Abstract
Enterohemorrhagic and enteropathogenic Escherichia coli (EHEC and EPEC) are enteric human pathogens that colonize the large and small intestines, respectively. To establish infection EHEC and EPEC must overcome innate host defenses, such as antimicrobial peptides (AMPs) produced by the intestinal epithelium. Gram-negative pathogens have evolved different mechanisms to resist AMPs, including outer-membrane proteases that degrade AMPs. We showed that the protease OmpT degrades the human AMP LL-37 more rapidly in EHEC than in EPEC. Promoter-swap experiments showed that this is due to differences in the promoters of the two genes, leading to greater ompT expression and subsequently greater levels of OmpT in EHEC. Here, we propose that the different ompT expression in EHEC and EPEC reflects the varying levels of LL-37 throughout the human intestinal tract. These data suggest that EHEC and EPEC adapted to their specific niches by developing distinct AMP-specific resistance mechanisms.
Keywords: :
Introduction
EHEC and EPEC are human diarrheal pathogens that cause characteristic attaching and effacing (A/E) lesions in the gut epithelium. As other A/E pathogens, EHEC and EPEC carry the Locus of Enterocyte Effacement (LEE) pathogenicity island that encodes various virulence factors, including a type III secretion system and effector proteins that are translocated into the epithelial cells to subvert host cell signaling and promote A/E lesion formation.Citation1 One of these effectors is the Translocated intimin receptor (Tir) that acts as a receptor for the outer-membrane (OM) adhesin intimin. This interaction is essential for intimate adhesion to intestinal epithelial cells. By adhering intimately to the intestinal mucosa, both EHEC and EPEC face the release of AMPs from epithelial cells. Although EHEC and EPEC have many virulence factors in common, they colonize distinct niches of the human gastrointestinal tract. EHEC colonizes the large intestine, whereas EPEC predominantly colonizes the proximal small intestine (duodenum and jejunum).Citation2
AMPs are important components of innate immune defenses and serve as endogenous antibiotics. AMPs are synthesized as inactive precursors that are processed into biologically active peptides by posttranslational proteolysis. Although mature AMPs are small (< 50 amino acid residues) and exhibit a cationic charge, they are diverse in amino acid sequence and adopt different three-dimensional structures. In mammals, there are two major groups of AMPs, the cathelicidins and the defensins. The number of cathelicidin genes varies between mammalian species. Humans have a single cathelicidin gene (CAMP) that is abundantly expressed in the skin, in neutrophils and in multiple mucosal epithelia. CAMP encodes hCAP18, an inactive precursor, which upon secretion is processed into the N-terminal cathelin prodomain and the C-terminal LL-37 active peptide. LL-37, which begins with 2 leucines and is 37 residues in length, adopts an amphipathic α-helical structure. In contrast to cathelicidins, there are numerous human defensin genes. Defensins are characterized by the presence of three intramolecular disulfide bonds. Based on the connectivity of these disulfide bonds, defensins are further divided into α- and β-defensins. In humans, six α-defensins have been identified.Citation3 Human neutrophil peptides 1–4 (HNP-1 through -4) are present in the azurophilic granules of neutrophils and human defensins 5 and 6 (HD-5 and -6) are secreted by Paneth cells of the small intestine. Human β-defensins 1–4 (HBD-1 through -4) are found predominantly in a variety of mucosal epithelia and tissues, including the colon.Citation4
The exact mechanisms of action of AMPs are still unclear. They have both direct bactericidal and immuno-modulatory functions. AMPs exert their bactericidal activity by interacting with the negatively charged bacterial membrane through electrostatic interactions and then disrupting the cytoplasmic membrane, which leads to bacterial cell lysis.Citation5 The in vivo bactericidal activity of AMPs has been questioned due to the facts that the AMP-mediated killing is salt sensitive and the AMP concentration at most sites is below the minimum inhibitory concentration (MIC). Nonetheless, it appears likely that the abundance of enteric α-defensins allows direct bacterial killing at sites close to the epithelium of the small intestine, where pathogens such as EPEC adhere intimately, but not necessarily in the lumen that is colonized by the microbiota. More recently, many immuno-modulatory functions have been attributed to AMPs. By interacting with a variety of host cell receptors, AMPs influence neutrophil recruitment, cytokine release, antigen presentation, and angiogenesis and wound healing.Citation6-Citation8 Most likely, the anti-infective activity of AMPs results from both direct bacterial killing and indirect immuno-modulatory effects.
