Abstract
The emergence of the pandemic H1N1 strain of influenza in 2009 was associated with a unique w-shaped age-related susceptibility curve, with higher incidence of morbidity and mortality among young persons and lower incidence among older persons, also observed during the 1918 influenza pandemic. Pre-existing H1N1 antibodies were not cross-reactive with the prior seasonal vaccine, forcing influenza experts to scramble to develop a new vaccine specific for the pandemic virus. We hypothesized that response to T-cell epitopes that are cross-conserved between pandemic H1N1 and the 2008 seasonal influenza vaccine strains might have contributed to partial protection from clinical illness among older adults, despite the lack of cross-reactive humoral immunity. Using immunoinformatics tools, we previously identified hemagglutinin and neuraminidase epitopes that were highly conserved between seasonal and pandemic H1N1. Here, we validated predicted CD4+ T-cell epitopes for their ability to bind HLA and to stimulate interferon-γ production in peripheral blood mononuclear cells from a cohort of donors presenting with influenza-like illness during the 2009 pandemic and a separate cohort immunized with trivalent influenza vaccine in 2011. A limited-epitope heterologous DNA-prime/peptide-boost vaccine composed of these sequences stimulated immune responses and lowered lung viral loads in HLA DR3 transgenic mice challenged with pandemic 2009 H1N1 influenza. Cross-priming with conserved influenza T-cell epitopes such as these may be critically important to T cell-mediated protection against pandemic H1N1 in the absence of cross-protective antibodies.
Introduction
As demonstrated by the 2009 H1N1 pandemic, novel influenza viruses can spread rapidly when limited immunity exists within the worldwide population. Due to the variability of influenza virus strains from year to year, a new vaccine based on the circulating strains (and their HA and NA proteins) is made available as often as every year, if necessary. These reformulated versions of the seasonal influenza virus are not always protective, due to a range of factors including genetic drift. There is an urgent need to develop novel influenza vaccines that will provide more universal protection against influenza infection in a shorter time frame than is currently feasible.
Because of the unique epidemiology of 2009 H1N1 influenza, we hypothesized that T-cell responses to cross-reactive epitopes might have contributed to diminished reports of influenza-like illnesses and confirmed infection among older adults in the absence of cross-reactive humoral immunity. There is considerable support for this hypothesis in published studies involving exposure or vaccination and heterotypic challenge in animal models.Citation1-Citation4 One explanation for the unusual age distribution is that, similar to the 1918 epidemic and again as occurred in 1977,Citation5,Citation6 older individuals might have established a cross-reactive cell-mediated immune response to novel H1N1 due to having been vaccinated or exposed to circulating influenza H1N1.Citation7 While cross-reactive T-cell responses (in the absence of a cross-reactive humoral immune response) may not have provided complete protection against infection, it is possible that the severity of the illness was reduced, leading to a lower hospitalization rate and lower reports of H1N1 in this age group, as was observed in a case-control study from Mexico.Citation8
We previously performed an immunoinformatics analysis to define cross-conserved HA and NA CD4+ T-cell epitopes between the emerging pandemic strain and the prior seasonal influenza vaccine strains, using the A/California/04/2009 sequence as a departure point. The analysis was performed in March 2009, immediately following publication of the virus sequence.Citation9,Citation10 Here we present results from in vitro studies confirming these predictions. The cross-conserved epitopes bound to multiple HLA and were antigenic in recall assays using peripheral blood mononuclear cells (PBMCs) from a patient presenting with influenza-like illness during the recent pandemic and in a cohort of 2011 trivalent influenza virus (TIV) vaccinees. Vaccination with cross-conserved influenza epitopes stimulated de novo T-cell responses that lower viral burden in HLA transgenic mice, despite an absence of vaccine-induced antibodies. These studies appear to support the hypothesis that cross-protective T-cell responses might have played a role in reducing influenza morbidity and mortality in humans.
Results
HLA binding properties of cross-conserved H1N1 influenza epitopes
In a previous study, we identified peptide sequences from 2008–2009 influenza A seasonal vaccine HA and NA antigens that are cross-conserved with the corresponding antigens from 2009 pandemic H1N1 and potentially immunogenic, using the EpiMatrix T cell epitope mapping algorithm.Citation9 These sequences were assayed in vitro for their capacity to bind multiple HLA types, including DRB1*0101, DRB1*0301, DRB1*0401, DRB1*0701, DRB1*1101, and DRB1*1501. Ninety percent of the peptides bound as predicted to at least 3 HLA alleles, 70% to at least 4 HLA alleles, 60% to at least 5, and 10% to all 6. Surprisingly, peptide HA_112–129 did not bind any HLA despite immunoinformatic predictions it would bind all assayed alleles except DRB1*0701. In our experience testing thousands of peptides predicted to bind HLA, this is a singularly unusual finding. It may be explained by self-association, which would interfere with HLA interactions no matter what allele is assayed. Given this highly unusual result, we exclude the peptide from the following summary of binding results for the group of peptides.
