes and their interaction with MOF has also been reported in mammals. However, while the mammalian MSL-associated MOF acetylates nucleosomal histone H4 almost exclusively on lysine 16, NSL-associated MOF exhibits a relaxed specificity and also acetylates nucleosomal histone H4 on lysines 5 and 8.Citation35,Citation38 Future studies will be crucial in revealing the interplay between the MSL and the NSL complexes in transcription regulation in Drosophila and mammals.
The enrichment of the MSL complex towards the 3′ end of genes has led to the proposal that the MSL complex could facilitate elongation of transcription.Citation39 Most target genes seem to be actively transcribed, reaffirming previous reports that MSL complex recruitment depends on passage of the transcription machinery, but not on the type of promoter or directionality of transcription.Citation40 At the level of polytene chromosome stainings, it is not possible to detect MSL complex recruitment on autosomal translocations on the X chromosome.Citation41,Citation42 However, it was shown recently by ChIP analysis that active autosomal transgenes inserted on the X chromosome are indeed capable of MSL recruitment.Citation43 These results suggest that in an X chromosomal context transcription activity does contribute to MSL complex recruitment, such that also active autosomal genes benefit, if inserted on the X chromosome.
Since not all actively transcribed genes are bound by the MSL complex, transcription per se may not be the only signal for MSL recruitment and dosage compensation. This notion is further supported by the observation that MSL-1 binding profiles at different developmental stages are similar.Citation33 Therefore, the decision of which genes will be subject to dosage compensation is very likely taken early during development and most of them are bound regardless of developmental changes.Citation44 However, a recent report showing that many X chromosomal genes bind MSL complexes only too transiently for the interaction to be detected by conventional methods, make it still conceivable that transcription activation is a prerequisite but may not be sufficient for MSL recruitment.Citation45
Is it possible that the X chromosome possesses special sequences that help to recruit MSL complexes? Despite the numerous known MSL targets a universal targeting sequence motif has been difficult to identify. Early in situ hybridization studies on Drosophila polytene chromosomes showed that a (dC − dA)n.(dG − dT)n sequence is enriched on the X chromosome.Citation46 More recently whole genome sequence analysis revealed that the X chromosome can be distinguished from the other chromosomes based on the C/An and G/Tn repeats as a sequence signature.Citation47
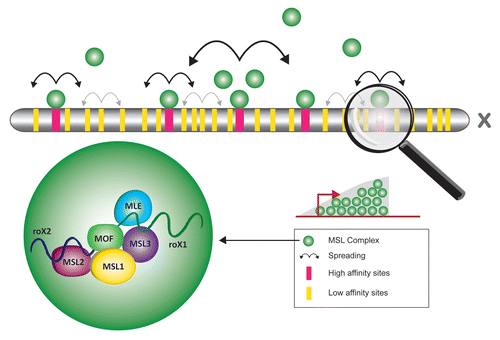
A number of high affinity binding sites (HAS) on the X chromosome have been defined on the basis that partial complexes of MSL-1-MSL-2 could still bind these sites even in the absence of the rest of the MSL components, enforcing the idea that sequence recognition motifs for MSL targeting do exist. Interestingly, the two roX genes located on the X chromosome act as high affinity sites and play an important role in assembly and spreading of the MSL complex () (reviewed in ref. Citation2). HAS were also previously called chromatin entry sites (CES) as according to the currently accepted model, MSL complexes bind to these discrete number of sites along the X chromosome and spread into the local chromatinCitation48 (). It has been a reasonable hypothesis that high affinity sites should have the highest probability for carrying the MSL targeting sequence motifs and recent efforts have been focused on analysis of these sequences. A “one-hybrid” assay approach suggested degenerate sequence motifs that represent weak DNA determinants whose cumulative contributions are required to form a HAS for MSL-2 targeting.Citation49 A ChIP-on-chip and ChIP-seq approach identified roughly 150 putative HAS and a GA-rich or TC-rich MSL recognition element (MRE) that seems to be of functional relevance.Citation30 A parallel study identified 131 HAS carrying GA- and CA-rich motifs.Citation34 Most recently, oligonucleotide profiling, a newly developed method for DNA sequence analysis, observed lower complexity of the X chromosome in Drosophila in comparison with the autosomes.Citation50,Citation51 A repetitive sequence motif [G(CG)N4] was found to be specifically enriched in regions targeted by MSL.Citation49,Citation50 In conclusion, it appears that the X chromosome is enriched for different types of sequence motifs such as low complexity dinucleotide (GA/TC)n-like repeats within the HAS and [G(CG)N4>]-like sequences within the low affinity sites.Citation50 It is possible that a perfect sequence satisfying all the targeting criteria does not exist. At this stage, it seems likely that the bona fide targeting of MSL complexes to the X chromosome is a result of the combination of different factors including active transcription and sequence motifs, which are insufficient for complex recruitment when present alone. It is also becoming clear that the local chromatin context may also be a defining factor in DCC recruitment and its specificity.
Histone Modification Crosstalk on the Male X
It has become increasingly evident in the past several years that factors, in addition to the DNA sequence motifs, could be required for the faithful targeting of MSL to the X chromosome and dosage compensation. Local chromatin context may constitute one obvious factor. Posttranslational modifications of histones within the nucleosomes can have a huge impact on chromatin structure and gene transcription, and represent a wealth of epigenetic information.Citation52 Examples of covalent modifications of histone tails include acetylation, phosphorylation, sumoylation, ubiquitination and methylation.
