Abstract
Nucleotide based vaccines represent an enticing, novel approach to vaccination. We have developed a novel immunization technology, RNActive® vaccines, that have two important characteristics: mRNA molecules are used whose protein expression capacity has been enhanced by 4 to 5 orders of magnitude by modifications of the nucleotide sequence with the naturally occurring nucleotides A (adenosine), G (guanosine), C (cytosine), U (uridine) that do not affect the primary amino acid sequence. Second, they are complexed with protamine and thus activate the immune system by involvement of toll-like receptor (TLR) 7. Essentially, this bestows self-adjuvant activity on RNActive® vaccines. RNActive® vaccines induce strong, balanced immune responses comprising humoral and cellular responses, effector and memory responses as well as activation of important subpopulations of immune cells, such as Th1 and Th2 cells. Pre-germinal center and germinal center B cells were detected in human patients upon vaccination. RNActive® vaccines successfully protect against lethal challenges with a variety of different influenza strains in preclinical models. Anti-tumor activity was observed preclinically under therapeutic as well as prophylactic conditions. Initial clinical experiences suggest that the preclinical immunogenicity of RNActive® could be successfully translated to humans.
Introduction
Since their inception by Edward Jenner roughly two hundred years ago, vaccines have achieved tremendous successes such as the eradication of diseases like smallpox and the containment of many childhood diseases. They are among the most cost-effective treatments that exist today. Despite these huge successes though, there is increasing scepticism toward vaccines in the Western world, partly because their success let people forget the disastrous consequences of the diseases successfully contained and made them focus on unintended effects. In addition, the recent swine flu epidemic demonstrated that present technology may not allow to produce vaccines in time to contain threatening diseases.
Although vaccines have been in widespread use for the last hundred years, vaccines against only comparatively few pathogens are available today. It has generally been difficult to make vaccines against many bacterial or parasitic diseases, e.g., such as S. aureus infections, tuberculosis or malaria, which pose an increasing risk due to increasing resistances against antibiotics and antiparasite drugs. For many viral diseases, vaccines are completely lacking, e.g., cytomegalo virus and Dengue virus, not to cite the desperate need for a vaccine against HIV. Besides research and development, investments of hundreds of million dollars are required for the set-up of production facilities well before licensure which constitutes a huge business risk.
As a consequence, the necessity to move beyond largely empirical approaches to vaccines research and development has spurred interest in novel approaches such as reverse, structural and synthetic vaccinology.Citation1 Nucleotide based vaccines appear well-suited to feed the needs of the aforementioned approaches, offering a comparatively simple and inexpensive basis for vaccination that would allow to take advantage of modern (protein) engineering methods. However, despite intensive research in the last decades, DNA vaccines have not yet achieved the break-through in humans. Here, we describe how vaccines based on messenger RNA (mRNA) might represent a suitable alternative for nucleotide based vaccination.
mRNA as the Basis for Vaccination
Early reports describing local protein expression after injection of mRNACitation2 were quickly followed by efforts to exploit this approach for vaccination. It was shown that subcutaneous injection of liposome-encapsulated mRNA, but not naked mRNA encoding the nucleoprotein (NP) of influenza virus elicited NP-specific cytotoxic T cells (CTLs).Citation3 Antigen-specific antibodies could be induced with mRNA encoding human carcinoembryonic antigen (CEA) by repeated intramuscular injection upon challenge with CEA positive tumor cells, but an anti-tumor effect was not described.Citation4 A humoral and cellular (cytolytic) immune response could principally be achieved after intradermal injection into the ear with a protamine-complexed mRNA.Citation5 In addition, vaccines could be successfully built on other principles using RNA as their basis, including replicon based approaches and transfection of dendritic cells pulsed in vitro with mRNA.Citation6-Citation11 More detailed analyses of the mechanisms underlying the observed immune responses indicated that naked mRNA resulted in a T-helper 2 cell (Th2) response,Citation12 whereas protamine/ RNA complexes acted as danger signal that activated mouse cells through a MyD88-dependent pathway involving Toll-like receptor 7 (TLR7) and TLR8.Citation13-Citation15 The complexes formed by protamine and irrelevant mRNA induced comparable anti-tumor effects to the oligonucleotide CpG after intratumoral injection, but importantly they did so also after injection at a distant site. While administration of CpG caused a substantial increase in spleen size, the protamine/ mRNA-complexes were indistinguishable from buffer controls in this respect which already indicated that mRNA based vaccines might exhibit a very good safety profile.
These studies were certainly very encouraging, but they also made clear that two obstacles would have to be overcome to generate a successful mRNA-based vaccine: measures to increase the protein expression encoded by a given mRNA as well as ways to elicit a balanced, long-lasting immune response comprising strong humoral and cellular responses would have to be found.
Moving Beyond Wild-Type mRNA: Creation of RNActive® Vaccines with Self-Adjuvanted, Highly Expression Enhanced, Modified mRNA
mRNA represents the minimal genetic vector, it contains only the elements directly required for expression of the encoded protein. In the minimal structure, a protein-encoding open reading frame (ORF) is flanked at the 5′- and 3′-end by two elements essential for the function of mature eukaryotic mRNA: the “cap”, a 7-methyl-guanosine residue bound to the 5′-end of the RNA via a 5′-5′ triphosphate bond, and a poly(A) tail at the 3′-end ().Citation16,Citation17 This basic structure is transcribed in vitro from a plasmid DNA template that contains at least a bacteriophage promoter and the ORF, optionally a poly(d[A/T]) sequence transcribed into poly(A) and a unique restriction site for linearization of the plasmid to ensure defined termination of transcription (the cap is not encoded by the template). In addition to these structures, protein expression can be affected by the 5′- untranslated region between cap and ORF and the 3′- untranslated region that resides between ORF and polyA-tail.Citation11 Using our proprietary mRNA technology, coding and non-coding parts of the molecule are constantly modified by in silico and experimental methods to increase both the level and duration of protein expression. Importantly, only the naturally occurring nucleotides A, G, C, and U are used in this process. The mRNAs designed this way are then subjected to an also proprietary purification process which again increases total protein expression.
Figure 1. Schematic representation of mRNA and expression levels reached by modifactions thereof. (A) The principle structure of an mRNA-molecule consists of a cap-region, followed by an (optional) 5′-untranslated region, the open reading frame, an (optional) 3′-untranslated region and the poly-A-tail. Sequence modifications of each depicted subunit of an mRNA molecule that only comprise the naturally occurring nucleotides A, G, C, U and that do not affect the primary amino acid sequence encoded by the open reading frame constitute the basis of modified mRNA-molecules used in RNActive® vaccines. Details in ref. Citation20 (B) Different generations of PpLuc-coding mRNAs produced over the last years were electroporated into HeLa cells (generation 1 to 4) and compared for their in vitro expression of luciferase. The luciferase level was determined at 6, 24, and 48 h or 72 h post transfection. The expression power of generation 4 and 5 was compared in human dermal fibroblasts after lipofection. The dynamic range of the assay was not sufficient to compare all mRNA molecules in one experiment.