Of the 54 peptide-HLA binding interactions assayed, 18.5% were very high affinity, 22.2% high affinity, 22.2% moderate affinity, 9.3% low affinity, 1.9% very low affinity, and no measurable affinity was observed for 25.9% (). The concordance of computational predictions and binding assay results was evaluated with classification of peptide-HLA binding pairs as either true positive, false positive, true negative, or false negative. A non-binder was defined as a confirmed true negative prediction if the peptide had an EpiMatrix Z-score that was lower than the defined cut-off (1.64) for its associated HLA allele. Positive predictions were defined as epitopes scoring ≥1.64 on the EpiMatrix Z-scale and binding HLA at any affinity. Overall, the concordance with predictions (both positive and negative) was 76%. With respect to each allele assayed, the values are 78% for DRB1*0101, 89% for DRB1*0301, 67% for DRB1*0401, 78% for DRB1*0701, 89% for DRB1*1101, and 56% for DRB1*1501. These HLA-binding and epitope prediction results are consistent with previously published studies using the same algorithms and assay conditions.Citation11
Figure 1. HLA DR binding affinities for immunoinformatic-predicted H1N1 influenza cross-conserved epitopes. Peptide identifiers and sequences are noted in the first and second columns, respectively. IC50 values in μM units were calculated from curves fitted to dose-dependence competition binding data for each peptide-HLA DR allele pair. Peptide binding affinity is shown according to the following classification: IC50 < 1 µM (black), 1 µM < IC50 < 10 µM (dark gray), 10 µM < IC50 < 50 µM (gray), 50 µM < IC50 < 100 µM (light gray), IC50 > 100 µM (lightest gray). IC50 values too high to accurately measure under binding conditions tested are considered non-binders (NB; shown in white cells).
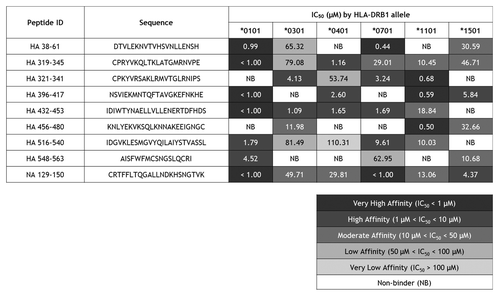
Epitope-specific IFNγ responses: Acute ILI subjects
Ex vivo IFNγ ELISpot assays were performed using PBMCs from six patients ages 18 to 65 y old, who were admitted to the Rhode Island Hospital with influenza-like illness between November 2009 and February 2010 (). RT-PCR detection of influenza in nasal washes confirmed pandemic H1N1 infection in one subject and no influenza infection in the others. ELISpot responses were considered positive when (1) the number of IFNγ spot-forming cells exceeded 50 per million PBMCs cultured, (2) spot counts were at least twice background, and (3) spot counts were statistically different from “no stimulus” measurements (P < 0.05). In the one pandemic H1N1 case, significant influenza-specific IFNγ recall responses were observed when PBMCs were stimulated ex vivo with a pool of epitope peptides and with individual peptides in an ELISpot assay (). No peptide-specific ex vivo interferon-γ (IFNγ) responses were detected in PBMC samples taken from the five other subjects (data not shown). Although three of these subjects received the 2008–2009 TIV vaccine, the absence of an ex vivo response was not unexpected as we previously found that vaccine-specific T cells are found at low frequency and a period of expansion in culture is required to detect them.Citation11
Table 1. Characteristics of cohort presenting with ILI during 2009 H1N1 pandemic
Figure 2. Ex vivo human IFNγ responses elicited by cross-conserved epitopes in H1N1 influenza infection. Cross-conserved epitopes were assayed for T cell reactivity by IFNγ ELISpot assay, using PBMCs isolated from patients that presented with influenza-like illness at Rhode Island Hospital. Data from the only H1N1 influenza-positive subject are presented. ELISpot responses were considered positive if three criteria were met: (1) spot-forming cells (SFC) per million PBMCs were at least 50 over background; (2) SFC per million PBMCs were at least 2-fold over background; and (3) antigen-stimulated SFC numbers were statistically different (Student’s t-test; *p < 0.05, **p < 0.01) from non-stimulated counts. Data are the mean SFC over background per million PBMCs that secrete IFNγ in response to individual and pooled influenza HA and NA cross-conserved epitopes are presented. The 50 SFC over background per million PBMCs cutoff is denoted by the dotted line.
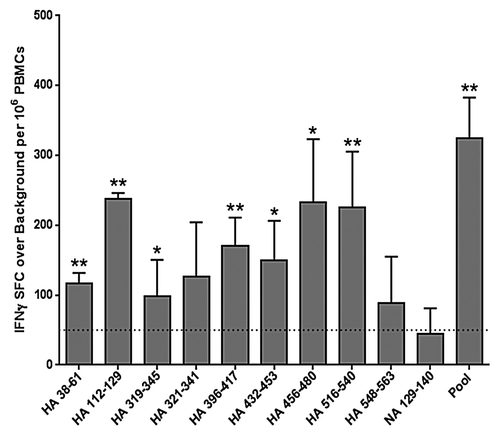
Epitope-specific IFNγ responses: 2011 TIV-immunized subjects
To better determine the antigenic potential of cross-conserved H1N1 peptides, we expanded low frequency influenza-specific T cells over a nine-day period using PBMC samples collected before and three weeks following 2011 TIV vaccination of 16 young and elderly adult subjects. Cells were expanded by stimulation with a pool of HA and NA cross-conserved peptides and then re-stimulated with pooled or individual peptides for measurement of IFNγ production by ELISpot assay. All subjects responded to the pool of cross-conserved peptides post-immunization by the criteria for a positive ELISpot response described above, apart from two subjects, one of which nearly met all three criteria (; Table S1). Two additional subjects did not respond prior to immunization but significant responses were observed in the group overall, suggesting that T-cell precursors for this set of epitopes do exist. The average spot forming cell count following T-cell expansion did not significantly increase post-immunization.
