Abstract
The appearance of new viruses and the cost of developing certain vaccines require that new vaccination strategies now have to be developed. DNA vaccination seems to be a particularly promising method. For this application, plasmid DNA is injected into the subject (man or animal). This plasmid DNA encodes an antigen that will be expressed by the cells of the subject. In addition to the antigen, the plasmid also encodes a resistance to an antibiotic, which is used during the construction and production steps of the plasmid. However, regulatory agencies (FDA, USDA and EMA) recommend to avoid the use of antibiotics resistance genes. Delphi Genetics developed the Staby® technology to replace the antibiotic-resistance gene by a selection system that relies on two bacterial genes. These genes are small in size (approximately 200 to 300 bases each) and consequently encode two small proteins. They are naturally present in the genomes of bacteria and on plasmids. The technology is already used successfully for production of recombinant proteins to achieve higher yields and without the need of antibiotics. In the field of DNA vaccines, we have now the first data validating the innocuousness of this Staby® technology for eukaryotic cells and the feasibility of an industrial production of an antibiotic-free DNA vaccine. Moreover, as a proof of concept, mice have been successfully vaccinated with our antibiotic-free DNA vaccine against a deadly disease, pseudorabies (induced by Suid herpesvirus-1).
Introduction
Today vaccination is an uncontested way of fighting disease. It has enabled the control of several diseases, including diphtheria, tetanus, poliomyelitis and mumps (at least in certain parts of the world). However, new viruses are appearing, with the characteristic of being able to mutate their genetic composition quickly (AIDS, SARS, Avian Flu H5N1, Swine Flu H1N1, and so on and so forth). In addition, the cost of developing vaccines precludes the targeting of all diseases. New vaccination strategies are therefore necessary in order to enable a response that is prompt and more appropriate than the current methods. DNA vaccination would seem to be one of the particularly promising methods at this time.
In the Eighties, Dubensky et al. reported for the first time that an in vivo DNA injection enabled a production of insulin.Citation1 Then, in the Nineties the injection of purified plasmid DNA into muscles of mice enabled detectable markers such as β-galactosidase.Citation2 The use of such methods was quickly envisaged for the purpose of accelerating the development of new vaccines. However, this method then lost its interest because of its low effectiveness in tests on large primates even though it had proved to be very effective in treating smaller animals.Citation3 In the course of the last ten years, great attention has been paid to the DNA delivery methods. It is indeed this stage that seemed to be limiting. Several avenues have been explored and have enabled new DNA delivery methods to be developed, one of which was electroporation, which seems currently to be the best injection method available.Citation4,Citation5 In vivo electrotransfer involves plasmid injection and application of high voltage pulses that, on one hand, transitorily disturb membranes and thus increase cells permeability and, on the other hand, promote electrophoresis of negatively charged DNA.Citation6 This new injection method has rekindled interest in DNA vaccination, especially considering its multiple advantages compared with the production of protein antigens: stability of the DNA (easier transport and storage), identical production process for all vaccines (DNA production), no problems relating to post-translational modifications (glycosylation), shortened development and production time when combating a pandemic, easy vaccine adaptation to a new serotype variant and “design” facility of multivalent vaccines (the presence of several genes is not a problem, unlike the presence of several different proteins with different biochemical characteristics).
Moreover, the use of plasmid DNA is regarded as safe in terms of integration and autoimmune reaction.Citation7-Citation9 Several clinical trials for human vaccines are in progress (about a hundred in all) but none of them has yet been approved and marketed. On the other hand, to our knowledge, there are three DNA vaccines in existence that have been approved for animal use and several others are in progress. The first on the market was a vaccine against Egyptian horse fever marketed in 2009 (West Nile virus, Fort Dodge Animal Health). The two others are vaccines for salmon (Aqua Health Ltd) and for dogs (vaccine against melanoma, Merial).