A large body of evidence compiled in the recent years has demonstrated the impact of histone acetylation on transcriptional activity (reviewed in refs. Citation53–Citation55). Gene activation for dosage compensation involves the MSL-associated MOF acetyltransferase activity on H4K16 (histone H4 lysine 16), which represents a hallmark of the male X chromosome.Citation32,Citation56–Citation58 The mechanism by which H4K16ac contributes to dosage compensation remains an active area of study.Citation32,Citation36,Citation45,Citation59 One possible mechanism could be mediated by the structural changes imposed by the presence of this modification on nucleosomes, thus creating an open chromatin environment for transcription associated complexes.Citation60 A recent genome-wide study showed increased DNA accessibility at active promoters and chromosomal regions that are hyperacetylated at H4K16.Citation59 A similar function is attributed to H3S10phCitation61 and H3K4me2.Citation62 Interestingly, data begins to accumulate elaborating on the involvement of the MOF and histone H4K16Ac in a variety of processes during evolution ranging from embryonic developmentCitation63,Citation64 and DNA damage repairCitation65–Citation68 to general transcription regulation.Citation69 Future studies should elaborate the role of MSL proteins beyond dosage compensation.
Interestingly, the peak of MSL binding at the 3′ end of the dosage compensated genes also coincides with trimethylation of lysine 36 on histone H3 (H3K36me3),Citation70,Citation71 a well-established mark for active genes.Citation72,Citation73 A crosstalk between H3K36me3 and H4K16ac was suggested based on the observation that downregulation of Hypb, the enzyme required for the methylation of H3K36me3 in Drosophila, leads to reduced H4K16 acetylation on X-linked genes.Citation70
MSL-3 binds H3K36me3 nucleosomes in in vitro assays.Citation71 However, recent structural work suggests co-recognition of DNA and a histone H4 tail monomethylated on lysine 20 (H4K20me) by the MSL-3 chromodomain.Citation74 In a parallel study, MSL-3 affinity for H4K20me was confirmed.Citation67 However, MSL-3/nucleic acid interaction was not reported.Citation67 Interestingly, in both reports H3K36me3 peptide was shown not to be the preferred substrate for the MSL-3 chromodomain.Citation74,Citation75 How the presence of H3K36me3 on active X-linked genes integrates into the targeting model needs to be further addressed. At the same time, there is no functional evidence yet for the role of H4K20me in the dosage compensation mechanism. Future studies will be instrumental in clarifying these questions.
Phosphorylation of histone H3 on serine 10 (H3S10ph) mediated by JIL-1 kinase has also been associated with the MSL complex.Citation61,Citation76,Citation77 JIL-1 and H3S10ph have been shown to be enriched on the male X chromosome.Citation61,Citation76,Citation77 However, JIL-1 null mutants lead to lethality in both sexes suggesting an additional, broader role for this enzymatic activity.Citation61,Citation78,Citation79 The mechanism by which JIL-1-mediated H3S10 phosphorylation leads to transcriptional defects in interphase cells remains a matter of some controversy. On the one hand, H3S10ph has been shown to facilitate RNA polymerase II release from promoter-proximal pausing in Drosophila.Citation80 The histone crosstalk between H3S10ph and H4K16ac has also been proposed to generate a histone code that mediates transcription elongation in mammals.Citation81 The latter is reaffirmed by a recent study showing that the 14-3-3 protein localizes to active genes in a JIL-1-dependent manner, binds H3 when phosphorylated and interacts with the Elp3 acetyltransferase (acetylates H3 on lysine 9, H3K9ac),Citation82 which functions during transcription elongation.Citation83 However, other studies support a different model in which Pol II-mediated transcription does not require H3S10ph and suggest that the transcriptional defects observed in the absence of JIL-1/H3S10ph are a result of structural alterations of chromatin.Citation84,Citation85 Interestingly, JIL-1-mediated ectopic H3S10 phosphorylation is sufficient to induce a change in higher-order chromatin structure from a “heterochromatin-like” state to a more open “euchromatic” state.Citation86 Furthermore, in the absence of JIL-1, levels of H3K9ac are significantly reduced leading to ectopic spreading of the major heterochromatin markers H3K9me2, HP1a87 and Su(var)3–7.Citation88
Conventional core histones can be replaced by histone variants which have different structural, mobility and stability characteristics, as well as altered susceptibility to modifications, and hence can affect the overall dynamics of chromatin (reviewed in ref. Citation89). Interestingly, histone H3.3 has shown to be frequently replaced on active genes and is enriched on the dosage compensated male X chromosome.Citation90
The primary role of the MSL complex appears to be X chromosomal regulation, but it seems that its function is facilitated by a number of more general factors. These include the DNA supercoiling factor SCF,Citation91 ISWI,Citation92 NURF301,Citation93 HP1,Citation94 UNRCitation95 and Su(var)3–7,Citation96 which display complex genetic interactions with the MSL complex. However, the mechanistic details of these interactions and how they mediate the formation of proper chromatin architecture along the male X chromosome to facilitate dosage compensation are largely unknown. Nevertheless, one emerging theme is the requirement for a permissive chromatin structure to be established and maintained to allow and facilitate the hypertranscription underlying the dosage compensation process. Furthermore, these histone modifications have a great impact on chromatin folding and flexibility, which in turns is essential for the genome architecture and function (reviewed in ref. Citation97). Further studies are required to elucidate the complex interplay between the various histone modifications and their role for chromatin organization and how they impact dosage compensation. A recently developed genetic system in Drosophila provides an exciting tool for studying the “histone code” and histone-dependent chromatin assembly in vivo.Citation98
Chromosomal Context and Dosage Compensation
In recent years it has become increasingly evident that nuclear three-dimensional structure and genome organization during interphase are of functional importance.Citation99 It has been shown that gene positioning within the nucleus in relation to other genes and subnuclear compartments contributes to transcriptional control, modulating activity and ensuring maximal expression in some cases or repression in others (reviewed in ref. Citation100). Since dosage compensation and transcription activation are intrinsically linked, it is not surprising that evidence has started to accumulate indicating a role of nuclear architecture in dosage compensation.