Figure 3. Antigen-specific human IFNγ responses elicited by cross-conserved epitopes before and after 2010–2011 TIV vaccination. Cross-conserved epitopes were assayed for T cell reactivity by cultured IFNγ ELISpot assay using PBMCs isolated from normal human donors before and three weeks after TIV vaccination (n = 16). Assays were performed following a nine-day T cell expansion after stimulation with cross-conserved influenza epitopes. The numbers of SFC over background per million PBMCs that secrete IFNγ in response to individual and pooled influenza HA and NA cross-conserved epitopes are presented. Individual subject average responses are represented by dots and the average response across subjects by white bars. The 50 SFC over background per million PBMCs cutoff is denoted by the dotted line.
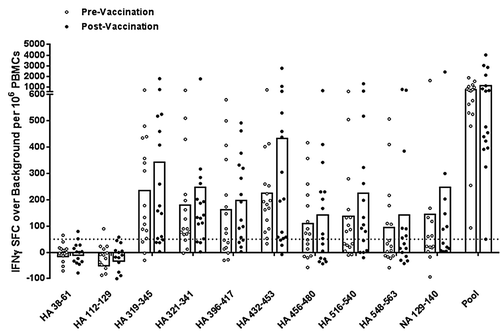
In individual peptide stimulations, eight out of ten (80%) epitopes stimulated IFNγ production greater than 50 spot forming cells per million PBMCs prior to immunization and vaccination boosted these epitope-specific responses. Differences between pre- and post-immunization IFNγ production were not significant by the Wilcoxon matched-pairs signed rank test, except for HA_456–480 (P < 0.05). While we expected to observe significant differences between these time points, it is very likely that they were lost in the course of in vitro expansion and that stimulation at a lower peptide concentration would preserve differences.
Two peptides (HA_38–61 and HA_112–129) stimulated no significant responses in any subjects. The small cohort size of the study with limited HLA diversity may explain this result. Alternatively, these sequences may stimulate type 1 helper T cell cytokines other than IFNγ, such as interleukin-2 and tumor necrosis α. Thus, positive responses may be observed in a larger cohort with broader HLA coverage, comparing multiple cytokine responses. Notably, while both these peptides stimulated no IFNγ response among vaccinated subjects even after a period of T-cell expansion, robust ex vivo responses were observed in the one subject infected during the 2009 pandemic (). While trends, let alone conclusions, cannot be drawn from a single subject, we can speculate that live virus infection stimulates different T cell specificities than TIV vaccination.
Multi-epitope DNA vaccine construction
Following in vitro antigenicity testing, we set out to evaluate the in vivo immunogenicity of cross-conserved H1N1 influenza epitopes using a heterologous DNA-prime/peptide-boost vaccination strategy. To design the DNA vaccine, epitope sequences were initially joined end to end in a random order. To avoid production of neo-epitopes at epitope junctions, the VaccineCAD algorithm was used to re-arrange epitopes in an order that diminishes potential junctional immunogenicity. This algorithm iteratively rearranges strings of epitopes while assessing junctional immunogenicity, prioritizing those sequences that contain the fewest junctional epitopes. The default order contained significant predicted immunogenicity at a single junction (EpiMatrix score ~10). Re-ordering of epitopes by VaccineCAD yielded a sequence with minimized junctional immunogenicity (EpiMatrix scores < −3), well below the threshold for potential immunogenicity.
Immunogenicity and efficacy of cross-conserved H1N1 influenza epitopes
We evaluated the cross-conserved H1N1 influenza epitopes for immunogenicity and efficacy in HLA DR3 transgenic mice. The HLA transgenic mouse model represents an improvement over wild-type mice (and other animal models), because animal and human MHC present different T-cell epitopes.Citation12 MHC class II-mediated cellular immunity in the mouse MHC II knockout, HLA DR3 knock-in strain is completely restricted by human HLA and not by mouse MHC.Citation13 Mice were injected intramuscularly with the DNA vaccine delivered by electroporation, three times, over two-week intervals. Two weeks later, they were boosted twice subcutaneously with corresponding epitope peptides formulated in incomplete Freund’s adjuvant (IFA), with immunostimulatory CpG ODN 1826, CL097 and muramyl dipeptide adjuvants, over a two-week interval. A control group of mice received empty vector in the DNA-prime phase and peptide-free IFA emulsion in the boost phase.