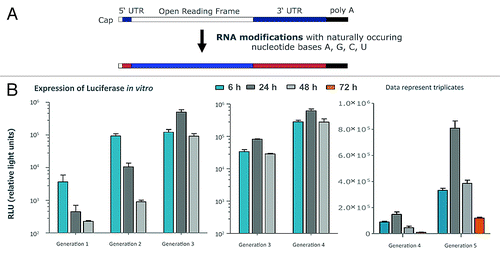
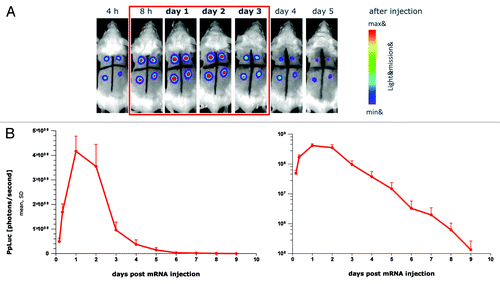
Over the years, this process resulted in an increase of protein expression by 4–5 orders of magnitude as assessed by luciferase expression in various test systems (). The kinetics of protein expression could be changed dramatically. While protein expression peaked after around 6–8 h in early experiments, similarly also reported by others using differing constructs,Citation18 the recently developed constructs peak after 24 h with an expression at 48 h that matches or exceeds the level reached after 6 h. The most recent generation presently in preclinical assessment represents a big leap forward, with the 72 h expression level slightly exceeding the 6 h expression level of the previous generation. Importantly, these results were also reflected in in vivo experiments in which luciferase expression could be clearly visualized for 5 d (). The quantitative assessment showed a peak expression after 24–48 h with strong expression still present after 72 h and expression still detectable up to 9 d (). The antigen dose and the duration of its presentation are considered critical factors for generating strong and sustained antigen-specific immune responses.Citation19 Thus, the protein expression achieved with our modified mRNA-constructs starts to mimic that seen after viral infections such as influenza virus infection which may be advantageous for the induction of antigen-specific immune responses.
Figure 2. Protein expression in vivo is strongly prolonged using CureVac’s proprietary mRNA technology and lasts for many days. Firefly luciferase-encoding mRNA, optimized for translation and stability, was injected intradermally into a BALB/c mouse (4 injection sites). At various time points after mRNA injection, luciferase expression was visualized in the living animal by optical imaging. (A) Visualization of luciferase expression at selected time points, showing maximal protein levels 24 to 48 h after mRNA injection. (B) Quantitative expression of luciferase over time until 9 d after mRNA injection. Results are shown on a linear scale (left-hand panel) or on a semi-logarithmic scale (right-hand panel). The figure is adapted with permission from ref. Citation11, details therein.
Adjuvanticity of mRNA Complexed with Protamine
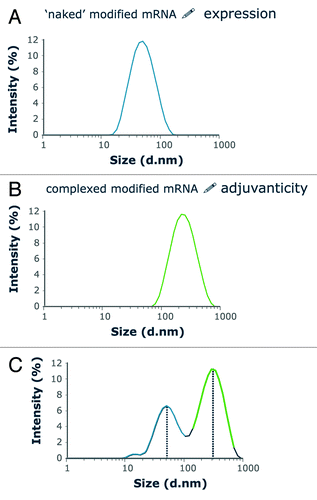
The “naked” mRNA described above achieved high antigen expression, but only weak immunostimlation.Citation20 Previous evidence had demonstrated that complexation of mRNA with protamine helped to arise Th1 responses against antigens with the possible involvement of TLR7/TLR8.Citation12,Citation14 Zeta-sizer analysis indicated that mRNA/ protamine complexes form particles of around 250–350 nm, whereas the “naked”, modified mRNA forms smaller particles around 50 nm (). However, complexes are very tight, so that the adjuvant effect comes at the cost of very weak antigen expression.Citation11,Citation20 Furthermore, the immune stimulating effects of mRNA/protamine complexes strongly depend on the ratio between protamine and mRNA. To achieve the two objectives of good antigen expression and adjuvanticity, an optimal mixture of the modified “naked” mRNA with preformed complexes of the very same mRNA with protamine was defined. Size distribution analysis of this two component mixture revealed that the two particle sizes described above were still detectable in the mixture (). Fluorescence correlation spectroscopy revealed that particles remain separate and that the mRNA complexed to protamine does not interchange with the “naked” mRNA in solution.Citation20 The two components are taken up by endocytic, yet distinct pathways into the cell as no colocalizaton was foundCitation11,Citation20. The naked mRNA appears to be taken up by scavenger receptors residing in caveolae, while the mRNA/protamine complex appears to remain in a different endocytic compartment.Citation11,Citation21 This would be consistent with the weak antigen expression by the complexes and also with stimulation of TLR7 which resides in an endosomal location.Citation22 Others reported that uptake by immature dendritic cells is largely driven by macropinocytosis and stops upon dendritic cell maturation.Citation23
Figure 3. Size analysis of RNActive® vaccines as shown by a vaccine encoding Ppluc that was produced using the RNActive® technology. The size distribution of particles composing the vaccine solution was analyzed using a Malvern Zetasizer Scattering Instrument. Closely similar data were generated with several different mRNAs (A) The naked mRNA has a size of around 50 nm. (B) mRNA is complexed with protamine at a mass ratio of mRNA:protamine of 2:1. The resulting particles are distinctly larger than the ones consisting of naked mRNA. (C) The size distribution analysis of a complete RNActive® vaccine shows that the particle sizes of the two individual components are maintained in the RNActive® vaccines. Further analysis with fluorescent correlation spectroscopy demonstrated that the protamine/ mRNA complexes are very tight, so that naked, free mRNA does not exchange with the protamine-complexed mRNA. Figure was adapted with permission from ref. Citation20, details therein.
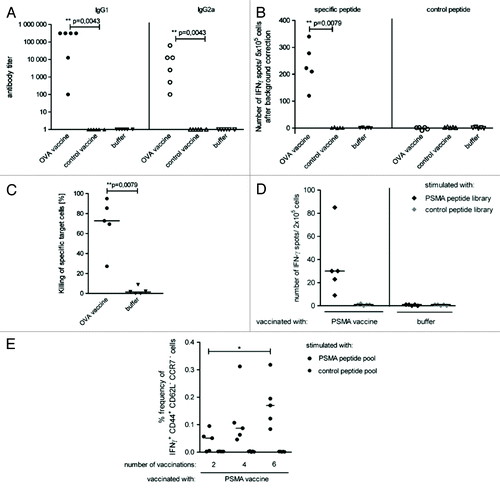
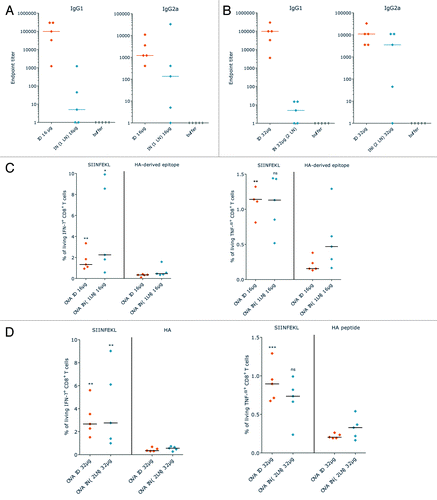
An analysis of the translational and immunostimulatory capacity of this two component mixture prepared with PpLuc mRNA demonstrated that the high and stable antigen expression by the “naked” mRNA component is maintained, whereas adjuvanticity is achieved by the immunostimulatory effect of the mRNA/protamine complexes.Citation20 The immunostimulatory effect of this self-adjuvanted, two-component vaccine which we termed RNActive®-vaccine, is lost in TLR7−/− mice, indicating a pivotal role of TLR7 in the mechanism of this vaccine (see also ). Vaccines encoding ovalbumin or prostate specific membrane antigen (PSMA) were produced to analyze the immune response induced by this novel vaccination approach. Consistently, RNActive®-vaccines prepared this way induced “balanced” immune responses: the ovalbumin vaccine induced a strong humoral immune response resulting in high IgG1 and IgG2a antibody titers which suggests that both Th2 and Th1 responses were elicited (). The T-cell response comprises both IFNγ-secreting, functional, cytolytic CD8+ T cells () as well as IFNγ-secreting CD4+ T cells () as evidenced by vaccination with an ovalbumin or PSMA encoding RNActive®-vaccine, respectively. Repeated vaccination substantially increased the frequency of IFNγ-secreting CD8+ T cells without increasing the frequency of CD4+ regulatory T cells (data not shown). Importantly, RNActive®-vaccines induce not just a strong effector immune response, but also a strong memory T-cell response, here elicited by repeated vaccination with a PSMA encoding RNActive®-vaccine ().