Presently, the resistant gene is used as a selection marker for the construction of the productive strain and the production of the plasmid DNA. However, it is recommended by regulatory agencies (FDA, USDA, EMA) for more than 15 years to avoid the use of antibiotics as selection marker (EMA document, reference CPMP/BWP/3088/99, 2001 and EMEA/CHMP/GTWP/65260/2008, 2008). These recommendations are understandable not only because of the risks of allergic reactions that antibiotics resistances represent, but also because of the risk of selection of antibiotics resistant pathogenic bacteria. In the case of DNA vaccine, the refusal of antibiotic resistance gene use is completely relevant since this resistance gene forms an integral part of the product injected into the subject. We propose to replace this resistance to an antibiotic by the Staby® technology based on natural poison/antidote bacterial genes to insure the plasmid retention during cloning and DNA production.Citation10-Citation12
There is a large number of poison/antidote systems, including in bacteria used industrially for production of proteins used as medical products (antigen vaccines, etc).Citation13-Citation17 Among these systems, there is the ccd system composed of the ccdA (antidote) and ccdB genes (poison). The Staby® technology is based on the use of this ccd system. These bacterial genes are known since the Nineties.Citation18,Citation19 They are small in size (approximately 200 to 300 bases each), naturally present in the genomes of the bacteria and on the plasmids, and they encode two small proteins.Citation18,Citation19 In a natural state, they are organized as an operon: a promoter followed by the gene of the antidote (ccdA) and then by the gene of the poison (ccdB). The system regulating the expression (absence of RBS upstream of the poison ORF) ensures that the poison is produced only after the antidote.Citation19 The protein antidote alone or in complex with the poison is able to repress the transcription of both genes. The particular property of the ccd system is that it targets the DNA gyrase, a topoisomerase absent from the cells of higher eukaryotes. The poison is therefore not toxic for mammalian cells.Citation20 The Staby® technology has already been applied to the production of recombinant proteins in Escherichia coli. This enables the plasmid encoding for the protein of interest to be stabilized in a bacterial population without the use of an antibiotic-resistance gene. Due to this particularly efficient stabilization and to the saving of energy by avoiding the expression of resistance gene, the protein yield is increased significantly.Citation10 In order to obtain this stabilization, the ccdB gene is placed in the chromosome of the bacterium and the gene of the antidote is placed on the plasmid. Daughter cells not receiving the plasmid cannot survive (). Moreover, in the presence of the antidote, the expression of the poison is repressed, thereby preventing the selection of potential spontaneous mutant encoding inactive poison.
Figure 1. (A) The Staby® technology. The CcdA protein encoded by the plasmid negatively regulates the transcription of the ccdB gene in the bacterial chromosome. In absence of plasmid, the bacterium died by producing the CcdB protein. (B) Scheme of the pStabyCMV-2 containing the CMV promoter separated to the TK polyadenylate sequence by an EcoRV restriction site.
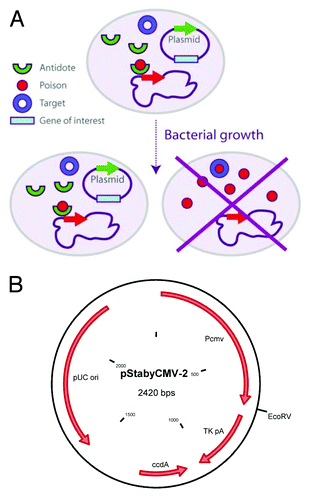
This technological base will enable us to build a new generation of DNA vaccines without antibiotic resistance gene. Indeed, the majority of DNA vaccines still encode a gene conferring resistance to an antibiotic, despite the recommendations of regulatory agencies.Citation21 In order to generate our new constructions, we modified the pStaby1.2 plasmid which contains the ccdA gene by replacing the prokaryotic promoter by the immediate early CMV promoter. The CMV promoter is followed by a thymidine kinase (TK) polyadenylate sequence and separated with the latter by a classical EcoRV cloning site (). In the present study, we investigated three questions related to the use of Staby® technology for the development and the production of DNA vaccines free of antibiotic resistance gene: (1) the potential toxicity of CcdA in eukaryotic cells; (2) the industrial production of plasmid free of antibiotic resistance gene; and (3) the possibility to develop safe and efficacious DNA vaccine free of antibiotic resistance gene. To address the latter hypothesis, we developed a DNA candidate vaccine against Suid herpesvirus-1 (SuHV-1), the causative agent of Aujeszky’s disease in pigs.Citation22 We took profit of the ability of this virus to cause a severe and lethal disease in mice (called pseudo-rabies) to test the efficacy of the candidate vaccine developed. All together, the results of the present study demonstrated the potential of the Staby® technology for the development and the production of DNA vaccines free of antibiotic resistance gene.