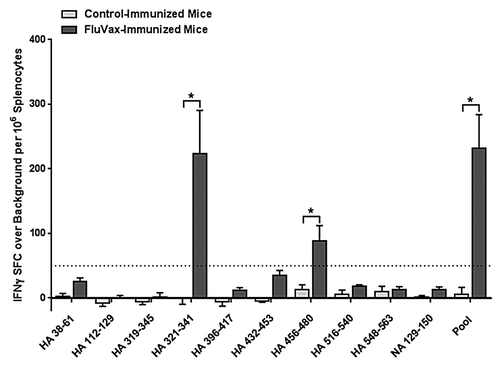
Three weeks following the final immunization, splenocytes were isolated to measure T-cell responses to individual epitopes from individual mice by IFNγ ELISpot assay (n = 6/group). ELISpot responses were considered positive according to the same criteria used in human studies described above. Immunization of DR3 transgenic mice stimulated statistically significant (Student t-test, P < 0.01) T-cell responses to the pool of epitopes and to two out of 10 individual epitopes (20%): HA_321–341 and HA_456–480 (). Out of the 10 epitopes evaluated, seven are predicted to bind HLA DR3Citation9; thus, two out of seven (29%) are true positive epitopes. By more relaxed criteria, epitopes HA_38–61 and HA_432–453 may also be considered positive as they stimulate responses that are >20 SFC per million splenocytes, > 2 × background and statistically distinct from background. Both these sequences are predicted to bind HLA DR3, thus raising the number of immunogenic epitopes to 40% of those tested and 57% positively predicted sequences. Three of the ten epitopes (HA_319–345, HA_396–417, HA_548–563) are not predicted to bind HLA DR3 and were not immunogenic in this study. In sum, seven out of 10 (70%) of immune responses measured are in concordance with the immunoinformatic predictions.
Figure 4. Cell-mediated response to immunization of HLA DR3 transgenic mice with cross-conserved H1N1 influenza class II HLA epitopes. Mice were primed with plasmid DNA vaccine and boosted with peptides comprising cross-conserved H1N1 influenza class II HLA epitopes or vaccine vehicle containing no epitopes. Epitope-specific cellular responses in splenocyte cultures for individual and pooled epitopes were measured by IFNγ ELISpot assay. Data are the mean spot forming cells (SFC) per million splenocytes ± standard deviation derived for 6 mice treated comparably. Individual epitope and pooled epitope responses in vaccinated mice showing statistical significance (Student’s t-test) when compared with controls are indicated: *p < 0.05. A solid line denotes the 50 SFC over background per million splenocytes cutoff.
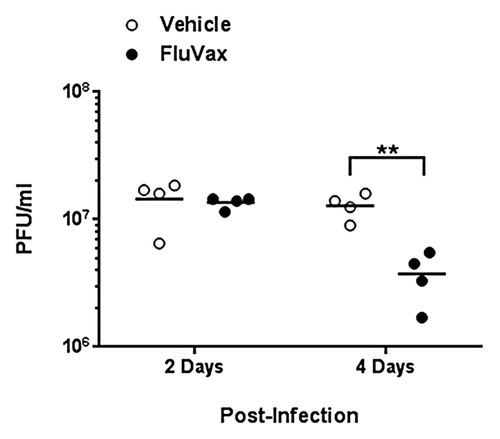
Next, we set out to evaluate vaccine efficacy against A/California/07/2009 challenge in DR3 transgenic mice. Three weeks following the final immunization, 16 mice in each group were challenged intranasally with 106 PFU of virus. Mice in both groups became ill and lost >20% of their pre-challenge weight by day 7 post-infection and were sacrificed. While it was not surprising that there was no difference in morbidity between immunized and naïve mice, as only two to four epitopes were immunogenic, we did detect differences in lung viral titers. Viral burden as measured by plaque assay differed significantly between the two groups four days post-infection (Student t-test, P < 0.01). Lung viral titers were 70% lower in immunized mice four days following infection; no significant difference was observed two days prior (). Notably, as vaccine-induced antibodies specific for neither whole HA antigen nor the peptide immunogens was observed by ELISA (data not shown), efficacy was apparently cell-mediated only. Thus, we conclude that this minimal-epitope based H1N1 influenza epitope-based vaccine is immunogenic and partially efficacious.
Figure 5. Epitope-driven vaccine reduces lung viral titers in pandemic H1N1 influenza infection of HLA DR3 transgenic mice. Mice that were primed with plasmid DNA vaccine and boosted with peptides comprising cross-conserved H1N1 influenza class II HLA epitopes or vaccine vehicle containing no epitopes were infected with 106 PFU A/California/07/2009. Data are the viral burden of four mice from each group at two and four days post-infection as determined by viral plaque assay. Individual titers are denoted by filled dots and average titers by horizontal lines. Titers in vaccinated mice showing statistical significance (Student t-test) when compared with controls are indicated: **p < 0.01
Discussion
Leading up to the present study, we used EpiMatrix, a T-cell epitope prediction and sequence comparison tool, to discover cross-conserved putative T-cell epitope sequences among the three hemagglutinin (HA) and neuraminidase (NA) proteins contained in 2008–2009 TIV and their counterparts in pandemic 2009 H1N1 influenza virus (A/California/04/2009).Citation9 We found greater than 50% conservation of helper T and CTL epitopes between A/California/04/2009 and TIV HA for “supertype” HLA alleles, whereas conservation was lower among NA epitopes. A total of 16 promiscuous helper T-cell epitopes are contained in the A/California/04/2009 HA sequence, of which nine (56%) were 100% conserved in the 2008–2009 influenza vaccine strain H1N1 HA; 81% were either identical or had one conservative amino acid substitution. Similarly, 50% of predicted CTL epitopes found in A/California/04/2009 HA also were found in TIV HA sequences.Citation3 Here, we confirmed immunoinformatic predictions in experiments designed to evaluate the capacity of the nine highest conserved helper T cell HA epitopes and the most conserved NA epitope to bind HLA and stimulate immune recall and de novo T-cell responses.