During the co-evolution of pathogens with the host immune system, bacterial pathogens have developed different strategies to resist innate immune defenses and survive the activity of AMPs. For example, bacterial pathogens produce capsule polysaccharides to shield their cell surface, they covalently modify their lipopolysaccharide or lipoteichoic acid to prevent AMP binding, they downregulate expression of AMPs by host cells and they produce proteases that degrade and inactivate AMPs.Citation9 These bacterial proteases are either secreted or localized at the OM of Gram-negative pathogens. OM proteases of the omptin family are present in many pathogens of the Enterobacteriaceae family.Citation10-Citation12 Omptin genes are present on mobile elements such as plasmids or cryptic prophages. Several studies have reported the critical role of omptins in the degradation of AMPs. Initially, the E. coli K12 omptin (OmpT) has been shown to cleave protamine.Citation13 More recently, EHEC OmpT was shown to cleave LL-37 at dibasic motifs present in the primary amino acid sequence, and the resulting proteolytic fragments displayed no bactericidal activity.Citation14 In addition, the CroP omptin of the murine A/E pathogen, Citrobacter rodentium, was shown to degrade murine cathelicidin-related antimicrobial peptide (mCRAMP).Citation15 In addition to inactivation of AMPs, omptins were shown to affect the host complement, coagulation and fibrinolysis systems. For example, the Pla omptin promotes dissemination of Yersinia pestis from the primary site of infection to lymph nodes by cleaving plasminogen into active plasmin.Citation16
Differential Expression of the EHEC and EPEC ompT Genes
We recently described differential expression of the ompT genes in EHEC and EPEC. OmpT is expressed at high levels at the OM of EHEC, but it is expressed at a much lower level in EPEC.Citation14 By using a promoter-swapping strategy, we demonstrated that differential expression of EHEC and EPEC ompT is strictly dependent on their respective promoters.Citation14 The EHEC and EPEC ompT genes are part of distinct cryptic prophages that have been inserted at different locations of the EHEC EDL933 (inserted at 1.75 Mbp) and EPEC E2348/69 (inserted at 0.48 Mbp) genomes. The intergenic regions upstream of the EHEC and EPEC ompT genes, which encompass the promoter sequences, are 481 bp and 657 bp in length, respectively. This is much longer than the typical E. coli intergenic region, which is 118 bp in average.Citation17 Although the proximal promoter sequences (-1 to –150 bp) are highly conserved, distal promoter sequences (upstream of -150 bp to next open-reading frame) largely diverge and are particularly AT-rich. To better understand the role of the proximal promoter sequence, we expressed EHEC ompT from the strictly conserved promoter sequence (-1 to –150 bp). Surprisingly, this resulted in decreased ompT expression (unpublished data), suggesting that the largely divergent distal promoter sequence may contain binding sites for transcriptional activators. Overall, we have shown that transcriptional regulation of omptin genes differs not only between bacterial species but also between E. coli strains. It appears most likely that the EHEC and EPEC ompT promoters have complex regulation involving both small regulatory RNAsCitation18 and multiple transcriptional regulators. This conclusion is consistent with a previous study showing that the Shigella icsP gene, which encodes the IcsP omptin, has promoter elements located more than 1 kb upstream of the transcriptional start site.Citation19
EHEC and EPEC OmpT Have Inherently Similar Catalytic Activities
The lower expression of the EPEC ompT gene, compared with EHEC ompT, resulted in slower degradation of various substrates, including LL-37.Citation14 Degradation of LL-37 by wild-type EPEC was minimal, whereas LL-37 was readily degraded by wild-type EHEC, in an OmpT-dependent manner. Significant degradation of LL-37 by EPEC OmpT was only observed upon plasmid complementation of the EPEC ΔompT strain that resulted in the overexpression of the EPEC ompT gene. Since the EHEC and EPEC ompT open-reading frames contain 10 nucleotide mismatches that result in 5 amino acid mismatches, it was important to verify that EHEC and EPEC OmpT proteins have similar catalytic activities. To examine this point, we fused both open-reading frames to the promoter of croP, the C. rodentium ompT homologCitation15 and expressed these constructs in the C. rodentium ΔcroP mutant (). As shown in , both OmpT proteins were expressed at similar levels at the OM of C. rodentium. Both EHEC and EPEC OmpT degraded the synthetic FRET substrate and LL-37 at similar rates ( and ). These results clearly indicate that EHEC and EPEC OmpT have inherently similar catalytic activities despite the 5 amino acid mismatches.