Figure 7. Effect of pattern recognition receptors on RNActive vaccination. (A and B) Male TLR7−/− or TLR9−/− BALB/c mice (purchased from Bioindustry Division. Oriental Yeast Co., Tokyo, Japan) were vaccinated intradermally on day 0 and day 7 with 20 µg of RNActive vaccines encoding HA (PR8) or ovalbumin as control. Serum samples were taken 7 d (A) and 28 d (B) after the final vaccination and IgG1 and IgG2a antibodies against HA determined with ELISAs (methods in Petsch et al.Citation25). Data points represent single mice. (C) One month later, the same mice were vaccinated with 20 µg of RNActive encoding nucleoprotein (NP) from PR8 with the same vaccination schedule. Splenocytes from vaccinated mice were stimulated with a peptide (amino acids 147–155) from NP 7 d after the last vaccination and the cytolytic activity determined with an in vivo killing assay (described in ref. Citation20).
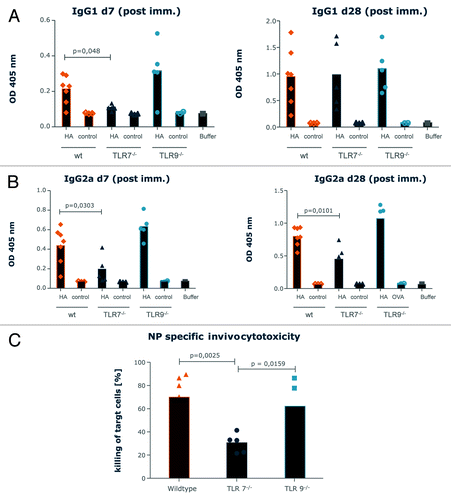
Figure 4. RNActive® vaccines induce effective B cell and T cell responses. Mice were vaccinated intradermally with an RNActive® vaccine encoding ovalbumin. (A) The presence of ovalbumin-specific antibodies was measured in serially diluted sera of vaccinated and control mice taken 11 d after the last vaccination and analyzed using ELISA. Data points represent antibody endpoint titers calculated for individual mice. (B) Ex vivo ELISpot analysis of the secretion of IFNγ in splenocytes from vaccinated and control mice. Cells were isolated on day 6 after the last vaccination and stimulated either with antigenic or with control peptide. The graph shows single data points for individual mice. (C) In vivo cytotoxicity against target cells loaded with the ovalbumin derived SIINFEKL peptide on day 5 after the last injection. The graph shows single data points for individual mice. (D) Ex vivo ELISpot analysis of the secretion of IFNγ in sorted CD4+ T cells from mice vaccinated with an RNActive® vaccine encoding prostate specific membrane antigen (PSMA) and control mice. Cells were isolated on day 6 after the last vaccination and stimulated either with PSMA-derived or control peptide library. (E) Frequencies of IFNγ+ CD44+ CD62L- CCR7- memory T cells in sorted CD8+ T cells from vaccinated and control mice. Cells were isolated on day 55 after last vaccination and stimulated ex vivo either with antigenic or with control peptide library and anti-CD28 antibody for 6 h. After intracellular staining of IFNγ secretion, cells were stained for surface markers of memory T cells. Adapted with permission from refs. Citation20 and Citation24.
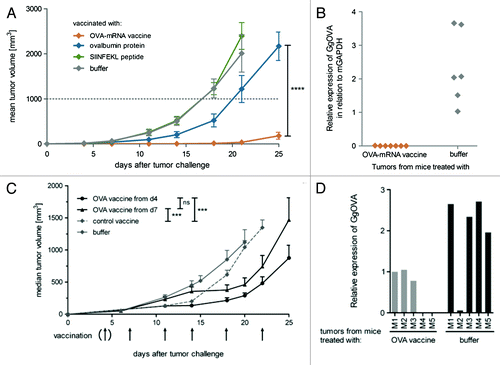
To study the anti-tumor, presumably T-cell dependent activity of our RNActive® -vaccines the E.G7-Ova model was employed. Mice were either vaccinated prophylactically with an ovalbumin encoding RNActive® vaccine and then challenged with ovalbumin expressing E.G7 tumor cells, or inoculated with E.G7-Ova tumor cells first and then immunized with the ovalbumin RNActive® vaccine once the tumor had become rather large (around 80 mm3). In the prophylactic model, the ovalbumin RNActive® vaccine protected against tumor growth for about three weeks before the tumor started to grow (). Likewise in the therapeutic model, even at the advanced tumor size treated, the RNActive® vaccine suppressed tumor growth for more than a week before it started to grow again (). When we assessed possible reasons for the observed tumor escape, we found that escaping tumors had largely downregulated or even eliminated ovalbumin expression (). Hence, RNActive® vaccines had successfully established cancer immunosurveillance and thus exerted an evolutionary pressure selecting cells either weakly or not expressing ovalbumin.
Figure 5. RNActive® vaccines in prophylactic and therapeutic tumor models. (A) C57BL/6 mice were immunized intradermally with an RNActive® vaccine encoding ovalbumin on day 1 and week 3. Six days after the last vaccination, C57BL/6 mice were challenged subcutaneously with 1 × 106 syngeneic E.G7-OVA tumor cells. Tumor growth was monitored by measuring the tumor size in three dimensions using calipers. (B) Outgrown tumors were excised and ovalbumin expression was quantified via RT-PCR relative to murine GAPDH from total RNA isolates. (C) C57BL/6 mice (n = 9) were challenged subcutaneously with 0.3 × 106 syngeneic E.G7- OVA tumor cells on day 0. Tumors were palpable on day 3. Mice were treated according to the schedule specified in the graph with either the RNActive® ovalbumin or with control vaccine (32 µg/vaccination) or with buffer. Therapy was started either on day 4 or on day 7. (D) Mice were treated as indicated in (C), but outgrown tumors were excised to quantify the expression of ovalbumin via RT-PCR. Figure adapted with permission from refs. Citation11 and Citation24.