Recognition of the cross-conserved epitopes by HLA and their immunoreactivity in exposed and in vaccinated subjects demonstrated in this study does not imply that they are necessarily protective sequences. Rather, it confirms the existence of cross-conserved T cells and suggests that further evaluation of the contributions of these cross-conserved epitopes to protection against infection, morbidity and mortality in the next pandemic, should one occur, would be entirely worthwhile. This is further supported by the vaccination and challenge study reported here. Despite the fact that immunized mice had a very restricted immune response (2 to 4 of 7 potential epitopes were immunogenic), reduced viral titers were observed. Our selection of HA and NA epitopes only for this study was based on our interest in determining whether cross-conserved T-cell responses to the proteins usually contained in seasonal influenza vaccine would protect against challenge. However, it is well known that conventional egg-based vaccine production results in a product that contains other influenza proteins. Immune response to residual internal proteins, which are highly cross-conserved between influenza strains, has been described.Citation14 HLA restriction of this animal model limited the number of epitopes that were potentially immunogenic as only two to four of seven induced T-cell response following vaccination. Had additional epitopes from internal proteins been included in this minimal epitope-based vaccine, greater protection against viral challenge may have been observed using this vaccine strategy, as internal antigens are well known to be cross-reactive in humans and animal models.Citation15,Citation16
This study is consistent with others showing that, in the absence of antibody responses, cellular immune response can provide effective protective immunity in animal models.Citation17-Citation19 With respect to pH1N1 infection, two independent studies demonstrated CTLs and CD4+ T cells raised against the seasonal H1N1 viruses, A/Brisbane/59/2007 and A/New Caledonia/20/99, respectively, were capable of responding against whole protein antigens from the pH1N1 virus.Citation20,Citation21 In addition, cross-reactive human T helper cell responses were observed for defined HLA-DR4 epitopes.Citation22 Moreover, ferrets immunized with pre-pandemic seasonal TIV were protected from influenza disease, though they lacked sterilizing immunity against pH1N1 infection.Citation23 These studies suggest that when antibodies may be unable to protect against variant virus challenge, T cells responding to conserved epitopes may act to mitigate disease severity. Putting the existing data on murine challenge and the current validation of cross-conserved epitopes into the human context, we would expect that priming with similar cross-conserved or “universal” influenza vaccine epitopes might improve the immune response to conventional influenza vaccines to provide enhanced protection against subsequent challenge. A number of “universal influenza vaccines” based on the idea that cross-conserved B-cell epitopes are currently in developmentCitation24-Citation26; here we show that cross-conserved T-cell epitopes should also be given serious consideration in vaccine design.
This study confirmed the immunogenicity of cross-reactive T-cell epitopes; a similar study performed with pandemic H1N1-naïve individuals also confirmed the antigenicity of the epitopes.Citation27 We do not presuppose that the T-cell epitopes will be sufficient; instead, it will be important to the field to determine if expanding T-cell response to cross-reactive epitopes results in a degree of cross-protection. The finding that T-cell responses to an extremely limited number (two to four) of cross-reactive epitopes may have the capacity to attenuate the course of novel H1N1-induced disease, in the absence of cross-reactive antibody response, may also lead to development of alternative approaches to “priming” immune responses and to improved means of limiting future flu pandemics, through the use of vaccines that diminish the number of individuals who develop clinical symptoms following infection.
Methods
Peptide synthesis
Peptides were synthesized using 9-fluoronylmethoxy-carbonyl (Fmoc) chemistry at 21st Century Biochemicals. Peptide purity was >80% as ascertained by analytical reversed phase HPLC. Peptide mass was confirmed by tandem mass spectrometry.
HLA binding
Peptides were assayed for HLA affinity in a competition binding assay based on Steere et al.Citation28 In 96-well plates, a test peptide and a reference peptide compete for binding to a purified class II HLA molecule (Benaroya Research Institute) for 24 h at 37 °C. Non-biotinylated test peptides were evaluated over a wide range of concentrations (0.01–200 μM) while biotinylated standard peptide was held at a fixed concentration (0.1 μM). Peptide-class II HLA complexes were then captured on ELISA plates using pan anti-HLA-DR antibody (L243, BioXCell). Plates were washed and incubated with Europium-labeled streptavidin (Perkin-Elmer) for one hour at room temperature. Europium activation buffer was added to develop the plates for 15–20 min at room temperature before they were read on a Time Resolved Fluorescence (TRF) plate reader. All assays were performed in triplicate. Dose dependence curves were generated by fitting data using the four-parameter logistic equation, and IC50 values were calculated in SigmaPlot 11.0 (Systat). Based on the IC50 values, peptide binding to a given HLA allele was classified as very high affinity (<1 µM), high affinity (1–10 µM), moderate affinity (10–50 µM), low affinity (50–100 µM), or very low affinity (>100 µM). Peptides that did not inhibit the binding of the biotinylated reference peptide at any concentration were considered non-binders. Binding assays were performed for six alleles: DRB1*0101, DRB1*0301, DRB1*0401, DRB1*0701, DRB1*1101, and DRB1*1501, providing a broad representation of class II HLA allele binding pockets.Citation29
PBMC collection, culture and characterization
Acute influenza-like illness (ILI) subjects
During the 2009–2010 influenza season, patients who presented to the emergency department at Rhode Island Hospital with acute influenza-like symptoms (defined as a fever combined with a cough) were recruited for enrollment. Six acute ILI study subjects between the ages of 18 and 65 were recruited and gave their consent to participate in the study (Lifespan IRB protocol 4074–08). Nasal washings were tested using reverse transcriptase polymerase chain reaction (RT PCR) for determination of influenza subtype. Blood samples were obtained and PBMCs were separated from whole blood by Ficoll density gradient centrifugation and PBMCs were resuspended in human R-10 media (RPMI + Glutamax-88%, Heat inactivated human AB serum-1%, L-Glutamine-<1%, Gentamycin-<1%). For all subjects, enzyme-linked immunospot (ELISpot) assays were performed ex vivo. Donor HLA class II types were determined using the One Lambda Micro SSPTM High Resolution HLA class II kit at the Hartford Hospital Transplant Immunology Laboratory. Human subject studies were performed in accordance with NIH regulations and with the approval of the Lifespan and Ethical and Independent Review Services institutional review boards.