Figure 1. EHEC and EPEC OmpT degrade peptides at similar rates (A) Schematic of EHEC and EPEC ompT expressed from the croP promoter. Plasmids pEHompT and pEPompT are derived from pWSK129 and contain the C. rodentium croP promoter fused to the ompT open-reading frames of EHEC or EPEC, respectively. (B) Amounts of EHEC and EPEC OmpT produced by C. rodentium ΔcroP cells transformed with pEHompT or pEPompT were analyzed by western blotting using an antiserum developed against the CroP OM protease of C. rodentium (74% identical to OmpT).Citation14 (C) Cleavage of the synthetic FRET substrate containing the dibasic sequence RK in its center [2Abz-SLGRKIQI-K(Dnp)-NH2] was monitored over time by measuring fluorescence emission at 430 nm. (D) Proteolytic degradation of LL-37 by C. rodentium ΔcroP cells expressing EHEC or EPEC ompT. LL-37 was incubated for 1 h with the various bacterial strains. Aliquots were separated by Tris-Tricine SDS-PAGE (10–20% acrylamide) and gels were stained with Coomassie blue G-250. The control lane, in which no bacteria were added, contained 1.5 µg LL-37 in phosphate-buffered saline.
![Figure 1. EHEC and EPEC OmpT degrade peptides at similar rates (A) Schematic of EHEC and EPEC ompT expressed from the croP promoter. Plasmids pEHompT and pEPompT are derived from pWSK129 and contain the C. rodentium croP promoter fused to the ompT open-reading frames of EHEC or EPEC, respectively. (B) Amounts of EHEC and EPEC OmpT produced by C. rodentium ΔcroP cells transformed with pEHompT or pEPompT were analyzed by western blotting using an antiserum developed against the CroP OM protease of C. rodentium (74% identical to OmpT).Citation14 (C) Cleavage of the synthetic FRET substrate containing the dibasic sequence RK in its center [2Abz-SLGRKIQI-K(Dnp)-NH2] was monitored over time by measuring fluorescence emission at 430 nm. (D) Proteolytic degradation of LL-37 by C. rodentium ΔcroP cells expressing EHEC or EPEC ompT. LL-37 was incubated for 1 h with the various bacterial strains. Aliquots were separated by Tris-Tricine SDS-PAGE (10–20% acrylamide) and gels were stained with Coomassie blue G-250. The control lane, in which no bacteria were added, contained 1.5 µg LL-37 in phosphate-buffered saline.](/cms/asset/04d01263-827b-4280-a082-17cdf4498786/kgmi_a_10921656_f0001.gif)
OmpT Protects EHEC from AMP-Mediated Membrane Disruption
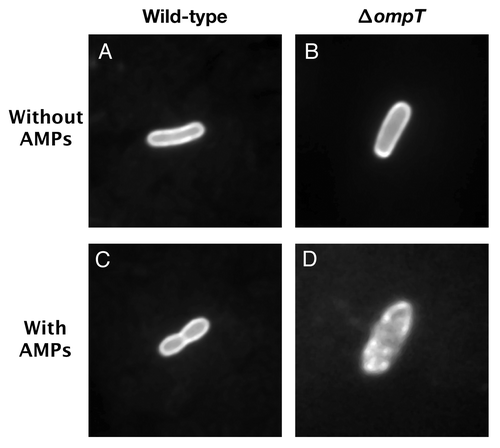
A fluorescence microscopy approach was developed to visualize the protective effect provided by OmpT against AMPs. Wild-type and ΔompT EHEC strains were transformed with plasmid pCAST-ChFP expressing the mCherry fluorescent protein (ChFP) fused to an OM targeting signal.Citation20 Cells were then incubated in the presence or absence of the synthetic α-helical AMP C18G, which was previously shown to be readily degraded by EHEC OmpT.Citation14,Citation21 In the absence of AMP, both the wild-type and ΔompT EHEC cells exhibited intense fluorescence outlines with uniform staining of the OM, indicating that ChFP localizes to the cell envelope (). Following exposure to the AMP C18G, the wild-type EHEC cells showed uniform membrane staining patterns, similar to untreated cells (). In contrast the ΔompT EHEC strain showed dramatic membrane perturbations characterized by regions of patchy fluorescence (). Altogether, these experiments clearly demonstrate that the presence of OmpT at the OM of EHEC cells protects the cell envelope from AMP-induced disruption.