Further analysis of the mechanism of action of RNActive® vaccines revealed that CD4+ T cells were essential during the induction phase of the immune response, whereas the anti-tumor immune response depended on CD8+ T cells. Interestingly, an analysis of changes within the tumor showed a higher frequency and persistence of CD8+ T cells after as few as two vaccinations.Citation20,Citation24 Close to 70 genes were upregulated in the tumors of vaccinated mice, among these NK-cell related genes, markers of activated, cytolytic T cells as well as those encoding chemokines, IFNγ and IFNγ-related genes.Citation24
Importantly, combination of RNActive® vaccines with other treatments such as anti-CTLA-4 antibodies resulted in largely synergistic effects.Citation24 Under conditions where the antibody was hardly active and the RNActive® vaccine only moderately active against the tumor, the combination was able to control tumor growth. When the long-term survivors were challenged with ovalbumin negative EL4 tumor cells (which are the parent cells to the ovalbumin positive E.G7-Ova cell line), the inoculated EL4 tumors were rejected by all long-term survivors. This suggests that the combination of RNActive® vaccines with the checkpoint inhibitor anti-CTLA4 not just resulted in a stronger anti-ovalbumin response, but also induced antigen spreading.
RNActive® Vaccines can be Used for Prophylaxis of Infectious Diseases
It was recently reported by others that an mRNA encoding a construct of influenza hemagglutinin (HA) did not result in a measurable anti-HA response upon intradermal injection.Citation18 This prompted us to ask whether RNActive® vaccines would face a similar problem thus rendering them inappropriate for the use as prophylactic vaccines. Hence, an RNActive® vaccine was generated encoding full-length HA from influenza virus A/PuertoRico/8/1934 (PR8HA) and injected intradermally into BALB/c mice in a prime-boost regimen at a 3 weeks intervalCitation25. Humoral responses were measured 4 weeks after the last injection. Both, HA-specific IgG1 and IgG2a antibodies as well as serum activity in HA inhibition (HI) assays could be achieved (). Average HI titers were ≥ 1:40, commonly defined as the protective limit in humans. Immunogenicity could be demonstrated in male and female animals, different mouse strains (C57BL/6, NMRI, DBA/2) as well as male Lewis rats (data not shown).
Figure 6. Immunological characterization of an RNActive® vaccine encoding hemagglutinin from influenza virus PR8. BALB/c mice were vaccinated intradermally with an RNActive® vaccine encoding hemagglutinin from influenza virus PR8 on day 1 and day 22. PR8HA-specific antibodies in the serum were quantified 4 weeks after the last immunization by IgG1- (A) and IgG2a-specific ELISA (B), and by hemagglutination inhibition (HI) assays (C). The dashed line in c indicates the conventionally defined protective HI titer of 1:40. (D) ELISPOT of IFNγ production in CD4+ T-cells sorted from vaccinated and buffer control mice on day 28. CD4+ T-cells were stimulated with a pool of five MHC class II–restricted peptides from HA (HA pept.) or as a control from the HIVpol protein (control; ctrl. pept.). Figure adapted with permission from ref. Citation25.
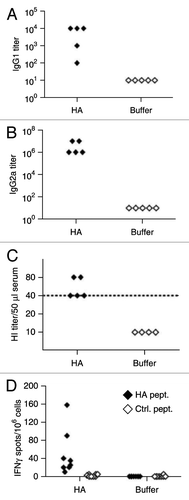
Together, these data indicated effective seroconversion and the presence of virus-neutralizing antibodies in all RNActive®-vaccinated animals. Additionally, CD4+ T-cell responses against a cocktail of major histocompatibility complex (MHC) class II-restricted PR8HA-derived peptides could be measured in an ex vivo interferon IFN-γ ELISPOT assay (). PR8HA mRNA-immunized BALB/c mice also showed a significantly higher cytotoxic activity of CD8+ T cells against an MHC class I-restricted PR8-HA peptide compared with buffer injected miceCitation25.
To assess the mechanism of action, we compared the antibody induction following vaccination of wild-type BALB/c mice with PR8HA to the one in TLR7−/− and TLR9−/− mice. Mice were vaccinated two times with a 3 weeks interval and antibodies were measured either 1 week or 4 weeks after the last vaccination. Seven days after the first priming vaccination, both IgG1 and IgG2 antibodies were clearly suppressed in TLR7−/− mice while there was no effect on the antibody levels of the two classes in TLR9−/− mice compared with wild-type mice (). For wild-type and TLR9−/− mice, the same results were obtained at the 4 weeks assessment. In contrast, TLR7−/− mice behaved differently: IgG2a levels remained suppressed in TLR7−/− mice, whereas IgG1 levels in TLR7−/− mice were the same as in wild-type mice and TLR9−/− mice at the 4 weeks assessment. This suggests that RNActive® dependent immune responses occur independently of TLR9 activation, but are strongly affected by TLR7 activation. For the IgG1 antibody response, i.e., a Th2 dependent response, this was only the case at the early and not the late time point, while the TLR7-dependent impact on IgG2a levels could be demonstrated at early as well as late time points. In line with a pronounced impact of TLR7 on the Th1 dependent immune response, we found that vaccination with an RNActive® vaccine encoding nucleoprotein (NP) from PR8 resulted in a much lower cytolytic activity upon stimulation with an NP-derived peptide in TLR7−/− mice than in wildtype and TLR9−/−mice ().
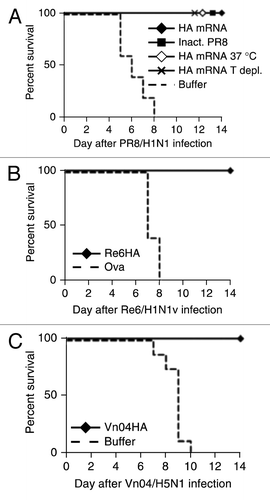
To investigate whether the immunological effect of RNActive® vaccines also translated in protection against viral infections, mice were vaccinated two times at a three weeks interval with RNActive® vaccines encoding HA from influenza strains PR8 (), swine flu () or bird flu () and subjected to homologous challenge with multiples of the LD50 of the respective viruses. In each case, 100% of the animals vaccinated with RNActive® vaccine () or inactivated PR8 virus () survived, whereas mice vaccinated with either an irrelevant vaccine () or buffer () were not protected. As shown in the challenge experiment with PR8 (), mice also survived the challenge when T cells were depleted prior to infection which showed that the protective effect was independent of T cells. Serum transfer experiments confirmed that the protective effect depended on neutralizing antibodies active in HI assays.Citation25 Importantly, vaccines stored at – 20 °C or 37 °C were equally able to protect mice against lethal challenge with PR8 virus suggesting that the cold chain is not required by RNActive® vaccines ().