2011 TIV-immunized subjects
Frozen PBMCs donated by 18 healthy adults (ages 22–77 y), before and three weeks after 2011 TIV immunization, were generously provided by VaxDesign. If applicable, the date of previous vaccination (prior to donation) was provided for each donor. T cell assays were performed following a period of culture to allow for expansion of antigen-specific T cells. Thawed whole PBMC populations were rested overnight and then stimulated with peptide antigen over nine days at 37 °C under a 5% CO2 atmosphere. In a 48-well plate, 5 × 106 cells in 150 μl Iscove's Modified Dulbecco's Medium (IMDM) were stimulated with a pool of HA and NA peptides at 10 μg/ml on Day 1. Three days later, IL-2 was added to 10 ng/ml and the culture volume raised to 300 μl. On Day 7, cells were supplemented with 10 ng/ml IL-2 by half media replacement. Finally, 2 days later, PBMCs were collected and washed in preparation for antigen re-stimulation to measure cytokine secretion by enzyme-linked immunospot (ELISpot) assay. Due to low cell recovery from two subjects, ELISpot assays were only performed on 16 of the 18 PBMC samples. Donor HLA class II types were determined using the One Lambda Micro SSPTM High Resolution HLA class II kit at the Hartford Hospital Transplant Immunology Laboratory. Human subject studies were performed in accordance with NIH regulations and with the approval of the Ethical and Independent Review Services institutional review board.
ELISpot assay
The frequency of epitope-specific PBMCs was determined by IFNγ ELISpot assay using the Mabtech IFNγ ELISpot Kit according to the manufacturer’s protocol. Washed PBMCs were added at 2.5 × 105 cells per well to 96-well plates pre-coated with anti-IFNγ antibody. Individual peptides were added at 10 μg/ml in triplicate wells. The pool of HA and NA peptides was added at 10 μg/ml, or 1 µg/ml per peptide. Triplicate wells were plated with PHA (2 μg/ml) as a positive control, and 6 wells with no peptide were used for background determination. Raw spot counts were recorded by ZellNet Consulting, Inc. using a Zeiss high-resolution automated ELISpot reader system and companion KS ELISpot software. Results were calculated as the average number of spots in the peptide wells, adjusted to spots per one million cells. A response was considered peptide-specific if the number of spots was at least twice background, greater than 50 spot forming cells per well (1 response per 20 000 PBMCs), and statistically different (P < 0.05) from that of the control wells by the Student t-test. ELISpot assays using splenic leukocytes from immunized mice were similarly performed using mouse IFNγ-specific reagents.
Multi-epitope DNA vaccine engineering
Epitope sequences were concatenated to form a multi-epitope gene containing 10 HLA class II epitopes. Epitopes were assembled in a random sequence at first and then re-ordered to avoid creation of novel epitopes at epitope junctions using the VaccineCAD algorithm.Citation30 No spacer sequences were required for insertion at epitope junctions to eliminate junctional immunogenicity. A histidine tag was incorporated downstream of the epitope sequences followed by two stop codons.
Genes were synthesized by GeneArt and subcloned at pre-determined flanking restriction sites downstream of the tissue plasminogen activator leader sequence in pNTC8682-eRNA41H (Nature Technology Corporation), a DNA vaccine vector that accommodates FDA recommendations for construction of plasmid DNA vaccines.Citation31
Plasmid DNA vaccine production
High purity plasmid for immunizations was prepared by Nature Technology Corporation, Inc. at research grade. Each plasmid underwent quality control testing including spectrophotometric concentration and A260/A280 ratio determination (1.97), restriction digest analysis to assure the presence of the multi-epitope genes, agarose gel electrophoresis determination of residual host RNA and DNA (none detected), and quantitative endotoxin testing (<2.0 EU/mg).
Peptide vaccine preparation
Peptides corresponding to epitopes in the DNA vaccine were formulated in incomplete Freund’s adjuvant (IFA) with 10 μg each of immunostimulatory CpG oligodeoxynucleotide 1826 (5′-TCCATGACGT TCCTGACGTT-3′), muramyl dipeptide (MDP) and CL097 (InvivoGen).