Figure 2. OmpT protects EHEC from C18G-induced membrane disruption. EHEC wild-type and ΔompT strains were transformed with plasmid pCAST–ChFP encoding the OM localized red fluorescent protein mCherry.Citation20 Bacterial cultures (1ml) grown to mid-log phase were resuspended in 50 µl N-minimal medium and incubated for 15 min at room temperature in the absence or presence of the synthetic α-helical AMP C18G (100 µM). Bacterial cells (3 µl) were spotted on agarose-coated glass slides and analyzed under oil immersion at 100 magnification with a Zeiss Axiovert 200 M fluorescence microscope. (A) EHEC wild-type cell in the absence of C18G. (B) EHEC ΔompT cell in the absence of C18G. (C) EHEC wild-type cell in the presence of C18G. (D) EHEC ΔompT cell in the presence of C18G.
AMPs Encountered by EHEC and EPEC
In the human small intestine, the niche colonized by EPEC, the α-defensins HD-5 and HD-6 are the most abundant AMPs. They are expressed and secreted in high amounts by Paneth cells located at the bottom of the intestinal crypts.Citation22 The cathelicidin LL-37 and β-defensins are only present in trace amounts in the small intestine.Citation23 In contrast, LL-37 and β-defensins are the main AMPs produced by the colonic epithelium in the large intestine, which is colonized by EHEC. Whereas the hBD-1 gene is constitutively expressed by the colonic mucosa, expression of other β-defensin genes is only induced in response to infectious or proinflammatory stimuli. Thus, EHEC and EPEC appear to encounter different sets of AMPs by colonizing their respective niches.
α-helical AMPs such as LL-37 or C18G are known to be highly susceptible to protease-mediated degradation. The fact that human LL-37 is relatively abundant in the large intestine is in good agreement with the high expression of OmpT by EHEC cells. Conversely, the presence of trace amounts of LL-37 in the small intestine is consistent with the low expression of OmpT by EPEC. Altogether, these considerations suggest that both bacterial species evolved resistance to LL-37 in a niche-dependent manner. However, it still remains unclear to what extent OmpT cleaves α- and β-defensins.
OmpT Only Degrades Reduced HNP-1
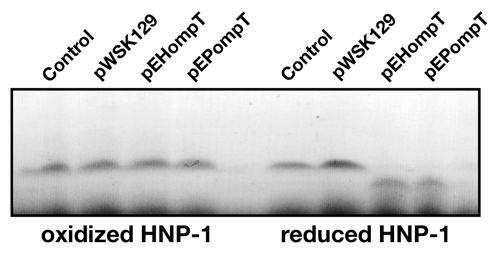
Mature defensins, which are stabilized by three disulfide bonds, have been shown to be highly resistant to proteolysis by host proteases such as MMP-7, trypsin and neutrophil elastase. Nonetheless, several proteases of host or bacterial origin were reported to degrade and inactivate defensins. For example, the host cathepsins B, L and S were shown to inactivate hBD-2 and hBD-3.Citation24 In addition, the cysteine proteases, known as gingipains, produced by Porphyromonas gingivalis degrade hBD-3.Citation25 The Proteus mirabilis ZapA and Burkholderia cenocepacia ZmpB zinc-dependent metalloproteases degraded hBD-1.Citation26,Citation27 Most mature defensins possess dibasic motifs in their primary sequences, raising the possibility that they may be recognized and cleaved by bacterial omptins such as OmpT. To assess the possibility that OmpT cleaves defensins, HNP-1 in its oxidized form was incubated for 1 h with the C. rodentium ΔcroP strains producing EHEC or EPEC OmpT. As shown in , no cleavage was observed, indicating that oxidized HNP-1 is resistant to OmpT-mediated proteolysis. To determine whether the disulfide bonds of HNP-1 were responsible for proteolysis resistance, HNP-1 was reduced by incubation with DTT and free cysteines were then blocked with iodoacetamide, prior to incubation with the strains expressing EHEC or EPEC OmpT. Most of reduced HNP-1 was cleaved in an OmpT-dependent manner (). These results indicate that OmpT can cleave reduced HNP-1, which possesses the dibasic motif RR in its amino acid sequence. They also suggest that the presence of disulfide bonds protects HNP-1 from degradation, possibly by hiding the RR motif in the protein core and preventing recognition by OmpT. Lastly, the activity of OmpT against other oxidized and reduced defensins remains to be tested.