Figure 8. Protective efficacy of mRNA vaccine against lethal virus challenge in BALB/c mice. (A) BALB/c mice (n = 5/group) were injected intradermally with 80 µg of PR8HA mRNA that was either frozen at −20°C before immunization (HA mRNA) or lyophilized and stored for 3 weeks at 37°C before immunization (HA mRNA 37°C). In one group of mice immunized with PR8HA mRNA (that had been frozen), T cells were depleted at days −1 and +3 with respect to challenge infection (HA T depl.). Control mice were injected intradermally with buffer or intramuscularly with 10 µg of inactivated PR8 virus (Inact. PR8). Vaccine or control injections were done on days 0 and 21. On day 56 mice were infected with 10x LD50 of PR8 virus and survival assessed. (B and C). BALB/c mice (n = 5/group for Re6; n = 8/group for Vn04) were injected intradermally with 80 µg of mRNA encoding HA from influenza strain A/Regensburg/D6/2009 (Re6/H1N1v) or A/Vietnam/1194/2004 (Vn04/H5N1), three independent experiments. Control mice were injected with (B) 80 µg of ovalbumin RNActive® vaccines or Ringer′s lactate buffer (C). Immunizations were done at day 0 and booster injections at day 21. Five weeks after booster injection mice were challenged with virus expressing the homologous HA. For Vn04 and PR98 10x LD50 were used as challenge dose. Due to technical limitations a 6.8x LD50 was used for the challenge with Re6. Statistical analysis was done using a log rank analysis (Mantel Cox test): (A) p = 0.0017, (B) p = 0.001, (C) p = < 0.0001. Further information in ref. Citation25.
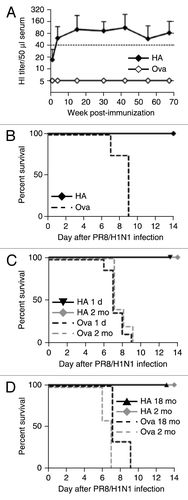
To assess whether vaccination with RNActive® vaccines resulted in long lasting immune effects, mice were vaccinated at week 1 and 2 at two mo of age and then monitored for their HI titers for up to 16 mo at monthly intervals (). HI titers of ≥ 1:40 were observed already at week 3 of the experiment and these titers were stably maintained throughout the course of the experiment. Mice were challenged with 10 × LD50 of PR8 virus at 18 mo of age (i.e., 16 mo after vaccination). All vaccinated animals were protected against lethal challenge infection (). Protection against influenza infection is still challenging in small children and elderly people. We therefore examined mRNA vaccine immunogenicity in very young (newborns > 1 d old) and very old (18 mo of age) BALB/c mice. Mice were immunized with 80 μg PR8HA or an irrelevant Ova mRNA vaccine on days 0 and 7. Five weeks after immunization both groups and control animals were challenged with 10 × LD50 of PR8 virus. Both, vaccinated newborn () and old () mice survived infectious challenge and suffered neither pronounced weight loss nor other signs of disease (data not shown), similarly to 2-mo-old adult mice. Hence, RNActive® vaccines may prevent or reduce disease caused by influenza virus infection at the extremes of age. More generally, RNActive® vaccines might overcome the problem of immosenescence. The reasons for this are not yet clear, but may result from successful TLR7 activation.
Figure 9. RNActive® vaccines elicit durable immune responses, circumvent immunosenescence in old mice, but are also immunogenic in newborn mice. (A) 8-week-old female BALB/c mice were injected intradermally on days 0 and 7 with 20 µg of PR8HA RNActive® (n = 5) or ovalbumin RNActive® (n = 4). For each condition, two independent experiments were performed. HI titers were monitored over a period of 70 weeks (16 mo) and plotted as mean + s.d. (B) Sixteen months after immunization, mice were challenged with 10 × LD50 of PR8 and survival was monitored. Newborn mice (1 d old, 1d, n = 9/group; three independent experiments) (C), aged (18 mo old, 18 mo, n = 3/group; one experiment) (D) or adult (2 mo old, 2 mo, n = 5/group) BALB/c mice were injected intradermally with 80 µg of PR8HA or ovalbumin RNActive® with an interval of 7 d. Five weeks after the second immunization, mice were challenged with 10 × LD50 of live PR8 virus and survival monitored for 14 d post-infection. Statistical analysis was done using a log rank analysis (Mantel Cox test): (B) p = 0.005, (C) 1 d: p = 0.0007; 2 mo: p = 0.0015, (D) 2 mo: p = 0.031; 18 mo: p = 0.01,
Prime-Only Efficacy of RNActive® Vaccines Encoding Different Influenza Antigens
To test whether a single vaccination was sufficient to achieve protection, mice were vaccinated with 80 µg of the RNActive® vaccine encoding HA from PR8. 100% of the vaccinated animals survived, but the temporary weight loss observed in the animals suggested that single vaccination was not sufficient to prevent clinical disease. Neuraminidase (NA) from PR8 was therefore included in the vaccine, since it is also present in approved influenza vaccines. A single administration of the NA vaccine was insufficient to give complete protection against influenza infection and achieved only 40% survival.Citation25 However, when both, HA and NA were targeted, a single vaccination sufficed to ensure 100% survival and prevent any weight loss, indicating that a combined vaccine could completely protect against clinical influenza infections.
Since the vaccination with an NP encoding RNActive® vaccine successfully induced T-cells, we asked whether this antigen which is more conserved between different influenza strainsCitation26-Citation28 could mediate protection against homologous and heterologous viral challenge. Mice were vaccinated three times with 80 μg of NP RNActive® at weeks 0, 3, and 6 and subsequently challenged with homologous PR8 (H1N1) or heterologous MB1 (H5N1) virus. This resulted in 100% survival upon lethal challenge with homologous PR8 virus and 80% survival upon lethal challenge with heterologous MB1 influenza (“bird flu”) virus 5 weeks after the last vaccination. T-cell depletion shortly before the challenge demonstrated that protection was mediated by T cells.
RNActive® Vaccines Are Also Active in Large Animals
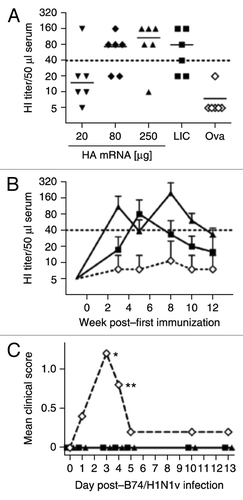
Ferrets are considered to be an appropriate animal species to predict immunogenicity of influenza vaccines in humans. The immunogenicity of HA derived from the clinical isolate Re6 (“swine flu”) was therefore encoded as RNActive® vaccine and analyzed in ferrets and compared with the licensed influenza vaccine Celvapan® that has a corresponding specificity. Ferrets were vaccinated i.d. at week 0 and week 1 with 20, 80, or 250 μg of the Re6HA RNActive® vaccine or 80 µg of an ovalbumin encoding RNActive® vaccine as negative control. Celvapan® was administered intramuscularly (i.m.) according to the recommended human schedule at week 0 and week 3. Two weeks after the last vaccination, ferrets immunized with 80 μg or 250 μg of Re6HA RNActive® vaccine showed HA-specific antibodies reaching median HI titers of ~1:40, comparable to the levels achieved with the licensed vaccine despite an only one week interval between prime and boost (). Animals vaccinated with ova mRNA did not raise significant HA-specific serum activity. In difference to the very stable HI titers in mice, HI titers decreased rather quickly at the same rates in all ferret groups including the Celvapan®-immunized groups (). A second boost in RNActive® vaccinated ferrets demonstrated boosterability, but again a subsequent decline of HI-titers was observed.