Mice
HLA DR3 transgenic mice were obtained from Dr. Chella David (Mayo Medical School) under commercial license. The mice express the HLA DR3α and β genes on a B.10-Ab0 mouse class II-negative background.Citation13 Experiments were conducted with mice 6 to 10 weeks old at the point of initiation. All studies were performed in full compliance with the standards of the TGA Sciences, Inc. Institutional Animal Care and Use Committee and in accordance with NIH publications entitled “Principles for Use of Animals” and “Guide for the Care and Use of Laboratory Animals.”
Vaccinations
Vaccine and placebo-treated mice (n = 22/group) were all female and 6–8 weeks old at the start of immunizations. DNA-prime vaccine was administered to mice intramuscularly by electroporation using the Ichor Medical Systems with 20 μL of 10 μg naked DNA in sterile PBS injected into the quadriceps muscle. For peptide-boost immunizations, each mouse was anesthetized with ketamine/xylazine and administered 100 μl IFA emulsion (50 μg peptide) subcutaneously by needle stick injection.
Virus preparation
Stocks of influenza A/California/07/2009 virus were propagated in the allantoic cavity of 9- to 11-d-old embryonated specific pathogen-free (SPF) hen’s eggs at 37 °C. The allantoic fluids from eggs inoculated with each virus was harvested 24 h post-inoculation and tested for hemagglutinating activity. Eggs inoculated with viruses were incubated at 33 °C and were harvested 3 d post-inoculation. Infectious allantoic fluids were pooled, divided into aliquots, and stored at −80 °C until used for studies. The viral titer was determined by serial titration in Madin-Darby canine kidney (MDCK) cells and calculated by the method developed by Reed and Muench.Citation32 Virus preparation was conducted using enhanced BSL-2 containment procedures in laboratories approved for use by the USDA and Centers for Disease Control and Prevention.
Respiratory challenge
Mice were dosed with 1 × 106 PFU A/California/07/2009 intranasally under ketamine/xylazine anesthesia. Mice were weighed daily post-infection and monitored for morbidity/mortality.
Viral plaque assay
Mouse lungs were obtained from naïve and vaccinated groups at two and four days post-infection (n = 4/group/time point). Lungs were weighed and homogenized at 100 mg/ml in DMEM and stored at −80 °C. MDCK cells at low (<10) or medium (<20) passage were used for all plaque assays. 750 000 cells were added to each well in 6-well plates and rested overnight at 37 °C under a 5% CO2 atmosphere. Ten-fold serial dilutions of lung homogenates in DMEM were made with viral dilutions loaded in duplicate on MDCK monolayers. Following a 1 h, room temperature incubation, viral suspensions were aspirated from wells and 2 ml warm L-15/Trypsin/agarose was poured onto cell monolayers. Following a 72 h incubation at 37 °C, agarose was carefully removed and plaques were stained with 1% Crystal Violet. Plaques were counted and PFU/ml calculated based on average of plaque assay duplicates.
Additional material
Download Zip (117.3 KB)Disclosure of potential conflicts of interest
Two of the contributing authors, De Groot AS and Martin WD, are senior officers and majority shareholders at EpiVax, Inc., a privately owned biotechnology company located in Providence, RI. Leonard Moise is employed by and holds stock options in EpiVax. These authors acknowledge that there is a potential conflict of interest related to their relationship with EpiVax and attest that the work contained in this research report is free of any bias that might be associated with the commercial goals of the company.
Acknowledgments
We are grateful to Dr Donald Drake (VaxDesign) for providing PBMC samples. This work was supported by NIH grant R21AI090359 (ADG).
References
- Perrone LA, Ahmad A, Veguilla V, Lu X, Smith G, Katz JM, Pushko P, Tumpey TM. Intranasal vaccination with 1918 influenza virus-like particles protects mice and ferrets from lethal 1918 and H5N1 influenza virus challenge. J Virol 2009; 83:5726 - 34; http://dx.doi.org/10.1128/JVI.00207-09; PMID: 19321609
- Ellebedy AH, Fabrizio TP, Kayali G, Oguin TH 3rd, Brown SA, Rehg J, Thomas PG, Webby RJ. Contemporary seasonal influenza A (H1N1) virus infection primes for a more robust response to split inactivated pandemic influenza A (H1N1) Virus vaccination in ferrets. Clin Vaccine Immunol 2010; 17:1998 - 2006; http://dx.doi.org/10.1128/CVI.00247-10; PMID: 20962210
- Bright RA, Carter DM, Crevar CJ, Toapanta FR, Steckbeck JD, Cole KS, Kumar NM, Pushko P, Smith G, Tumpey TM, et al. Cross-clade protective immune responses to influenza viruses with H5N1 HA and NA elicited by an influenza virus-like particle. PLoS One 2008; 3:e1501; http://dx.doi.org/10.1371/journal.pone.0001501; PMID: 18231588