Figure 3. Cleavage of reduced HNP-1 by OmpT. HNP-1 was purchased from Peptides international Inc. (Louisville, KT). HNP-1 was reduced by incubation at 50°C for 30 min with dithiothreitol (1 mM). Free cysteines were then blocked with iodoacetamide (17 mM) for 30 min prior to incubation with bacterial strains. Oxidized or reduced HNP-1 was incubated for 1 h with the C. rodentium ΔcroP strain containing the empty plasmid pWSK129 or plasmid pWSK129 expressing EHEC OmpT (pEHompT) or EPEC OmpT (pEPompT). Bacterial cells were pelleted by centrifugation and supernatants were resolved on Tris-Tricine SDS-PAGE (10–20% acrylamide). Gels were stained with Coomassie blue G-250. Control lanes, in which no bacteria were added, contained 1 µg of oxidized or reduced HNP-1 in phosphate-buffered saline.
Physiological Relevance of Reduced Defensins
Compared with other β-defensins, hBD-1 has specific features. It is constitutively expressed by epithelial cells throughout the body and has weak bactericidal activity. In a recent study, Schroder et al.,Citation28 found that reduction of the three disulfides of hBD-1 greatly enhances its antimicrobial activity against some microorganisms such as Gram-positive anaerobic bacteria and the pathogenic fungus Candida albicans, but not E. coli K12. The authors also showed co-localization of reduced hBD-1 and thioredoxin reductase in human colonic mucosa, suggesting that the thioredoxin system catalyzes reduction of HBD-1. Although it is still unclear whether these results are also true for other defensins, these new findings suggest that reduced defensins may have physiological functions not exhibited by oxidized defensins. In the light of the degradation of reduced HNP-1 by EHEC and EPEC OmpT (), it seems possible that degradation of reduced defensins by bacterial omptins is a physiologically relevant phenomenon. It would thus be interesting to test whether cleavage of reduced hBD-1 by OmpT plays a role in the lack of enhanced antimicrobial activity of this peptide against E. coli K12.
Concluding Remarks
By colonizing different niches of the human gastrointestinal tract, EHEC and EPEC are exposed to different host AMPs. Here we propose that EHEC and EPEC have adapted to these differences by evolving slightly different mechanisms to resist the bactericidal effects of specific AMPs. EHEC likely developed high ompT expression as a means to survive significant amounts of LL-37 in the large intestine. The mechanism by which EHEC resists to β-defensins in the large intestine remains to be identified. In contrast, the low ompT expression in EPEC is adequate to subdue the trace amounts of LL-37 in the small intestine. However, to resist α-defensins that are prevalent in the small intestine, EPEC is likely to have evolved other mechanisms that are not necessarily present in EHEC. These mechanisms remain to be characterized.
| Abbreviations: | ||
| AMP | = | antimicrobial peptide |
| EHEC | = | enterohemorrhagic Escherichia coli |
| EPEC | = | enteropathogenic Escherichia coli |
Acknowledgments
This work was supported by the Canadian Institutes of Health Research (CIHR, MOP-15551) and the Natural Sciences and Engineering Research Council (NSERC, 217482). S.G. is supported by a Canada Research Chair. We would like to thank Dr Shawn Lewenza for providing plasmid pCAST-ChFP.