In higher eukaryotes individual chromosomes occupy discrete chromosome territories in the nucleus forming subchromosomal domains of various size.Citation101,Citation102 Interestingly, gene-poor or inactive domains reside more internally while active ones tend to localize more peripherally and even loop out of their chromosomal territory and dynamically relocate to specialized subnuclear compartments in association with their activation status.Citation103–Citation106
The conventional perception of the nuclear periphery as a site of gene repression has been challenged by findings that physical interactions between actively transcribed genes and the nuclear pore complex (NPC) members can exist (reviewed in ref. Citation107). It has been proposed that the NPCs could represent platforms for the preassembled transcriptional machineries such as transcription factories.Citation108,Citation109 This would help create increased local concentration of transcription factors and enzymatic activities for the coordinated regulation of gene expression.Citation110,Citation111 NPCs have been implicated in transcriptional regulation through post-translational modifications of transcriptional factors such as phosphorylationCitation112 or sumoylation.Citation113 Facilitation of mRNA processing and export is obviously one of the most classical functions attributed to NPCs (reviewed in ref. Citation114). NPCs might also have a role in creating boundaries preventing the actively transcribed domains from the invasion of repressive signals from the surrounding peripheral lamina-associated heterochromatin. In addition, NPC components could also establish promoter-end gene loop formation, facilitating RNA Pol II recycling during transcription but also serving as a transcriptional memory allowing rapid reinduction of transcription.Citation115,Citation116 At the same time, chromatin structure can also affect nuclear envelope integrity and NPC formation.Citation117
Gene-to-pore association in yeast is mediated, at least partly, by DNA zip codes named gene recruitment sequences (GRSs).Citation118 Whether similar sequences exist and are required for gene targeting to the nuclear periphery in other organisms remains unclear. Adaptor or bridging proteins involved in the interaction between chromatin and the NPC have also been reported including EYN2 and Xmas-2,Citation119 the Htz1 histone variant,Citation120 mRNA-export receptorsCitation113,Citation121 and the CCCTC-binding factor CTCF.Citation122,Citation123 A number of nucleoporins have also been reported to be involved in gene-NPC interactions.Citation35,Citation124–Citation126 The fact that many NPC components such as Nup153,Citation127 and Megator (Mtor),Citation128,Citation129 shuttle between the nucleoplasm and the nuclear pores and form dynamic filamentous structures protruding into the nucleosplasm, has led to the suggestion that two pools of NPC proteins exist that may have different functions depending on their subnuclear localization. Several different Nups have been recently reported to associate with actively transcribed genes including a large number of nucleoplasm-residing genes.Citation130–Citation132 However, whether these interactions directly activate transcription or bind as a consequence of activation is still under investigation. It is possible that they may facilitate mRNA export providing a physical route to the periphery of the nucleus. A role for Nups in mediating dynamic looping outside the chromosomal territory and transient associations with the NPCs, has also been suggested.Citation113,Citation116
Intriguingly, the Drosophila MSL complex member MOF has been shown to co-purify with components of the NPC including Nup153 and Mtor.Citation35 RNAi-mediated knockdown of these components led to reduction of the typical MSL binding pattern on the X, suggesting a role in the dosage compensation process. In support of this idea it was shown that Nup153 and Mtor bind genome-wide in large domains, the so-called nucleoporin associated regions (NARs), which are heavily enriched for active transcription marks. Interestingly, up to 75% of the male X chromosome is enriched in these domains.Citation132 Taking into consideration the recently demonstrated recruitment of actively expressed genes by a common transcription factor (Klf-1) to “specialized” transcription factories in erythroid cells,Citation133 it is tempting to speculate that nucleoporins might act in a similar manner. Nucleoporins may mediate the coordinated expression of a number of genes genome-wide, including the extreme case of chromosome-wide upregulation of X-linked genes underlying dosage compensation in Drosophila males. Hyperactivation of the X chromosome seems to correlate with a particular chromosomal conformation where HAS reside in proximity to each other, forming a dosage-compensated chromosomal domain in an MSL-1-MSL-2-dependent manner.Citation134 A model has been proposed where MSL assembly from the sites of roX transcription in the interior of the chromosomal territory establishes a focal point of enrichment promoting radial spreading and a gradient of decreasing concentration of functional MSL complexes towards the periphery.Citation134
While many NARs seem to localize at the nuclear periphery, a subset of them are clearly nucleoplasmic showing that both soluble and peripheral bound pools contribute towards gene expression control.Citation132 It is possible that transcription control of a subset of genes occurs within nucleoporin-associated domains to allow the coupling of transcription and post-transcriptional events thus facilitating transcriptional output.Citation132 Future studies will be crucial in understanding how nuclear architecture influences gene expression and how this additional layer of control influences the X chromosome.
Concluding Remarks
Classical genetic studies have been instrumental in identifying and characterizing the dosage compensation complex members. Now, recent advances in the fields of biochemical and genomic analyses continue to reveal novel insights into the mechanisms by which dosage compensation is achieved. It is evident that MSL complex members not only take advantage of general factors to help regulate the X chromosome but are also involved in additional functions beyond X chromosomal regulation. Future studies promise to unravel how the MSL complex members balance their various roles and fine-tune gene expression.
Figures and Tables
Figure 1 Dosage compensation mechanisms in flies, worms and mammals. Dosage compensation process in Drosophila melanogaster is achieved by transcriptional upregulation of the male X chromosome (red) and is regulated by the MSL complex (red balls). In C. elegans the two X chromosome in hermaphrodites are repressed by half (green) and regulated by the DCC complex (blue balls). In mammals only one of the female X chromosomes is active (red) while the other is transcriptionally inactive (grey) and regulated by the presence of Xist RNA (grey blocks).
Figure 2 MSL proteins decorate the male X chromosome. Polytene stainings from third instar larvae immunostained with specific antibodies raised against MSL-1 and MOF. The figure shows enrichment of these proteins on the male X chromosome. DNA is stained with Hoechst322 (blue), MSL-1 (red), MOF (green). MOF protein can also been seen localized on autosomes albeit with lower intensity compared to the X chromosome.