Results
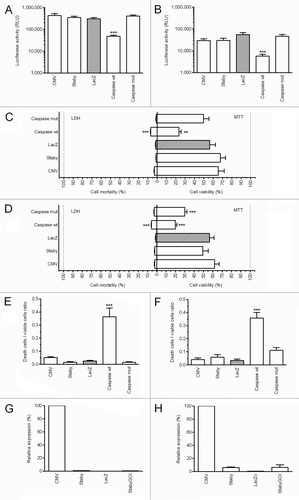
Since in our stabilization system, the antibiotic resistance gene is replaced by a gene encoding an antidote protein, our first objective was to evaluate the possible toxicity of the plasmid (pStabyCMV-2) and particularly the putative toxicity of the CcdA antidote protein. To reach this goal, 293T human cells and B16F10 murine cells were co-transfected with the pVAX2-Luc as reporter and either pStabyCMV-2 (Staby; encoding the ccdA gene under control of a prokaryotic promoter) or pcDNA3.3-CcdA (CMV; encoding the ccdA gene under control of a CMV promoter which is highly active in eukaryotes) or pcDNA3.3-LacZN (LacZ) or pCaspase3-wt (Caspasewt) or pCaspase3-mut (Caspasemut) ( and ). These last four plasmids were used as controls and replaced pStabyCMV-2 (equal molar quantities). 293T and B16F10 cells were efficiently transfected by lipofectamine 2000 as demonstrated by luciferase expression. The expression of luciferase was lower when these cells were transfected with the pCaspase3-wt. This can be explained by the toxicity of the protein encoded by this plasmid, resulting in a marked decrease of the number of living cells. This result was confirmed by the death to live cell ratio estimated using the LDH (lactate dehydrogenase) and MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) assays (). Together, the results demonstrated that pStabyCMV-2 is not toxic for eukaryotic cells. It is important to note that pStabyCMV-2 carries the ccdA sequence but it does not induce its expression in human or murine cells because there is no eukaryotic promoter that controls its expression. Interestingly, we also demonstrated that the pcDNA3.3-CcdA that encodes the CcdA antidote under control of a CMV promoter did not provoke any toxicity. In order to evaluate the level of ccdA mRNA in the transfected cells, qPCR experiments were performed. RNA was isolated from cells (293-T and B16F10) which were transfected with pcDNA3.3-CcdA, pStabyCMV-2, pcDNA3.3-LacZN or pStabyCMV-2-GOI (StabyGOI; pStabyCMV-2 plasmid containing a gene of a human transmembrane protein) and a qPCR analysis was performed. Values for ccdA were normalized to values for actin which is constitutively expressed in these cells. We observed a higher expression when the ccdA gene is under the CMV promoter (). If this value was designated as 100%, the level of ccdA mRNA when the ccdA gene is under a prokaryotic promoter was lower than 1% in human 293T cells and was lower than 7% in murine B16F10 cells. These results suggest that the use of ccdA is safe for eukaryotic cells when artificially overexpressed using the CMV promoter and a fortiori when using a prokaryotic promoter.
Figure 2. CcdA in vitro toxicity in 293T cells (A, C, E and G) and B16F10 cells (B, D, F and H). Both cells were cotransfected with the pVAX2-Luc as reporter and either pStabyCMV-2 (Staby) or pStabyCMV-2-GOI (StabyGOI) or pcDNA3.3-CcdA (CMV) or pcDNA3.3-LacZN (LacZ) or pCaspase3-wt (Caspasewt) or pCaspase3-mut (Caspasemut). (A, B) Cells containing the pCaspase3-wt show a lower expression of the Luc reporter gene suggesting toxicity. (C, D) LDH and MTT assays did not revealed any toxicity of the pStabyCMV-2 or pcDNA3.3-CcdA. (E, F) The death to live cell ratio, obtained from results of the LDH and MTT tests showed significant toxicity for the wild-type caspase-3 encoding plasmid only. (G, H) qPCRs show the innocuousness overexpression of the CcdA gene. Statistical analysis: One-way ANOVA with Tukey post-test. **p value < 0.01, ***p value < 0.001 compared with LacZ.
The second objective was to check the potential impact of Staby® selection system on the plasmid manufacturing process at an industrial scale. For this study, pStabyCMV-2-GOI was transformed into the E. coli CYS21 strain and used according to fedbatch fermentation processes. Fermentation yield obtained in this study was up to 1,350 mg plasmid/l (data not shown). This result shows that antibiotic free plasmid DNA containing the ccdA gene can be manufactured at large scale.
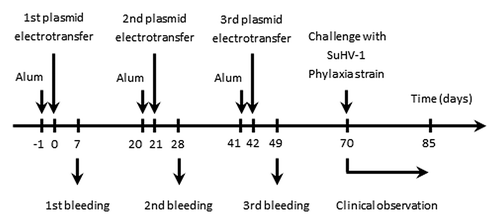
The third and last objective of the present study was to investigate the possibility to develop safe and efficacious DNA vaccines using Staby® technology. The induction of pseudo-rabies by SuHV-1 in mice was selected as an experimental model. With this goal in mind, a DNA candidate vaccine encoding glycoprotein D (gD) of SuHV-1 was produced. Its safety and efficacy was tested in mice as follow (). Mice were vaccinated by electrotransfer three times at three weeks interval, accompanied by three bleedings at day 7, 28 and 49 (the first electrotransfer being defined as day 0). This protocol did not induced detectable clinical signs, supporting the safety of the vaccination program.
Figure 3. Flowchart of the experiments performed to assess the safety and the efficacy of pStabyCMV-2-gD as a DNA candidate vaccine against Aujeszky’s disease. Mice (n = 10) were immunised by DNA electrotransfer of pStabyCMV-2-gD or pStabyCMV-2 (used as negative control). At the indicated times, blood samples were collected and analyzed for detection of anti-gD antibodies (see ). Seventy days after the first plasmid electrotransfer, mice were challenged by injection with the Phylaxia strain of SuHV-1 (see ).