Figure 10. Immune effects of RNActive® vaccines in ferrets and pigs and protective efficacy in pigs. (A, B) Immunogenicity of RNActive® vaccines was assessed in six-month-old male ferrets (n = 6/group) that were immunized intradermally with 20, 80 or 250 µg of Re6HA RNActive® vaccine or 80 µg of Ova RNActive® vaccine, or intramuscularly with 500 µl of the licensed vaccine Celvapan® (LIC) which contains 7.5 µg C7HA. mRNA was injected at weeks 0, 1, and 6. Celvapan® was injected at weeks 0 and 3. (A) HI titers were measured 2 weeks after the booster vaccination (week 3 for RNActive® vaccine, week 5 for Celvapan®). (B) The kinetics of HI titers was recorded over the whole experiment for groups treated with 250 µg Re6HA, 80 µg ovalbumin (ova) or Celvapan (symbols are the same in panels A and B). Data are expressed as mean + s.d. for clarity and represent two independent experiments. (C) 2-mo-old seronegative pigs (n = 5/group) were immunized on days 0 and 21 with 250 µg of each Re6HA, Re6NA, and PR8NP RNActive® vaccine, 500 µl Mutagrip 2011/2012, or buffer. On day 16 post-immunization, animals were infected with 106.5 TCID50 of A/Bayern/74/2009 (B74/H1N1v) virus. Clinical symptoms were measured in a blinded fashion and were recorded over the ensuing 13 d (*: impaired general condition 5/5 buffer-treated animals, **: impaired general condition 4/5 buffer-treated animals). Further details in Petsch et al.25.
To investigate whether RNActive® vaccines are immunogenic in animals with a weight more similar to humans, 3 mo-old female domestic pigs were immunized twice intradermally 3 weeks apart with 250 μg of RNActive® encoding HA from influenza strain A/California/7/2009 (“C7”). Blood samples were taken 1 week before the first immunization, on the day of the second immunization, as well as 2 weeks and 4 weeks after the second immunization. The pigs that initially weighed 20–25 kg had reached a weight of ~50 kg by this time. All but one animal exhibited a 4-fold or greater rise in HI titers with peak HI titers of ~1:1200.Citation25 Similar to the results in ferrets, the immunogenicity of the RNActive® vaccine in pigs met the requirements for licensure.
However, since not all pigs had been seronegative when included in the previous experiment, the protective efficacy of RNActive® vaccination in pigs was assessed in animals with proven seronegativity for influenza virus.Citation25 Female domestic pigs (20–25 kg initial body weight, 2 mo age) were vaccinated intradermally on days 0 and 21 with RNActive® vaccines encoding HA, NA (both influenza strain Re6, “swine flu”) and NP (PR8) (250 μg each) or with a human dose (500 μl) of the licensed vaccine Mutagrip (2011/2012, Sanofi Pasteur) or with buffer. The mRNA vaccine combination was chosen, since results in mice indicated a better protection for a combinatorial approach and since the licensed split virion vaccines such as Mutagrip also contain significant amounts of NA and NP proteins.Citation29 Animals were challenged intranasally with 106.5 median tissue culture infective doses (TCID50) of Influenza A/Bayern/74/2009 (“B74/H1N1v”). All pigs in the negative control group (buffer) showed mild to moderate clinical signs of disease, which were recorded in a blinded fashion 1–5 d post-infection, including disturbed general state of health and occasional sneezing or bleeding of the nose, whereas none of the immunized pigs showed signs of disease during this observation period (). Quantitative RT-PCR done on nasal swabs revealed reduced virus genome copies in mRNA-vaccinated (290- and 698-fold reduction at 6 and 7 d post-infection) and Mutagrip-vaccinated pigs (87- and 48-fold on 6 and days post-infection) compared with the buffer control groupCitation25 indicating faster clearance of the virus in immunized pigs. The experiment convincingly established efficacy of RNActive® vaccines also in large animals.
Intradermal vs. Intranodal Administration of RNActive® Vaccines
As already mentioned above, an HA construct encoded by mRNA prepared in a manner different from the one described here did not induce immune responses against HA after intradermal application.Citation18 Intranodal administration was required to achieve immunogenicity of HA mRNA, an approach that was also chosen to demonstrate activity of mRNA constructs encoding immunostimulatory molecules in addition to the antigen.Citation30,Citation31 To assess whether intranodal administration would be advantageous over intradermal administration also for RNActive® vaccines, a test experiment was performed and mice were immunized on days 0, 3, 6, and 9 with two doses (16 µg or 32 µg) of an ovalbumin encoding RNActive® vaccine. Due to volume restrictions, the higher dose was injected into two different lymph nodes. The humoral immune response was measured on day 15. Somewhat contrary to our expectation, intradermal administration resulted in a stronger IgG1 response at both dosages than intranodal administration (). A similar, though less pronounced trend was also observed for IgG2a antibodies at both doses. Differing from the humoral immune response, the frequency of IFNγ and TNFα secreting CD8+ T cells was similar between intradermal and intranodal administration at both, the low and the high dose (). In the mice vaccinated intranodally, anti-tumor activity after intranodal administration did not appear to have advantages over intradermal administration of RNActive® vaccines, but this observation awaits further testing.
Figure 11. Intradermal vs. intranodal administration of RNActive® vaccines. Mice were vaccinated with 16 or 32 µg of RNActive® vaccine encoding ovalbumin or HA from PR8 as control either intradermally or intranodally on days 0, 3, 6, and 9. A maximum volume of 10 µl could be injected into lymph nodes, for which reason the 32 µg dose was administered to two different lymph nodes. (A and B) On day 15, serum samples were taken and IgG1 and IgG2a antibody titers against ovalbumin were determined (method described in Fotin-Mleczek et al.Citation20). (A) 16 µg dose, (B) 32 µg dose. (C and D) Splenocytes were also isolated from vaccinated mice and the frequency of IFNγ+ or TNFα+ CD8+ T cells determined by intracellular cytokine staining after stimulation with the SIINFEKL peptide from ovalbumin or HA derived epitopes as control.Citation20,Citation25 (C) 16 µg dose, (D) 32 µg dose
Clinical Experiences with RNActive® Vaccines
Based on encouraging preclinical results, the decision was taken to advance the self-adjuvanted RNActive® vaccines to clinical testing. Two indications were chosen: castrate-resistant, non-metastatic or mildly symptomatic metastatic prostate carcinoma and stage IIIB/IV disease of NSCLC (non-small cell lung cancer) that was at least stable after first-line platinum-based chemotherapy. A cocktail of the four tumor associated antigens PSA (prostate specific antigen), PSCA (prostate stem cell antigen), PSMA (prostate specific membrane antigen) and STEAP1 (six transmembrane epithelial antigen of the prostate 1) was selected for the first-in-man phase I/ IIa study in prostate cancer patients and designated CV9103. For the NSCLC cocktail CV9201, five tumor-associated antigens were chosen: MAGE-C1, MAGE-C2, NY-ESO-1, survivin, and 5T4.Citation32,Citation33
In the prostate-carcinoma study with CV9103, an unexpectedly high level of cellular immunogenicity was observed. Antigen-specific T cells were detected in around 80% of prostate carcinoma patients independent of their HLA-background.Citation34 Importantly, the majority of immune responders, around 60%, reacted against multiple antigens. Immune responses were detected against all antigens independent of their cellular localization. Antigen-unspecific B cells were increased in around 75% of patients, a tendency for increased activation was also observed for natural killer (NK)-cells. Favorable clinical courses were observed (Kuebler et al., manuscript in preparation)
As in the prostate-carcinoma study, the number of vaccinations in the phase I/IIa trial NSCLC patients was limited to five intradermal immunizations, but due to the more advanced disease a more intensive vaccination schedule had to be chosen. Similar to CV9103, the NSCLC cocktail CV9201 showed a favorable safety profile. More specific information was gained on the B-cell activation. A significant increase of pre-germinal center B cells (pGCB) by a factor of at least 2 was observed in about 60% of the patients. This correlated with an increase of total CD4+ effector T cells during treatment. An antigen specific humoral and cellular immune response was determined in roughly two thirds of the treated patients. Antigen-specific B cells of a memory subtype were detected in one exemplary patient. Together, more than 80% of the treated NSCLC patients had a detectable antigen specific immune response and/or an increase in germinal center B cells (Sebastian et al. manuscript in preparation).