References
- Wong AR, Pearson JS, Bright MD, Munera D, Robinson KS, Lee SF, et al. Enteropathogenic and enterohaemorrhagic Escherichia coli: even more subversive elements. Mol Microbiol 2011; 80:1420 - 38; http://dx.doi.org/10.1111/j.1365-2958.2011.07661.x; PMID: 21488979
- Nataro JP, Kaper JB. Diarrheagenic Escherichia coli.. Clin Microbiol Rev 1998; 11:142 - 201; PMID: 9457432
- Lehrer RI, Lu W. α-Defensins in human innate immunity. Immunol Rev 2012; 245:84 - 112; http://dx.doi.org/10.1111/j.1600-065X.2011.01082.x; PMID: 22168415
- Selsted ME, Ouellette AJ. Mammalian defensins in the antimicrobial immune response. Nat Immunol 2005; 6:551 - 7; http://dx.doi.org/10.1038/ni1206; PMID: 15908936
- Brogden KA. Antimicrobial peptides: pore formers or metabolic inhibitors in bacteria?. Nat Rev Microbiol 2005; 3:238 - 50; http://dx.doi.org/10.1038/nrmicro1098; PMID: 15703760
- Bowdish DM, Davidson DJ, Hancock RE. Immunomodulatory properties of defensins and cathelicidins. Curr Top Microbiol Immunol 2006; 306:27 - 66; http://dx.doi.org/10.1007/3-540-29916-5_2; PMID: 16909917
- Menendez A, Brett Finlay B. Defensins in the immunology of bacterial infections. Curr Opin Immunol 2007; 19:385 - 91; http://dx.doi.org/10.1016/j.coi.2007.06.008; PMID: 17702560
- Nijnik A, Hancock RE. The roles of cathelicidin LL-37 in immune defences and novel clinical applications. Curr Opin Hematol 2009; 16:41 - 7; http://dx.doi.org/10.1097/MOH.0b013e32831ac517; PMID: 19068548
- Gruenheid S, Le Moual H. Resistance to antimicrobial peptides in Gram-negative bacteria. FEMS Microbiol Lett 2012; 330:81 - 9; http://dx.doi.org/10.1111/j.1574-6968.2012.02528.x; PMID: 22339775
- Kukkonen M, Korhonen TK. The omptin family of enterobacterial surface proteases/adhesins: from housekeeping in Escherichia coli to systemic spread of Yersinia pestis.. Int J Med Microbiol 2004; 294:7 - 14; http://dx.doi.org/10.1016/j.ijmm.2004.01.003; PMID: 15293449
- Hritonenko V, Stathopoulos C. Omptin proteins: an expanding family of outer membrane proteases in Gram-negative Enterobacteriaceae.. Mol Membr Biol 2007; 24:395 - 406; http://dx.doi.org/10.1080/09687680701443822; PMID: 17710644
- Haiko J, Suomalainen M, Ojala T, Lähteenmäki K, Korhonen TK. Invited review: Breaking barriers--attack on innate immune defences by omptin surface proteases of enterobacterial pathogens. Innate Immun 2009; 15:67 - 80; http://dx.doi.org/10.1177/1753425909102559; PMID: 19318417
- Stumpe S, Schmid R, Stephens DL, Georgiou G, Bakker EP. Identification of OmpT as the protease that hydrolyzes the antimicrobial peptide protamine before it enters growing cells of Escherichia coli.. J Bacteriol 1998; 180:4002 - 6; PMID: 9683502
- Thomassin JL, Brannon JR, Gibbs BF, Gruenheid S, Le Moual H. OmpT outer membrane proteases of enterohemorrhagic and enteropathogenic Escherichia coli contribute differently to the degradation of human LL-37. Infect Immun 2012; 80:483 - 92; http://dx.doi.org/10.1128/IAI.05674-11; PMID: 22144482
- Le Sage V, Zhu L, Lepage C, Portt A, Viau C, Daigle F, et al. An outer membrane protease of the omptin family prevents activation of the Citrobacter rodentium PhoPQ two-component system by antimicrobial peptides. Mol Microbiol 2009; 74:98 - 111; http://dx.doi.org/10.1111/j.1365-2958.2009.06854.x; PMID: 19708916