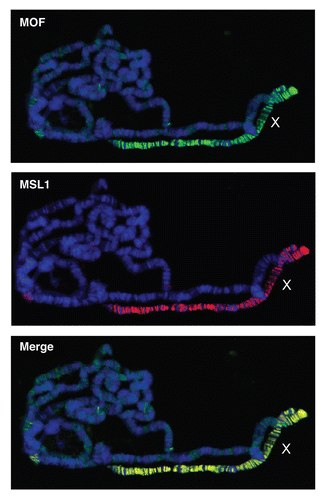
Figure 3 X chromosomal targeting of the male X chromosome in Drosophila melanogaster. The ribonucleoprotein containing MSL complex (green) targets the male X chromosome on several high (red) and low (yellow) affiinity sites. The high affinity sites such as the roX genes are thought to act as platforms for complex assembly from where the complex spreads (bold arrows) into the surrounding regions. Low affinity sites appear to have no or only limited spreading potential (grey arrows). On an individual gene level the MSL complex is enriched on the body of X linked genes peaking towards their 3′ end.
Acknowledgements
We are grateful to Vera Titschen for helping with the preparation of figures. We also thank Erinc Hallacli, Thomas Conrad, Matthew Turley and Juan M. Vaquerizas for critical reading of the manuscript and helpful suggestions.
References
- Hallacli E, Akhtar A. X chromosomal regulation in flies: when less is more. Chromosome Res 2009; 17:603 - 619
- Lucchesi JC. The structure-function link of compensated chromatin in Drosophila. Curr Opin Genet Dev 2009; 19:550 - 556
- Meyer BJ. Targeting X chromosomes for repression. Curr Opin Genet Dev 2010; 20:179 - 189
- Ercan S, Giresi PG, Whittle CM, Zhang X, Green RD, Lieb JD. X chromosome repression by localization of the C. elegans dosage compensation machinery to sites of transcription initiation. Nat Genet 2007; 39:403 - 408
- McDonel P, Jans J, Peterson BK, Meyer BJ. Clustered DNA motifs mark X chromosomes for repression by a dosage compensation complex. Nature 2006; 444:614 - 618
- Blauwkamp TA, Csankovszki G. Two classes of dosage compensation complex binding elements along Caenorhabditis elegans X chromosomes. Mol Cell Biol 2009; 29:2023 - 2031
- Jans J, Gladden JM, Ralston EJ, Pickle CS, Michel AH, Pferdehirt RR, et al. A condensin-like dosage compensation complex acts at a distance to control expression throughout the genome. Genes Dev 2009; 23:602 - 618
- Chow J, Heard E. X inactivation and the complexities of silencing a sex chromosome. Curr Op Cell Biol 2009; 21:359 - 366
- Tian D, Sun S, Lee JT. The long noncoding RNA, Jpx, is a molecular switch for X chromosome inactivation. Cell 2010; 143:390 - 403
- Muller J, Hart CM, Francis NJ, Vargas ML, Sengupta A, Wild B, et al. Histone methyltransferase activity of a Drosophila polycomb group repressor complex. Cell 2002; 111:197 - 208
- Silva J, Mak W, Zvetkova I, Appanah R, Nesterova TB, Webster Z, et al. Establishment of histone H3 methylation on the inactive X chromosome requires transient recruitment of Eed-Enx1 polycomb group complexes. Dev Cell 2003; 4:481 - 495
- Zhao J, Sun BK, Erwin JA, Song JJ, Lee JT. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science 2008; 322:750 - 756
- Jonkers I, Barakat TS, Achame EM, Monkhorst K, Kenter A, Rentmeester E, et al. RNF12 is an X-encoded dose-dependent activator of X chromosome inactivation. Cell 2009; 139:999 - 1011
- Shin J, Bossenz M, Chung Y, Ma H, Byron M, Taniguchi-Ishigaki N, et al. Maternal Rnf12/RLIM is required for imprinted X-chromosome inactivation in mice. Nature 2010; 467:977 - 981
- Schütt C, Nöthiger R. Structure, function and evolution of sex-determining systems in Dipteran insects. Development 2000; 127:667 - 677
- Bell LR, Horabin JI, Schedl P, Cline TW. Positive autoregulation of Sex-lethal by alternative splicing maintains the female determined state in Drosophila. Cell 1991; 65:229 - 239
- Salz HK, Erickson JW. Sex determination in Drosophila: The view from the top. Fly 2010; 4:60 - 70
- Kelley RL, Wang J, Bell L, Kuroda MI. Sex lethal controls dosage compensation in Drosophila by a nonsplicing mechanism. Nature 1997; 387:195 - 199
- Zhou S, Yang Y, Scott MJ, Pannuti A, Fehr K, Eisen A, et al. Male-specific lethal 2, a dosage compensation gene of Drosophila, undergoes sex-specific regulation and encodes a protein with a RING finger and a metallothionein-like cysteine cluster. EMBO J 1995; 14:2884 - 2895
- Bashaw GJ, Baker BS. The regulation of the Drosophila msl-2 gene reveals a function for Sex-lethal in translational control. Cell 1997; 89:789 - 798
- Beckmann K, Grskovic M, Gebauer Ft, Hentze MW. A dual inhibitory mechanism restricts msl-2 mRNA translation for dosage compensation in Drosophila. Cell 2005; 122:529 - 540
- Kelley RL, Solovyeva I, Lyman LM, Richman R, Solovyev V, Kuroda MI. Expression of Msl-2 causes assembly of dosage compensation regulators on the X chromosomes and female lethality in Drosophila. Cell 1995; 81:867 - 877
- Chang KA, Kuroda MI. Modulation of MSL1 Abundance in female drosophila contributes to the sex specificity of dosage compensation. Genetics 1998; 150:699 - 709
- Gladstein N, McKeon MN, Horabin JI. Requirement of male-specific dosage compensation in Drosophila females—implications of early X chromosome gene expression. PLoS Genet 2010; 6:1001041
- Bashaw GJ, Baker BS. The msl-2 dosage compensation gene of Drosophila encodes a putative DNA-binding protein whose expression is sex specifically regulated by Sex-lethal. Development 1995; 121:3245 - 3258