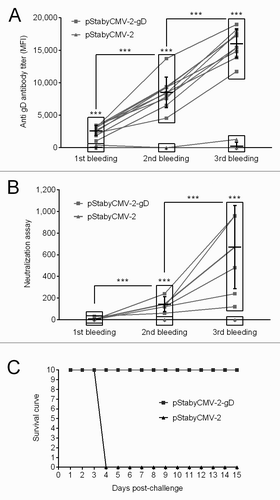
The immune response induced by the vaccine was investigated as follows. First, specific antibodies raised against SuHV-1 gD were quantified by indirect immunofluorescent staining of MAC-T cells transiently expressing gD. The sera of the animals were used as first antibodies. Stained cells were analyzed by flow cytometry and the mean fluorescent intensity channel (MFI) was recorded as a relative measure of specific anti-gD antibody concentration in the serum of vaccinated animals (). Specific antibodies were detected as early as 1 week after the first DNA immunization. Subsequent boosts drastically increased the concentration of antibodies. After the first boost we observed a 3-fold increased in the MFI and a further 2-fold increase after the second boost (). Second, neutralization assays were performed to investigate whether the antibodies produced were able to neutralize SuHV-1 infectivity. In the complement independent neutralization assay used, the sera obtained after the first immunization did not neutralize SuHV-1. In contrast, sera collected after the second and the third immunizations exhibited increasing concentrations of neutralizing antibodies for all mice of the group (). Finally, as the results of the neutralization assays suggested that the immune response conferred by pStabyCMV-2-gD vaccination could be protective, animals were exposed to a lethal challenge. Four weeks after the last immunization, mice were inoculated intramuscularly with the SuHV-1 Phylaxia strain (). Clinical examinations were performed for 15 days after viral inoculation. While none of the mice immunized with pStabyCMV-2-gD expressed pseudo-rabies clinical signs, all mice immunized with pStabyCMV-2 developed pseudo-rabies in a synchronized manner at the beginning of day 4 post-inoculation. These mice died or were euthanized for bioethic reasons during the same day. All together, the results present above demonstrate the potential of the Staby® technology for the development and the production of safe and efficacious DNA vaccines.
Figure 4. Evaluation of the immune response induced by pStabyCMV-2-gD in mice. (A) Specific antibodies raised against SuHV-1 gD were detected by indirect immunofluorescent staining of MAC-T cells transfected with pStabyCMV-2-gD using sample sera as primary antibodies. The MFI of labeled cells was measured by flow cytometry. (B) Neutralizing antibodies were quantified by complement independent neutralization assay. Symbol *** indicates statistical differences (p ≤ 0.01; paired Student’s t test; Graph Pad Software) observed for a specific time point between pStabyCMV-2-gD and pStabyCMV-2 groups (symbol above rectangle), or between different time points within the group of animals vaccinated with the pStabyCMV-2-gD. (C) Immunized animals were challenged with the Phylaxia strain. Time 0 represents the day of challenge.
Discussion
The last ten years, DNA vaccination has been a growing field of research. As explained above, the interest to DNA vaccine is linked to all its advantages over conventional vaccines: the stability of the DNA, the ease of development and production, the ability to induce a wider range of immune response types and the assurance to produce the antigen with post-translational modifications. However, conventional DNA vaccines represent a risk for public health as this kind of vaccine contains an antibiotic resistance gene. To avoid the spread of resistance genes in environment we propose to exchange these genes by the Staby® technology. This technology is based on the ccd system (ccdA/ccdB) naturally present in bacteria. The ccdB gene is inserted in the bacterial chromosome and codes for a poison while the ccdA gene is present on the plasmid and codes for the antidote. This system gives a very strong stability to the plasmid during cell growth and plasmid production.
We report in this study the development of a new plasmid vector designed for use in vaccination. This vector, pStabyCMV-2 contains the cytomegalovirus immediate early promoter (CMV) to produce the antigen with a high expression rate in eukaryotic cells and the antidote gene ccdA to produce the DNA vaccine plasmid without the use of antibiotics. As the pStabyCMV-2 will be the final product injected to the patient, it was obvious to test and to prove the safety of the CcdA protein in eukaryotic cells. We effectively showed that the stabilization technology and particularly the ccdA antidote gene present on the plasmid are safe for eukaryotic cells even when it is artificially overexpressed.
The Staby® technology has been proved compatible with any culture medium or process used for production. Here, we demonstrated that high yield of industrial plasmid DNA production is achievable using this plasmid stabilization technology.