Overall, both RNActive® vaccine cocktails showed very high immunogenicity rates in patients with prostate carcinoma or NSCLC. Importantly, a response against multiple antigens was seen in the majority of immune responders in both studies. Recent evidenceCitation35 suggests that this is indicative of a substantially improved overall survival in vaccinated patients. Manuscripts on the clinical and immunological details are presently prepared. Based on these encouraging data, a controlled phase II trial was initiated in a similar population of patients with prostate carcinoma.
Conclusion
For decades, efforts to generate nucleotide based vaccines have focused on DNA. However, in recent years, it has become clear that RNA can be safely worked with in laboratory and clinical settings and does not require the cold chain. Moreover, it can be produced in a highly flexible and versatile process that can be upscaled to industrial requirements with only comparatively modest investments. The format presented here, self-adjuvanted RNActive® vaccines, induces balanced immune responses comprising humoral and cellular responses, effector and memory responses as well as activation of important subpopulations of immune cells, such as Th1 and Th2 cells or pre-germinal center B cells and germinal center B cells. In contrast to DNA vaccines, self-adjuvanted RNActive® vaccines activate the immune system in a different way that pivotally involves activation of an important pattern recognition receptor, TLR7, by RNActive® vaccines. The first clinical studies performed indicate a favorable safety profile of RNActive® vaccines. Furthermore, the immunological effects induced in preclinical experiments by RNActive® vaccines could be translated to the human situation. It is hoped that the impressive activity observed in therapeutic models of tumor diseases as well as prophylactic models of infectious diseases will also be mirrored by equally impressive effects on human diseases. If successful, RNActive® vaccines will be able to do more than classical vaccines at considerably reduced costs. RNActive® vaccines would thus represent a truly disruptive technology with wide ranging implications for several clinical areas.
| Abbreviations: | ||
| A | = | adenosine |
| C | = | cytosine |
| C7 | = | influenza strain A/California/7/2009 |
| CEA | = | carcinoembryonic antigen |
| CTL | = | cytolytic T cells |
| DNA | = | deoxyribonucleic acid |
| G | = | guanosine |
| HA | = | hemagglutinin |
| HI | = | HA inhibition assay |
| i.d. | = | intradermally |
| i.m. | = | intramuscularly |
| i.n | = | intranodally |
| MHC | = | major histocompatibility complex |
| mRNA | = | messenger RNA |
| NA | = | neuraminidase |
| NK cells | = | natural killer cells |
| NP | = | nucleoprotein |
| NSCLC | = | non-small cell lung cancer |
| ORF | = | open reading frame |
| ova | = | ovalbumin |
| pGCB | = | pre-germinal center B cells |
| PSMA | = | prostate specific membrane antigen |
| RNA | = | ribonucleic acid |
| Th1 | = | T-helper 1 cell |
| Th2 | = | T-helper 2 cell |
| TLR7 | = | toll-like receptor 7 |
| TLR8 | = | toll-like receptor 8 |
| U | = | uridine |
| UTR | = | untranslated region |
Acknowledgments
The authors would like to acknowledge the long, fruitful collaboration with Prof. Lothar Stitz (Friedrich-Löffler-Insitut), without whose expertise and constant good advice it would not have been possible to achieve many of the results reported here.
Disclosure of Potential Conflicts of Interest
The authors are employees of CureVac GmbH, Tübingen, Germany.
References
- Rappuoli R, Mandl CW, Black S, De Gregorio E. Vaccines for the twenty-first century society. Nat Rev Immunol 2011; 11:865 - 72; PMID: 22051890
- Wolff JA, Malone RW, Williams P, Chong W, Acsadi G, Jani A, et al. Direct gene transfer into mouse muscle in vivo. Science 1990; 247:1465 - 8; http://dx.doi.org/10.1126/science.1690918; PMID: 1690918
- Martinon F, Krishnan S, Lenzen G, Magné R, Gomard E, Guillet JG, et al. Induction of virus-specific cytotoxic T lymphocytes in vivo by liposome-entrapped mRNA. Eur J Immunol 1993; 23:1719 - 22; http://dx.doi.org/10.1002/eji.1830230749; PMID: 8325342
- Conry RM, LoBuglio AF, Wright M, Sumerel L, Pike MJ, Johanning F, et al. Characterization of a messenger RNA polynucleotide vaccine vector. Cancer Res 1995; 55:1397 - 400; PMID: 7882341
- Hoerr I, Obst R, Rammensee HG, Jung G. In vivo application of RNA leads to induction of specific cytotoxic T lymphocytes and antibodies. Eur J Immunol 2000; 30:1 - 7; http://dx.doi.org/10.1002/1521-4141(200001)30:1<1::AID-IMMU1>3.0.CO;2-#; PMID: 10602021
- Pascolo S. Messenger RNA-based vaccines. Expert Opin Biol Ther 2004; 4:1285 - 94; http://dx.doi.org/10.1517/14712598.4.8.1285; PMID: 15268662
- Pascolo S. Vaccination with messenger RNA. Methods Mol Med 2006; 127:23 - 40; PMID: 16988444
- Ulmer JB, Mason PW, Geall A, Mandl CW. RNA-based vaccines. Vaccine 2012; 30:4414 - 8; http://dx.doi.org/10.1016/j.vaccine.2012.04.060; PMID: 22546329
- Gust TC, Zenke M. RNA transfer and its use in dendritic cell-based immunotherapy. Expert Opin Biol Ther 2005; 5:173 - 81; http://dx.doi.org/10.1517/14712598.5.2.173; PMID: 15757379
- Probst J, Fotin-Mleczek M, Schlake T, Thess A, Kramps T, Kallen KJ. Messenger RNA vaccines. Springer Verlag Wien, 2012.