- McDowell KA, Hilfiker A, Lucchesi JC. Dosage compensation in Drosophila: the X chromosome binding of MSL-1 and MSL-2 in female embryos is prevented by the early expression of the Sxl gene. Mech Dev 1996; 57:113 - 119
- Rastelli L, Richman R, Kuroda MI. The dosage compensation regulators MLE, MSL-1 and MSL-2 are interdependent since early embryogenesis in Drosophila. Mech Dev 1995; 53:223 - 233
- Uenoyama T, Uchida S, Fukunaga A, Oishi K. Studies on the sex-specific lethals of Drosophila melanogaster. IV. Gynadomorph analysis of thress male-specific lethals mle, msl-227 AND mle(3)132. Genetics 1982; 102:223 - 231
- Alekseyenko AA, Larschan E, Lai WR, Park PJ, Kuroda MI. High-resolution ChIP-chip analysis reveals that the Drosophila MSL complex selectively identifies active genes on the male X chromosome. Genes Dev 2006 2006; 20:848 - 857
- Alekseyenko AA, Peng S, Larschan E, Gorchakov AA, Lee OK, Kharchenko P, et al. A sequence motif within chromatin entry sites directs MSL establishment on the Drosophila X chromosome. Cell 2008; 134:599 - 609
- Gilfillan GD, Straub T, de Wit E, Greil F, Lamm R, van Steensel B, et al. Chromosome-wide gene-specific targeting of the Drosophila dosage compensation complex. Genes Dev 2006; 20:858 - 870
- Kind J, Vaquerizas JM, Gebhardt P, Gentzel M, Luscombe NM, Bertone P, et al. Genome-wide analysis reveals MOF as a key regulator of dosage compensation and gene expression in Drosophila. Cell 2008; 133:813 - 828
- Legube G, McWeeney SK, Lercher MJ, Akhtar A. X-chromosome-wide profiling of MSL-1 distribution and dosage compensation in Drosophila. Genes Dev 2006; 20:871 - 883
- Straub T, Grimaud C, Gilfillan GD, Mitterweger A, Becker PB. The chromosomal high-affinity binding sites for the Drosophila dosage compensation complex. PLoS Genet 2008; 4:1000302
- Mendja S, Taipale M, Kind J, Holz H, Gebhardt P, Schelder M, et al. Nuclear pore components are involved in the transcriptional regulation of dosage compensation in Drosophila. Mol Cell 2006; 21:811 - 823
- Prestel M, Feller C, Straub T, Mitloehner H, Becker PB. The activation potential of MOF is constrained for dosage compensation. Mol Cell 2010; 38:815 - 826
- Raja SJ, Charapitsa I, Conrad T, Vaquerizas JM, Gebhardt P, Holz H, et al. The nonspecific lethal complex is a transcriptional regulator in Drosophila. Mol Cell 2010; 38:827 - 841
- Cai Y, Jin J, Swanson SK, Cole MD, Choi SH, Florens L, et al. Subunit composition and substrate specificity of a MOF-containing histone acetyltransferase distinct from the male-specific lethal (MSL) complex. J Biol Chem 2010; 285:4268 - 4272
- Smith ER, Allis CD, Lucchesi JC. Linking global histone acetylation to the transcription enhancement f X-chromosomal genes in Drosophila males. J Biol Chem 2001; 276:31483 - 31486
- Kind J, Akhtar A. Cotranscriptional recruitment of the dosage compensation complex to X-linked target genes. Genes Dev 2007; 21:2030 - 2040
- Fagegaltier D, Baker BS. X Chromosome sites autonomously recruit the dosage compensation complex in Drosophila males. PLoS Biol 2004; 2:341
- Oh H, Bone JR, Kuroda MI. Multiple Classes of MSL binding sites target dosage Compensation to the X chromosome of Drosophila. Curr Biol 2004; 14:481 - 487
- Gorchakov AA, Alekseyenko AA, Kharchenko P, Park PJ, Kuroda M. Long-range spreading of dosage compensation in Drosophila captures transcribed autosomal genes inserted on X. Genes Dev 2009; 23:2266 - 2271
- Kotlikova IV, Demakova OV, Semeshin VF, Shloma VV, Boldyreva LV, Kuroda MI, et al. The Drosophila dosage compensation complex binds to polytene chromosomes independently of developmental changes in transcription. Genetics 2006; 172:963 - 974
- Gelbart ME, Larschan E, Peng S, Park PJ, Kuroda MI. Drosophila MSL complex globally acetylates H4K16 on the male X chromosome for dosage compensation. Nat Struct Mol Biol 2009; 16:825 - 832
- Pardue ML, Lowenhaupt K, Rich A, Nordheim A. (dCdA)n.(dG-dT)n sequences have evolutionarily conserved chromosomal locations in Drosophila with implications for roles in chromosome structure and function. EMBO J 1987; 6:1781 - 1789
- Stenberg P, Pettersson F, Saura A, Berglund A, Larsson J. Sequence signature analysis of chromosome identity in three Drosophila species. BMC Bioinformatics 2005; 6:158 - 1
- Kelley RL, Meller VH, Gordadze PR, Roman G, Davis RL, Kuroda MI. Epigenetic spreading of the Drosophila dosage compensation complex from roX RNA genes into flanking chromatin. Cell 1999; 98:513 - 522
- Gilfillan GD, Koenig C, Dahlsveen IK, Prakoura N, Straub T, Lamm R, et al. Cumulative contributions of weak DNA determinants to targeting the Drosophila dosage compensation complex. Nucleic Acids Res 2007; 35:3561 - 3572
- Gallach M, Arnau V, Aldecoa R, Marin I. A sequence motif enriched in regions bound by the Drosophila dosage compensation complex. BMC Genomics 2010; 11:169 - 1
- Gallach M, Arnau V, Marin I. Global patterns of sequence evolution in Drosophila. BMC Genomics 2007; 8:408