As the CcdA protein is not toxic for eukaryotic cells and that the industrial production is feasible, we designed an antibiotic-free DNA vaccine against Aujeszky’s disease. Currently, vaccination against Aujeszky’s disease is performed with different types of vaccines – inactivated, attenuated, subunits and recombinant. From all the glycoproteins of SuHV-1, gD was selected as candidate antigen since it has been shown to play an essential role in viral entry and to represent a major target for neutralizing antibodies, protecting mice and swine from Aujeszky’s disease.Citation23-Citation26
This study confirms that mice can be effectively protected against SuHV-1 infection by electrotransfer of a plasmid encoding gD as the single antigen, in contrast to a combination of plasmids coding for the three major SuHV-1 glycoproteins.Citation27-Citation29 Moreover the immunization program used in the present study relied on a strategy to reduce the quantity of injected DNA by using electroporation as a delivery method. We used 20 µg /mouse to induce complete immune protection as compared with previous studies that did not employ electroporation and used as much as 100 µg plasmid per mouse.Citation27,Citation28 One study that reported the use of gene gun for the delivery of a DNA vaccine used even lower amount of plasmid (3 µg), but even though it was able to induce neutralizing antibodies the titers were low (reciprocal dilution of sera containing neutralizing antibodies was 30 after 2 immunizations, while in our system we observed that a minimum dilution of 120 was needed to induce protection) and no protection was reported.Citation30 A recent study in pigs confirmed that electroporation can improve the performance of DNA vaccine coding for glycoprotein B of SuHV-1.Citation31
The present study demonstrated that electrotransfer of a Staby plasmid encoding SuHV-1 gD gene is effective in inducing a humoral immune response (as revealed by indirect immunofluorescent assay (MFI) and neutralization assay) and more importantly in conferring an immune protection against a lethal challenge.
Materials and Methods
pStabyCMV-2 (2,421 bp) contains the ccdA antidote gene under the control of a weak constitutive prokaryotic promoter while pcDNA3.3-CcdA (5,617 bp) encodes the ccdA gene under the control of the CMV promoter.Citation12 pcDNA3.3-LacZN (8,467 bp) encodes β-galactosidase and was used as a control for the transfection lethality. Two plasmids (6,808 bp each) encoding the caspase-3 wt (highly apoptotic) or mutated (with a less pronounced effect) were used as positive controls of toxicity. These plasmids encoding caspases were a generous gift from Dr Kris Huygen.Citation32 pVAX2-Luc (4,626 bp) encoding luciferase under the control of the CMV promoter was used to check the efficacy of each transfection. The pVAX2-Luc and the pVAX2-empty plasmids (2,933 bp) were kindly provided by Dr Pascal Bigey (Paris, France).Citation33 Plasmids were prepared using Qiagen Endofree Plasmid Maxi or Giga Kit according to the manufacturer's protocol. All plasmid dilutions were done in PBS and stored at -20 °C before use.
Two types of eukaryotic cells were used: 293T human embryonic kidney cells (8,000 cells per well) and B16F10 murine melanoma cells (4,000 cells per well). Cells were plated in 96-well plates 24 h before transfection. Cells were cotransfected with 0.02 pmol of pVAX2-Luc and 0.02 pmol of pcDNA3.3-CcdA, pStabyCMV-2, pcDNA3.3-LacZN, pCaspase3-wt or pCaspase3-mut. A pVAX2-empty plasmid was added to reach the same total plasmid quantities (i.e., 0.2 µg) for each condition. Cotransfection of the three plasmids was performed using lipofectamine 2000 following the manufacturer’s recommendations (Invitrogen). The luciferase expression was first measured using OneGlo (Promega). Then, in parallel, MTT and LDH cytotoxicity assays were performed 72 h after transfection. MTT measures the mitochondrial succinate deshydrogenase activity and provides an evaluation of cell viability.Citation34 LDH measures lactate dehydrogenase which is released by damaged cells and provides therefore an evaluation of cell mortality.Citation35 For qPCR analysis of ccdA expression, 293T and B16F10 (80,000 and 40,000 cells per well, respectively) were plated using 12-well plates and transfected as described above. Total RNA was isolated and purified using trizol and PureLinkRNA mini kit (Ambion) 72 h after transfection. Total ARN was treated by TURBO DNase (Ambio) and used for reverse transcription with Maxima Reverse Transcriptase (Thermo Scientific) and oligo (dT) primer. A quantitative PCR (qPCR) assay was performed using the LightCycler 480 instruments (Roche) with LightCycler 480 Probes Master. For ccdA amplification, we used primers 5′-GTAAGCACAACCATGCAGAATGA-3′, 5′-CCTTCCTGATTTTCCGCTTTC-3′ and Taq Man probe 5′-CCCGTCGTCTGCGTGCCG-3′. Values for ccdA were normalized to values for actin. We used Pre-developed TaqMan Assay Reagents control Kit (Applied Biosystems) for human (293T cells) or mouse (B16F10 cells) actin amplification. Values obtained from the pcDNA3.3-CcdA samples were designated as 100%. Experiments were performed in triplicate.