- Sodeinde OA, Subrahmanyam YV, Stark K, Quan T, Bao Y, Goguen JD. A surface protease and the invasive character of plague. Science 1992; 258:1004 - 7; http://dx.doi.org/10.1126/science.1439793; PMID: 1439793
- Blattner FR, Plunkett G 3rd, Bloch CA, Perna NT, Burland V, Riley M, et al. The complete genome sequence of Escherichia coli K-12. Science 1997; 277:1453 - 62; http://dx.doi.org/10.1126/science.277.5331.1453; PMID: 9278503
- Guillier M, Gottesman S. Remodelling of the Escherichia coli outer membrane by two small regulatory RNAs. Mol Microbiol 2006; 59:231 - 47; http://dx.doi.org/10.1111/j.1365-2958.2005.04929.x; PMID: 16359331
- Castellanos MI, Harrison DJ, Smith JM, Labahn SK, Levy KM, Wing HJ. VirB alleviates H-NS repression of the icsP promoter in Shigella flexneri from sites more than one kilobase upstream of the transcription start site. J Bacteriol 2009; 191:4047 - 50; http://dx.doi.org/10.1128/JB.00313-09; PMID: 19363111
- Lewenza S, Mhlanga MM, Pugsley AP. Novel inner membrane retention signals in Pseudomonas aeruginosa lipoproteins. J Bacteriol 2008; 190:6119 - 25; http://dx.doi.org/10.1128/JB.00603-08; PMID: 18641140
- Darveau RP, Blake J, Seachord CL, Cosand WL, Cunningham MD, Cassiano-Clough L, et al. Peptides related to the carboxyl terminus of human platelet factor IV with antibacterial activity. J Clin Invest 1992; 90:447 - 55; http://dx.doi.org/10.1172/JCI115880; PMID: 1644916
- Bevins CL, Salzman NH. Paneth cells, antimicrobial peptides and maintenance of intestinal homeostasis. Nat Rev Microbiol 2011; 9:356 - 68; http://dx.doi.org/10.1038/nrmicro2546; PMID: 21423246
- Hase K, Eckmann L, Leopard JD, Varki N, Kagnoff MF. Cell differentiation is a key determinant of cathelicidin LL-37/human cationic antimicrobial protein 18 expression by human colon epithelium. Infect Immun 2002; 70:953 - 63; http://dx.doi.org/10.1128/IAI.70.2.953-963.2002; PMID: 11796631
- Taggart CC, Greene CM, Smith SG, Levine RL, McCray PB Jr., O’Neill S, et al. Inactivation of human beta-defensins 2 and 3 by elastolytic cathepsins. J Immunol 2003; 171:931 - 7; PMID: 12847264
- Maisetta G, Brancatisano FL, Esin S, Campa M, Batoni G. Gingipains produced by Porphyromonas gingivalis ATCC49417 degrade human-β-defensin 3 and affect peptide’s antibacterial activity in vitro. Peptides 2011; 32:1073 - 7; http://dx.doi.org/10.1016/j.peptides.2011.02.003; PMID: 21335044
- Belas R, Manos J, Suvanasuthi R. Proteus mirabilis ZapA metalloprotease degrades a broad spectrum of substrates, including antimicrobial peptides. Infect Immun 2004; 72:5159 - 67; http://dx.doi.org/10.1128/IAI.72.9.5159-5167.2004; PMID: 15322010
- Kooi C, Sokol PA. Burkholderia cenocepacia zinc metalloproteases influence resistance to antimicrobial peptides. Microbiology 2009; 155:2818 - 25; http://dx.doi.org/10.1099/mic.0.028969-0; PMID: 19542010
- Schroeder BO, Wu Z, Nuding S, Groscurth S, Marcinowski M, Beisner J, et al. Reduction of disulphide bonds unmasks potent antimicrobial activity of human β-defensin 1. Nature 2011; 469:419 - 23; http://dx.doi.org/10.1038/nature09674; PMID: 21248850