- Shilatifard A. Chromatin modifications by methylation and ubiquitination: Implications in the regulation of gene expression. Ann Rev Biochem 2006; 75:243 - 269
- Anamika K, Krebs AR, Thompson J, Poch O, Devys D, Tora L. Lessons from genome-wide studies: an integrated definition of the coactivator function of histone acetyl transferases. Epigenetics Chromatin 2010; 3:18
- Carrozza MJ, Utley RT, Workman JL, Cote J. The diverse functions of histone acetyltransferase complexes. Trends Genet 2003; 19:321 - 329
- Verdone L, Agricola E, Caserta M, Di Mauro E. Histone acetylation in gene regulation. Brief Funct Genomic Proteomic 2006; 5:209 - 221
- Akhtar A, Becker PB. Activation of transcription through histone H4 acetylation by MOF, an acetyltransferase essential for dosage compensation in Drosophila. Mol Cell 2000; 5:367 - 375
- Bone JR, Kuroda MI. Dosage compensation regulatory proteins and the evolution of sex chromosomes in Drosophila. Genetics 1996; 144:705 - 713
- Hilfiker A, Hilfiker-Kleiner D, Pannuti A, Lucchesi JC. mof, a putative acetyl transferase gene related to the Tip60 and MOZ human genes and to the SAS genes of yeast, is required for dosage compensation in Drosophila. EMBO J 1997; 16:2054 - 2060
- Bell O, Schwaiger M, Oakeley EJ, Lienert F, Beisel C, Stadler MB, et al. Accessibility of the Drosophila genome discriminates PcG repression, H4K16 acetylation and replication timing. Nat Struct Mol Biol 2010; 17:894 - 900
- Shogren-Knaak M, Ishii H, Sun JM, Pazin MJ, Davie JR, Peterson CL. Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science 2006; 311:844 - 847
- Jin Y, Wang Y, Johansen Jr, Johansen KM. Jil-1, a Chromosomal kinase implicated in regulation of chromatin structure, associates with the male specific lethal (Msl) dosage compensation complex. J Cell Biol 2000; 149:1005 - 1010
- Jenuwein T, Allis CD. Translating the Histone Code. Science 2001; 293:1074 - 1080
- Thomas T, Dixon MP, Kueh AJ, Voss AK. Mof (MYST1 or KAT8) is essential for progression of embryonic development past the blastocyst stage and required for normal chromatin architecture. Mol Cell Biol 2008; 28:5093 - 5105
- Gupta A, Guerin-Peyrou TG, Sharma GG, Park C, Agarwal M, Ganju RK, et al. The mammalian ortholog of Drosophila MOF that acetylates histone H4 lysine 16 is essential for embryogenesis and oncogenesis. Mol Cell Biol 2008; 28:397 - 409
- Li X, Corsa CA, Pan PW, Wu L, Ferguson D, Yu X, et al. MOF and H4 K16 acetylation play important roles in DNA damage repair by modulating recruitment of DNA damage repair protein Mdc1. Mol Cell Biol 2010; 30:5335 - 5347
- Sharma GG, So S, Gupta A, Kumar R, Cayrou C, Avvakumov N, et al. MOF and histone H4 acetylation at lysine 16 are critical for DNA damage response and double-strand break repair. Mol Cell Biol 2010; 30:3582 - 3595
- Taipale M, Rea S, Richter K, Vilar A, Lichter P, Imhof A, et al. hMOF histone acetyltransferase is required for histone H4 lysine 16 acetylation in mammalian cells. Mol Cell Biol 2005; 25:6798 - 6810
- Gupta A, Sharma GG, Young CS, Agarwal M, Smith ER, Paull TT, et al. Involvement of human MOF in ATM function. Mol Cell Biol 2005; 25:5292 - 5305
- Smith ER, Cayrou C, Huang R, Lane WS, Cĉté J, Lucchesi J. A human protein complex homologous to the Drosophila MSL complex is responsible for the majority of histone H4 acetylation at lysine 16. Mol Cell Biol 2005; 25:9175 - 9188
- Bell O, Conrad T, Kind J, Wirbelauer C, Akhtar A, Schübeler D. Transcription-coupled methylation of histone H3 at lysine 36 regulates dosage compensation by enhancing recruitment of the MSL complex in Drosophila melanogaster. Mol Cell Biol 2008; 28:3401 - 3409
- Larschan E, Alekseyenko AA, Gortchakov AA, Peng S, Li B, Yang P, et al. MSL complex is attracted to genes marked by H3K36 trimethylation using a sequence-independent mechanism. Molecular Cell 2007; 28:121 - 133
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, et al. High-resolution profiling of histone methylations in the human genome. Cell 2007; 129:823 - 837
- Pokholok DK, Harbison CT, Levine S, Cole M, Hannett NM, Lee TI, et al. Genome-wide map of nucleosome acetylation and methylation in yeast. Cell 2005; 122:517 - 527
- Kim D, Blus BJ, Chandra V, Huang P, Rastinejad F, Khorasanizadeh S. Corecognition of DNA and a methylated histone tail by the MSL3 chromodomain. Nat Struct Mol Biol 2010; 17:1027 - 1029
- Moore SA, Ferhatoglu Y, Jia Y, Al-Jiab RA, Scott MJ. Structural and biochemical studies on the chromobarrel domain of male specific lethal 3 (MSL3) reveal a binding preference for mono or dimethyl lysine 20 on histone H4. J Biol Chem 2010; 285:40879 - 40890
- Deng H, Zhang W, Bao X, Martin J, Girton J, Johansen J, et al. The JIL-1 kinase regulates the structure of Drosophila polytene chromosomes. Chromosoma 2005; 114:173 - 182
- Jin Y, Wang Y, Walker DL, Dong H, Conley C, Johansen J, et al. JIL-1: a novel chromosomal tandem kinase implicated in transcriptional regulation in Drosophila. Molecular Cell 1999; 4:129 - 135