The industrial fermentation process was realized with E. coli CYS21 strain (containing the ccdB gene in its chromosome; Delphi Genetics) transformed with pStabyCMV-2-GOI.
SuHV-1 gD ORF (Gene bank sequence ID 2952521) was cloned into pStabyCMV-2 resulting in pStabyCMV-2-gD DNA. Bovine mammary epithelial cells (MAC-T) were used for transfection experiments. SuHV-1 Phylaxia strain was propagated and titrated in Swine testis cells (ST).
DNA immunization was performed as follows. Six weeks old female Balb/c mice were injected with alum (10 mg/ml, 50 µl/mouse; Thermo Scientific) in both tibialis cranialis the day before DNA immunization. The following day, 20 µg of DNA plasmid diluted in 50 µl of PBS was injected into tibialis cranialis (10 µg per leg). Then, we placed the leg between plate electrodes and we delivered 8 square-wave electric pulses (200 V/cm, 20 ms, 2 Hz). Conductive gel was used to ensure electrical contact with the skin (EKO ultrasound transmission gel). The pulses were delivered by a Cliniporator system (IGEA) using 4 mm plate electrodes (IGEA) as previously described.Citation36 All mice were immunized three times at an interval of three weeks. Mouse blood was collected from the caudal vein one week after each immunization. Blood samples were incubated overnight at 4°C, centrifuged for 10 min at 1,000 g and the supernatants were transferred into sterile tubes and stored at -20°C until use.
Anti-gD antibodies were detected by indirect immunofluorescent staining of MAC-T cells transiently expressing gD. MAC-T cells were transfected with 2 µg of plasmid (pStabyCMV-2-gD and pStabyCMV-2) in the presence of polyethylenimine according to the manufacturer’s instructions (Polysciences). Twenty-four hours post transfection staining with mouse sera at a 1:150 dilution was performed, followed by goat anti-mouse Alexa 488 secondary antibody. The mean fluorescent channel of positive cells was detected by flow cytometry.
Anti-gD neutralizing antibodies were analyzed by a complement independent neutralization assay. Mice sera were inactivated for 30 min at 56°C and then serially diluted 2-fold in 96-well plates, mixed with 7 plaque-forming units (pfu) of SuHV-1 Phylaxia strain and incubated for 2 h at 37°C. After incubation the sera-virus mix was added to 1x104 ST cells/well in 96-well plates and incubated for 4 d at 37°C (8 replicates). After four days, cell monolayers were examined for cytopathic effect. Neutralization titers were calculated as the reciprocal highest serum dilution preventing cytopathic effect.
In order to test the protection induced by immunization, the mice were challenged by intramuscular injection into the quadriceps of 2,800 pfu of SuHV-1 Phylaxia strains. Challenged mice were examined twice daily. According to bioethical rules, mice that expressed pruritus for more than 12 h and/or performed auto mutilation were euthanized. The experiments performed in the present study were approved by the bioethical comity of the Université de Liège (Ethical protocol number 1194).
| Abbreviations: | ||
| gD | = | glycoprotein D |
| LDH | = | Lactate dehydrogenase |
| MFI | = | mean fluorescent intensity |
| MTT | = | 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide |
| pfu | = | plaque-forming units |
| SuHV-1 | = | Suid herpesvirus 1 |
| TK | = | thymidine kinase |
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
This work is part of DNAVAC, a certified BioWin project financed by Walloon region (DG06). AV and HN are members of the BELVIR consortium (IAP, phase VII) sponsored by Belgian Science Policy Office (BELSPO). Gaëlle Vandermeulen is a postdoctoral researcher of the Fonds de la Recherche Scientifique-FNRS.
References
- Dubensky TW, Campbell BA, Villarreal LP. Direct transfection of viral and plasmid DNA into the liver or spleen of mice. Proc Natl Acad Sci U S A 1984; 81:7529 - 33; http://dx.doi.org/10.1073/pnas.81.23.7529; PMID: 6095303
- Wolff JA, Malone RW, Williams P, Chong W, Acsadi G, Jani A, et al. Direct gene transfer into mouse muscle in vivo. Science 1990; 247:1465 - 8; http://dx.doi.org/10.1126/science.1690918; PMID: 1690918
- Liu MA, Ulmer JB. Human clinical trials of plasmid DNA vaccines. Adv Genet 2005; 55:25 - 40; http://dx.doi.org/10.1016/S0065-2660(05)55002-8; PMID: 16291211
- Heller R, Shirley S, Guo S, Donate A, Heller L. Electroporation Based Gene Therapy–from the Bench to the Bedside. Conference proceedings: Annual International Conference of the IEEE Engineering in Medicine and Biology Society IEEE Engineering in Medicine and Biology Society Conference 2011; 2011:736-8.