- Schlake T, Thess A, Fotin-Mleczek M, Kallen KJ. Developing mRNA-vaccine technologies. RNA Biol 2012; 9:1319 - 30; http://dx.doi.org/10.4161/rna.22269; PMID: 23064118
- Carralot JP, Probst J, Hoerr I, Scheel B, Teufel R, Jung G, et al. Polarization of immunity induced by direct injection of naked sequence-stabilized mRNA vaccines. Cell Mol Life Sci 2004; 61:2418 - 24; http://dx.doi.org/10.1007/s00018-004-4255-0; PMID: 15378210
- Scheel B, Braedel S, Probst J, Carralot JP, Wagner H, Schild H, et al. Immunostimulating capacities of stabilized RNA molecules. Eur J Immunol 2004; 34:537 - 47; http://dx.doi.org/10.1002/eji.200324198; PMID: 14768059
- Scheel B, Teufel R, Probst J, Carralot JP, Geginat J, Radsak M, et al. Toll-like receptor-dependent activation of several human blood cell types by protamine-condensed mRNA. Eur J Immunol 2005; 35:1557 - 66; http://dx.doi.org/10.1002/eji.200425656; PMID: 15832293
- Scheel B, Aulwurm S, Probst J, Stitz L, Hoerr I, Rammensee HG, et al. Therapeutic anti-tumor immunity triggered by injections of immunostimulating single-stranded RNA. Eur J Immunol 2006; 36:2807 - 16; http://dx.doi.org/10.1002/eji.200635910; PMID: 17013976
- Banerjee AK. 5′-terminal cap structure in eucaryotic messenger ribonucleic acids. Microbiol Rev 1980; 44:175 - 205; PMID: 6247631
- Wickens M. How the messenger got its tail: addition of poly(A) in the nucleus. Trends Biochem Sci 1990; 15:277 - 81; http://dx.doi.org/10.1016/0968-0004(90)90054-F; PMID: 1974368
- Kreiter S, Selmi A, Diken M, Koslowski M, Britten CM, Huber C, et al. Intranodal vaccination with naked antigen-encoding RNA elicits potent prophylactic and therapeutic antitumoral immunity. Cancer Res 2010; 70:9031 - 40; http://dx.doi.org/10.1158/0008-5472.CAN-10-0699; PMID: 21045153
- Kuhn AN, Diken M, Kreiter S, Vallazza B, Türeci O, Sahin U. Determinants of intracellular RNA pharmacokinetics: Implications for RNA-based immunotherapeutics. RNA Biol 2011; 8:35 - 43; http://dx.doi.org/10.4161/rna.8.1.13767; PMID: 21289486
- Fotin-Mleczek M, Duchardt KM, Lorenz C, Pfeiffer R, Ojkić-Zrna S, Probst J, et al. Messenger RNA-based vaccines with dual activity induce balanced TLR-7 dependent adaptive immune responses and provide antitumor activity. J Immunother 2011; 34:1 - 15; http://dx.doi.org/10.1097/CJI.0b013e3181f7dbe8; PMID: 21150709
- Lorenz C, Fotin-Mleczek M, Roth G, Becker C, Dam TC, Verdurmen WP, et al. Protein expression from exogenous mRNA: uptake by receptor-mediated endocytosis and trafficking via the lysosomal pathway. RNA Biol 2011; 8:627 - 36; http://dx.doi.org/10.4161/rna.8.4.15394; PMID: 21654214
- Hornung V, Guenthner-Biller M, Bourquin C, Ablasser A, Schlee M, Uematsu S, et al. Sequence-specific potent induction of IFN-alpha by short interfering RNA in plasmacytoid dendritic cells through TLR7. Nat Med 2005; 11:263 - 70; http://dx.doi.org/10.1038/nm1191; PMID: 15723075
- Diken M, Kreiter S, Selmi A, Britten CM, Huber C, Türeci O, et al. Selective uptake of naked vaccine RNA by dendritic cells is driven by macropinocytosis and abrogated upon DC maturation. Gene Ther 2011; 18:702 - 8; http://dx.doi.org/10.1038/gt.2011.17; PMID: 21368901
- Fotin-Mleczek M, Zanzinger K, Heidenreich R, Lorenz C, Thess A, Duchardt KM, et al. Highly potent mRNA based cancer vaccines represent an attractive platform for combination therapies supporting an improved therapeutic effect. J Gene Med 2012; 14:428 - 39; http://dx.doi.org/10.1002/jgm.2605; PMID: 22262664
- Petsch B, Schnee M, Vogel AB, Lange E, Hoffmann B, Voss D, et al. Protective efficacy of in vitro synthesized, specific mRNA vaccines against influenza A virus infection. Nat Biotechnol 2012; 30:1210 - 6; http://dx.doi.org/10.1038/nbt.2436; PMID: 23159882
- McMichael AJ, Gotch FM, Noble GR, Beare PA. Cytotoxic T-cell immunity to influenza. N Engl J Med 1983; 309:13 - 7; http://dx.doi.org/10.1056/NEJM198307073090103; PMID: 6602294
- Ulmer JB, Donnelly JJ, Parker SE, Rhodes GH, Felgner PL, Dwarki VJ, et al. Heterologous protection against influenza by injection of DNA encoding a viral protein. Science 1993; 259:1745 - 9; http://dx.doi.org/10.1126/science.8456302; PMID: 8456302
- Rimmelzwaan GF, Fouchier RA, Osterhaus AD. Influenza virus-specific cytotoxic T lymphocytes: a correlate of protection and a basis for vaccine development. Curr Opin Biotechnol 2007; 18:529 - 36; http://dx.doi.org/10.1016/j.copbio.2007.11.002; PMID: 18083548
- Chaloupka I, Schuler A, Marschall M, Meier-Ewert H. Comparative analysis of six European influenza vaccines. Eur J Clin Microbiol Infect Dis 1996; 15:121 - 7; http://dx.doi.org/10.1007/BF01591484; PMID: 8801083
- Van Lint S, Goyvaerts C, Maenhout S, Goethals L, Disy A, Benteyn D, et al. Preclinical evaluation of TriMix and antigen mRNA-based antitumor therapy. Cancer Res 2012; 72:1661 - 71; http://dx.doi.org/10.1158/0008-5472.CAN-11-2957; PMID: 22337996
- Kreiter S, Diken M, Selmi A, Diekmann J, Attig S, Hüsemann Y, et al. FLT3 ligand enhances the cancer therapeutic potency of naked RNA vaccines. Cancer Res 2011; 71:6132 - 42; http://dx.doi.org/10.1158/0008-5472.CAN-11-0291; PMID: 21816907
- Sebastian M, von Boehmer L, Zippelius A, Mayer F, Reck M, Atanackovic D, et al. Messenger RNA vaccination in NSCLC: Findings from a phase I/IIa clinical trial. J Clin Oncol 2011; 29:2584
- Sebastian M, von Boehmer L, Zippelius A, Mayer F, Reck M, Atanackovic D, et al. Messenger RNA vaccination and B-cell responses in NSCLC patients. J Clin Oncol 2012; 30:2573; PMID: 22565006
- Kübler H, Maurer T, Stenzl A, Feyerabend S, Steiner U, Schostak M, et al. Final analysis of a phase I/IIa study with CV9103, an intradermally administered prostate cancer immunotherapy based on self-adjuvanted mRNA. J Clin Oncol 2011; 29:4535
- Walter S, Weinschenk T, Stenzl A, Zdrojowy R, Pluzanska A, Szczylik C, Staehler M, Brugger W, Dietrich PY, Mendrzyk R, et al. Multipeptide immune response to cancer vaccine IMA901 after single-dose cyclophosphamide associates with longer patient survival. Nat Med 2012; 18:1254 - 61; http://dx.doi.org/10.1038/nm.2883; PMID: 22842478