- Wang Y, Zhang W, Jin Y, Johansen Jr, Johansen KM. The JIL-1 tandem kinase mediates histone H3 phosphorylation and is required for maintenance of chromatin structure in Drosophila. Cell 2001; 105:433 - 443
- Zhang W, Jin Y, Ji Y, Girton J, Johansen J, Johansen KM. Genetic and phenotypic analysis of alleles of the Drosophila chromosomal JIL-1 kinase reveals a functional requirement at multiple developmental stages. Genetics 2003; 165:1341 - 1354
- Ivaldi MS, Karam CS, Corces VG. Phosphorylation of histone H3 at Ser10 facilitates RNA polymerase II release from promoter-proximal pausing in Drosophila. Genes Dev 2007; 21:2818 - 2831
- Zippo A, Serafini R, Rocchigiani M, Pennacchini S, Krepelova A, Oliviero S. Histone crosstalk between H3S10ph and H4K16ac generates a histone code that mediates transcription elongation. Cell 2009; 138:1122 - 1136
- Karam CS, Kellner WA, Takenaka N, Clemmons AW, Corces VG. 14-3-3 mediates histone cross-talk during transcription elongation in Drosophila. PLoS Genet 2010; 6:1000975 - 1
- Winkler GS, Kristjuhan A, Erdjument-Bromage H, Tempst P, Svejstrup JQ. Elongator is a histone H3 and H4 acetyltransferase important for normal histone acetylation levels in vivo. Proc Natl Acad Sci USA 2002; 99:3517 - 3522
- Cai W, Bao X, Deng H, Jin Y, Girton J, Johansen J, et al. RNA polymerase II-mediated transcription at active loci does not require histone H3S10 phosphorylation in Drosophila. Development 2008; 135:2917 - 2925
- Ciurciu A, Komonyi O, Boros IM. Loss of ATAC-specific acetylation of histone H4 at Lys12 reduces binding of JIL-1 to chromatin and phosphorylation of histone H3 at Ser10. J Cell Sci 2008; 121:3366 - 3372
- Deng H, Bao X, Cai W, Blacketer MJ, Belmont AS, Girton J, et al. Ectopic histone H3S10 phosphorylation causes chromatin structure remodeling in Drosophila. Development 2008; 135:699 - 705
- Zhang W, Deng H, Bao X, Lerach S, Girton J, Johansen Jr, et al. The JIL-1 histone H3S10 kinase regulates dimethyl H3K9 modifications and heterochromatic spreading in Drosophila. Development 2006; 133:229 - 235
- Deng H, Cai W, Wang C, Lerach S, Delattre M, Girton J, et al. JIL-1 and Su(var)3-7 interact genetically and counteract each other's effect on position-effect variegation in Drosophila. Genetics 2010; 185:1183 - 1192
- Schulze SR, Wallrath LL. Gene regulation by chromatin structure: Paradigms established in Drosophila melanogaster. Ann Rev Entomol 2006; 52:171 - 192
- Mito Y, Henikoff JG, Henikoff S. Genome-scale profiling of histone H3.3 replacement patterns. Nat Genet 2005; 37:1090 - 1097
- Furuhashi H, Nakajima M, Hirose S. DNA super-coiling factor contributes to dosage compensation in Drosophila. Development 2006; 133:4475 - 4483
- Deuring R, Fanti L, Armstrong JA, Sarte M, Papoulas O, Prestel M, et al. The ISWI chromatin-remodeling protein is required for gene expression and the maintenance of higher order chromatin structure in vivo. Mol Cell 2000; 5:355 - 365
- Bai X, Larschan E, Kwon SY, Badenhorst P, Kuroda MI. Regional control of chromatin organization by noncoding roX RNAs and the NURF remodeling complex in Drosophila melanogaster. Genetics 2007; 176:1491 - 1499
- Spierer A, Seum C, Delattre M, Spierer P. Loss of the modifiers of variegation Su(var)3-7 or HP1 impacts male X polytene chromosome morphology and dosage compensation. J Cell Sci 2005; 118:5047 - 5057
- Patalano S, Mihailovich M, Belacortu Y, Paricio N, Gebauer F. Dual sex-specific functions of Drosophila Upstream of N-ras in the control of X chromosome dosage compensation. Development 2009; 136:689 - 698
- Spierer A, Begeot F, Spierer P, Delattre M. SU(VAR)3-7 Links Heterochromatin and Dosage Compensation in Drosophila. PLoS Genet 2008; 4:1000066
- Li Q, Barkess Gi, Qian H. Chromatin looping and the probability of transcription. Trends Genet 2006; 22:197 - 202
- Gunesdogan U, Jackle H. Herzig A. A genetic system to assess in vivo the functions of histones and histone modifications in higher eukaryotes. EMBO Rep 2010; 11:772 - 776
- Goetze S, Mateos-Langerak J, van Driel R. Three-dimensional genome organization in interphase and its relation to genome function. Semin Cell Dev Biol 2007; 18:707 - 714
- Koehler A, Hurt E. Gene Regulation by nucleoporins and links to cancer. Mol Cell 2010; 38:6 - 15
- Bolzer A, Kreth G, Solovei I, Koehler D, Saracoglu K, Fauth C, et al. Three-dimensional maps of all chromosomes in human male fibroblast nuclei and prometaphase rosettes. PLoS Biol 2005; 3:157
- Cremer T, Cremer C. Chromosome territories, nuclear architecture and gene regulation in mammalian cells. Nat Rev Genet 2001; 2:292 - 301
- Brown KE, Baxter J, Graf D, Merkenschlager M, Fisher AG. Dynamic repositioning of genes in the nucleus of lymphocytes preparing for cell division. Mol Cell 1999; 3:207 - 217
- Chambeyron S, Bickmore WA. Does looping and clustering in the nucleus regulate gene expression?. Curr Op Cell Biol 2004; 16:256 - 262
- Grimaud C, Becker PB. Form and function of dosage-compensated chromosomes—a chicken-and-egg relationship. Bioessays 2010; 32:709 - 717