- Vandermeulen G, Athanasopoulos T, Trundley A, Foster K, Préat V, Yáñez-Muñoz RJ, et al. Highly potent delivery method of gp160 envelope vaccine combining lentivirus-like particles and DNA electrotransfer. J Control Release 2012; 159:376 - 83; http://dx.doi.org/10.1016/j.jconrel.2012.01.035; PMID: 22310089
- Satkauskas S, André F, Bureau MF, Scherman D, Miklavcic D, Mir LM. Electrophoretic component of electric pulses determines the efficacy of in vivo DNA electrotransfer. Hum Gene Ther 2005; 16:1194 - 201; http://dx.doi.org/10.1089/hum.2005.16.1194; PMID: 16218780
- Liu MA, Wahren B, Karlsson Hedestam GB. DNA vaccines: recent developments and future possibilities. Hum Gene Ther 2006; 17:1051 - 61; http://dx.doi.org/10.1089/hum.2006.17.1051; PMID: 17032152
- Moss RB. Prospects for control of emerging infectious diseases with plasmid DNA vaccines. J Immune Based Ther Vaccines 2009; 7:3; http://dx.doi.org/10.1186/1476-8518-7-3; PMID: 19735569
- Schalk JA, Mooi FR, Berbers GA, van Aerts LA, Ovelgönne H, Kimman TG. Preclinical and clinical safety studies on DNA vaccines. Hum Vaccin 2006; 2:45 - 53; http://dx.doi.org/10.4161/hv.2.2.2620; PMID: 17012886
- Peubez I, Chaudet N, Mignon C, Hild G, Husson S, Courtois V, et al. Antibiotic-free selection in E. coli: new considerations for optimal design and improved production. Microb Cell Fact 2010; 9:65; http://dx.doi.org/10.1186/1475-2859-9-65; PMID: 20822537
- Sodoyer R, Courtois V, Peubez I, Mignon C. Antibiotic-Free Selection for Bio-Production: Moving Towards a New “Golden Standard”. In: Pana M, ed. Antibiotic Resistant Bacteria - a Continuous Challenge in the New Millennium: InTech, 2012:531-48.
- Szpirer CY, Milinkovitch MC. Separate-component-stabilization system for protein and DNA production without the use of antibiotics. Biotechniques 2005; 38:775 - 81; http://dx.doi.org/10.2144/05385RR02; PMID: 15945374
- Cherny I, Gazit E. The YefM antitoxin defines a family of natively unfolded proteins: implications as a novel antibacterial target. J Biol Chem 2004; 279:8252 - 61; http://dx.doi.org/10.1074/jbc.M308263200; PMID: 14672926
- Guglielmini J, Szpirer C, Milinkovitch MC. Automated discovery and phylogenetic analysis of new toxin-antitoxin systems. BMC Microbiol 2008; 8:104; http://dx.doi.org/10.1186/1471-2180-8-104; PMID: 18578869
- Hayes F, Van Melderen L. Toxins-antitoxins: diversity, evolution and function. Crit Rev Biochem Mol Biol 2011; 46:386 - 408; http://dx.doi.org/10.3109/10409238.2011.600437; PMID: 21819231
- Hazan R, Sat B, Engelberg-Kulka H. Escherichia coli mazEF-mediated cell death is triggered by various stressful conditions. J Bacteriol 2004; 186:3663 - 9; http://dx.doi.org/10.1128/JB.186.11.3663-3669.2004; PMID: 15150257
- Pandey DP, Gerdes K. Toxin-antitoxin loci are highly abundant in free-living but lost from host-associated prokaryotes. Nucleic Acids Res 2005; 33:966 - 76; http://dx.doi.org/10.1093/nar/gki201; PMID: 15718296
- Bernard P, Kézdy KE, Van Melderen L, Steyaert J, Wyns L, Pato ML, et al. The F plasmid CcdB protein induces efficient ATP-dependent DNA cleavage by gyrase. J Mol Biol 1993; 234:534 - 41; http://dx.doi.org/10.1006/jmbi.1993.1609; PMID: 8254658
- Salmon MA, Van Melderen L, Bernard P, Couturier M. The antidote and autoregulatory functions of the F plasmid CcdA protein: a genetic and biochemical survey. Mol Gen Genet 1994; 244:530 - 8; http://dx.doi.org/10.1007/BF00583904; PMID: 8078480
- Bernard P. Positive selection of recombinant DNA by CcdB. Biotechniques 1996; 21:320 - 3; PMID: 8862819
- Vandermeulen G, Marie C, Scherman D, Preat V.. New Generation of Plasmid Backbones Devoid of Antibiotic Resistance Marker for Gene Therapy Trials. Molecular therapy. the journal of the American Society of Gene Therapy 2011; 19:1942 - 9; http://dx.doi.org/10.1038/mt.2011.182