- Volpi EV, Chevret E, Jones T, Vatcheva R, Williamson J, Beck S, et al. Large-scale chromatin organization of the major histocompatibility complex and other regions of human chromosome 6 and its response to interferon in interphase nuclei. J Cell Sci 2000; 113:1565 - 1576
- Akhtar A, Gasser SM. The nuclear envelope and transcriptional control. Nat Rev Genet 2007; 8:507 - 517
- Menon BB, Sarma NJ, Pasula S, Deminoff SJ, Willis KA, Barbara KE, et al. Reverse recruitment: The Nup84 nuclear pore subcomplex mediates Rap1/Gcr1/Gcr2 transcriptional activation. Proc Natl Acad Sci USA 2005; 102:5749 - 5754
- Sutherland H, Bickmore WA. Transcription factories: gene expression in unions?. Nat Rev Genet 2009; 10:457 - 466
- Misteli T. Concepts in nuclear architecture. Bioessays 2005; 27:477 - 487
- Papantonis A, Cook PR. Genome architecture and the role of transcription. Cur Opin Cell Biol 2010; 22:271 - 276
- Sarma NJ, Haley TM, Barbara KE, Buford TD, Willis KA, Santangelo GM. Glucose-responsive regulators of gene expression in Saccharomyces cerevisiae function at the nuclear periphery via a reverse recruitment mechanism. Genetics 2007; 175:1127 - 1135
- Dieppois G, Stutz F. Connecting the transcription site to the nuclear pore: a multi-tether process that regulates gene expression. J Cell Sci 2010; 123:1989 - 1999
- Carmody SR, Wente SR. mRNA nuclear export at a glance. J Cell Sci 2009; 122:1933 - 1937
- Lainé JP, Singh BN, Krishnamurthy S, Hampsey M. A physiological role for gene loops in yeast. Genes Dev 2009; 23:2604 - 2609
- Tan-Wong SM, Wijayatilake HD, Proudfoot NJ. Gene loops function to maintain transcriptional memory through interaction with the nuclear pore complex. Genes Dev 2009; 23:2610 - 2624
- Titus LC, Dawson TR, Rexer DJ, Ryan KJ, Wente SR. Members of the RSC chromatin-remodeling complex are required for maintaining proper nuclear envelope structure and pore complex localization. Mol Biol Cell 2010; 21:1072 - 1087
- Ahmed S, Brickner DG, Light WH, Cajigas I, McDonough M, Froyshteter AB, et al. DNA zip codes control an ancient mechanism for gene targeting to the nuclear periphery. Nat Cell Biol 2010; 12:111 - 118
- Kurshakova MM, Krasnov AN, Kopytova DV, Shidlovskii YV, Nikolenko JV, Nabirochkina EN, et al. SAGA and a novel Drosophila export complex anchor efficient transcription and mRNA export to NPC. EMBO J 2007; 26:4956 - 4965
- Brickner DG, Cajigas I, Fondufe-Mittendorf Y, Ahmed S, Lee PC, Widom J, et al. H2A.Z-mediated localization of genes at the nuclear periphery confers epigenetic memory of previous transcriptional state. PLoS Biol 2007; 5:81
- Dieppois G, Iglesias N, Stutz F. Cotranscriptional recruitment to the mRNA export receptor Mex67p contributes to nuclear pore anchoring of activated genes. Mol Cell Biol 2006; 26:7858 - 7870
- Hou C, Dale R, Dean A. Cell type specificity of chromatin organization mediated by CTCF and cohesin. Proc Natl Acad Sci USA 2010; 107:3651 - 3656
- Ohlsson R, Lobanenkov V, Klenova E. Does CTCF mediate between nuclear organization and gene expression?. Bioessays 2010; 32:37 - 50
- Casolari JM, Brown CR, Komili S, West J, Hieronymus H, Silver PA. Genome-wide localization of the nuclear transport machinery couples transcriptional status and nuclear organization. Cell 2004; 117:427 - 439
- Luthra R, Kerr SC, Harreman MT, Apponi LH, Fasken MB, Ramineni S, et al. Actively transcribed GAL genes can be physically linked to the nuclear pore by the SAGA chromatin modifying complex. J Biol Chem 2007; 282:3042 - 3049
- Schmid M, Arib G, Laemmli C, Nishikawa J, Durussel T, Laemmli UK. Nup-PI: the nucleopore-promoter interaction of genes in yeast. Mol Cell 2006; 21:379 - 391
- Rabut G, Lénárt P, Ellenberg J. Dynamics of nuclear pore complex organization through the cell cycle. Cur Op Cell Biol 2004; 16:314 - 321
- Zimowska G, Aris JP, Paddy MR. A Drosophila Tpr protein homolog is localized both in the extrachromosomal channel network and to nuclear pore complexes. J Cell Sci 1997; 110:927 - 944
- Zimowska G, Paddy MR. Structures and dynamics of Drosophila Tpr inconsistent with a static, filamentous structure. Exp Cell Res 2002; 276:223 - 232
- Capelson M, Liang Y, Schulte R, Mair W, Wagner U, Hetzer MW. Chromatin-bound nuclear pore components regulate gene expression in higher eukaryotes. Cell 2010; 140:372 - 383
- Kalverda B, Pickersgill H, Shloma VV, Fornerod M. Nucleoporins directly stimulate expression of developmental and cell cycle genes inside the nucleoplasm. Cell 2010; 140:360 - 371
- Vaquerizas JM, Suyama R, Kind J, Miura K, Luscombe NM, Akhtar A. Nuclear pore proteins nup153 and megator define transcriptionally active regions in the Drosophila genome. PLoS Genet 2010; 6:1000846
- Schoenfelder S, Sexton T, Chakalova L, Cope NF, Horton A, Andrews S, et al. Preferential associations between co-regulated genes reveal a transcriptional interactome in erythroid cells. Nat Genet 2010; 42:53 - 61
- Grimaud C, Becker PB. The dosage compensation complex shapes the conformation of the X chromosome in Drosophila. Genes Dev 2009; 23:2490 - 2495