- Mettenleiter TC. Aujeszky’s disease (pseudorabies) virus: the virus and molecular pathogenesis--state of the art, June 1999. Vet Res 2000; 31:99 - 115; http://dx.doi.org/10.1051/vetres:2000110; PMID: 10726640
- Eloit M, Gilardi-Hebenstreit P, Toma B, Perricaudet M. Construction of a defective adenovirus vector expressing the pseudorabies virus glycoprotein gp50 and its use as a live vaccine. J Gen Virol 1990; 71:2425 - 31; http://dx.doi.org/10.1099/0022-1317-71-10-2425; PMID: 2172456
- Marchioli CC, Yancey RJ Jr., Petrovskis EA, Timmins JG, Post LE. Evaluation of pseudorabies virus glycoprotein gp50 as a vaccine for Aujeszky’s disease in mice and swine: expression by vaccinia virus and Chinese hamster ovary cells. J Virol 1987; 61:3977 - 82; PMID: 2824827
- van Rooij EM, Rijsewijk FA, Moonen-Leusen HW, Bianchi AT, Rziha HJ. Comparison of different prime-boost regimes with DNA and recombinant Orf virus based vaccines expressing glycoprotein D of pseudorabies virus in pigs. Vaccine 2010; 28:1808 - 13; http://dx.doi.org/10.1016/j.vaccine.2009.12.004; PMID: 20018271
- Wathen LM, Platt KB, Wathen MW, Van Deusen RA, Whetstone CA, Pirtle EC. Production and characterization of monoclonal antibodies directed against pseudorabies virus. Virus Res 1985; 4:19 - 29; http://dx.doi.org/10.1016/0168-1702(85)90017-6; PMID: 3002067
- Hong W, Xiao S, Zhou R, Fang L, He Q, Wu B, et al. Protection induced by intramuscular immunization with DNA vaccines of pseudorabies in mice, rabbits and piglets. Vaccine 2002; 20:1205 - 14; http://dx.doi.org/10.1016/S0264-410X(01)00416-9; PMID: 11803083
- Yoon HA, Aleyas AG, George JA, Park SO, Han YW, Kang SH, et al. Differential segregation of protective immunity by encoded antigen in DNA vaccine against pseudorabies virus. Immunol Cell Biol 2006; 84:502 - 11; http://dx.doi.org/10.1111/j.1440-1711.2006.01463.x; PMID: 16869937
- van Rooij EM, Haagmans BL, de Visser YE, de Bruin MG, Boersma W, Bianchi AT. Effect of vaccination route and composition of DNA vaccine on the induction of protective immunity against pseudorabies infection in pigs. Vet Immunol Immunopathol 1998; 66:113 - 26; http://dx.doi.org/10.1016/S0165-2427(98)00186-X; PMID: 9860185
- Hwang DY, Lee JB, Kim TJ, Song JY, Hyun BH, Song CS, et al. Induction of immune responses to glycoprotein gD of Aujeszky’s disease virus with DNA immunization. J Vet Med Sci 2001; 63:659 - 62; http://dx.doi.org/10.1292/jvms.63.659; PMID: 11459012
- Le Moigne V, Cariolet R, Béven V, Keranflec’h A, Jestin A, Dory D. Electroporation improves the immune response induced by a DNA vaccine against pseudorabies virus glycoprotein B in pigs. Res Vet Sci 2012; 93:1032 - 5; http://dx.doi.org/10.1016/j.rvsc.2011.09.020; PMID: 22051145
- Gartner T, Romano M, Suin V, Kalai M, Korf H, De Baetselier P, et al. Immunogenicity and protective efficacy of a tuberculosis DNA vaccine co-expressing pro-apoptotic caspase-3. Vaccine 2008; 26:1458 - 70; http://dx.doi.org/10.1016/j.vaccine.2007.12.056; PMID: 18280621
- Bloquel C, Bejjani R, Bigey P, Bedioui F, Doat M, BenEzra D, et al. Plasmid electrotransfer of eye ciliary muscle: principles and therapeutic efficacy using hTNF-alpha soluble receptor in uveitis. FASEB J 2006; 20:389 - 91; PMID: 16352651
- van Meerloo J, Kaspers GJ, Cloos J. Cell sensitivity assays: the MTT assay. Methods Mol Biol 2011; 731:237 - 45; http://dx.doi.org/10.1007/978-1-61779-080-5_20; PMID: 21516412
- Decker T, Lohmann-Matthes ML. A quick and simple method for the quantitation of lactate dehydrogenase release in measurements of cellular cytotoxicity and tumor necrosis factor (TNF) activity. J Immunol Methods 1988; 115:61 - 9; http://dx.doi.org/10.1016/0022-1759(88)90310-9; PMID: 3192948
- Vandermeulen G, Richiardi H, Escriou V, Ni J, Fournier P, Schirrmacher V, et al. Skin-specific promoters for genetic immunisation by DNA electroporation. Vaccine 2009; 27:4272 - 7; http://dx.doi.org/10.1016/j.vaccine.2009.05.022; PMID: 19450641