- Laurie KL, Carolan LA, Middleton D, Lowther S, Kelso A, Barr IG. Multiple infections with seasonal influenza A virus induce cross-protective immunity against A(H1N1) pandemic influenza virus in a ferret model. J Infect Dis 2010; 202:1011 - 20; http://dx.doi.org/10.1086/656188; PMID: 20715930
- Pearce DC, Pallaghy PK, McCaw JM, McVernon J, Mathews JD. Understanding mortality in the 1918-1919 influenza pandemic in England and Wales. Influenza Other Respi Viruses 2011; 5:89 - 98; http://dx.doi.org/10.1111/j.1750-2659.2010.00186.x; PMID: 21306572
- Gaydos JC, Top FH Jr., Hodder RA, Russell PK. Swine influenza a outbreak, Fort Dix, New Jersey, 1976. Emerg Infect Dis 2006; 12:23 - 8; http://dx.doi.org/10.3201/eid1201.050965; PMID: 16494712
- Greenbaum JA, Kotturi MF, Kim Y, Oseroff C, Vaughan K, Salimi N, Vita R, Ponomarenko J, Scheuermann RH, Sette A, et al. Pre-existing immunity against swine-origin H1N1 influenza viruses in the general human population. Proc Natl Acad Sci U S A 2009; 106:20365 - 70; http://dx.doi.org/10.1073/pnas.0911580106; PMID: 19918065
- Garcia-Garcia L, Valdespino-Gómez JL, Lazcano-Ponce E, Jimenez-Corona A, Higuera-Iglesias A, Cruz-Hervert P, Cano-Arellano B, Garcia-Anaya A, Ferreira-Guerrero E, Baez-Saldaña R, et al. Partial protection of seasonal trivalent inactivated vaccine against novel pandemic influenza A/H1N1 2009: case-control study in Mexico City. BMJ 2009; 339:b3928; http://dx.doi.org/10.1136/bmj.b3928; PMID: 19808768
- De Groot AS, Ardito M, McClaine EM, Moise L, Martin WD. Immunoinformatic comparison of T-cell epitopes contained in novel swine-origin influenza A (H1N1) virus with epitopes in 2008-2009 conventional influenza vaccine. Vaccine 2009; 27:5740 - 7; http://dx.doi.org/10.1016/j.vaccine.2009.07.040; PMID: 19660593
- Garten RJ, Davis CT, Russell CA, Shu B, Lindstrom S, Balish A, Sessions WM, Xu X, Skepner E, Deyde V, et al. Antigenic and genetic characteristics of swine-origin 2009 A(H1N1) influenza viruses circulating in humans. Science 2009; 325:197 - 201; http://dx.doi.org/10.1126/science.1176225; PMID: 19465683
- Moise L, Terry F, Ardito M, Tassone R, Latimer H, Boyle C, Martin WD, De Groot AS. Universal H1N1 influenza vaccine development: Identification of consensus class II hemagglutinin and neuraminidase epitopes derived from strains circulating between 1980 and 2011. [Epub ahead of print] Hum Vaccin Immunother 2013; 9; http://dx.doi.org/10.4161/hv.25598; PMID: 23846304
- Rötzschke O, Falk K, Deres K, Schild H, Norda M, Metzger J, Jung G, Rammensee HG. Isolation and analysis of naturally processed viral peptides as recognized by cytotoxic T cells. Nature 1990; 348:252 - 4; http://dx.doi.org/10.1038/348252a0; PMID: 1700304
- Kong YC, Lomo LC, Motte RW, Giraldo AA, Baisch J, Strauss G, Hämmerling GJ, David CS. HLA-DRB1 polymorphism determines susceptibility to autoimmune thyroiditis in transgenic mice: definitive association with HLA-DRB1*0301 (DR3) gene. J Exp Med 1996; 184:1167 - 72; http://dx.doi.org/10.1084/jem.184.3.1167; PMID: 9064334
- Co MD, Orphin L, Cruz J, Pazoles P, Green KM, Potts J, Leporati AM, Babon JA, Evans JE, Ennis FA, et al. In vitro evidence that commercial influenza vaccines are not similar in their ability to activate human T cell responses. Vaccine 2009; 27:319 - 27; http://dx.doi.org/10.1016/j.vaccine.2008.09.092; PMID: 18977404
- Lee LY, Ha LA, Simmons C, de Jong MD, Chau NV, Schumacher R, Peng YC, McMichael AJ, Farrar JJ, Smith GL, et al. Memory T cells established by seasonal human influenza A infection cross-react with avian influenza A (H5N1) in healthy individuals. J Clin Invest 2008; 118:3478 - 90; PMID: 18802496
- Yewdell JW, Bennink JR, Smith GL, Moss B. Influenza A virus nucleoprotein is a major target antigen for cross-reactive anti-influenza A virus cytotoxic T lymphocytes. Proc Natl Acad Sci U S A 1985; 82:1785 - 9; http://dx.doi.org/10.1073/pnas.82.6.1785; PMID: 3872457
- Epstein SL, Lo CY, Misplon JA, Bennink JR. Mechanism of protective immunity against influenza virus infection in mice without antibodies. J Immunol 1998; 160:322 - 7; PMID: 9551987
- Price GE, Soboleski MR, Lo CY, Misplon JA, Pappas C, Houser KV, Tumpey TM, Epstein SL. Vaccination focusing immunity on conserved antigens protects mice and ferrets against virulent H1N1 and H5N1 influenza A viruses. Vaccine 2009; 27:6512 - 21; http://dx.doi.org/10.1016/j.vaccine.2009.08.053; PMID: 19729082
- Alexander J, Bilsel P, del Guercio MF, Stewart S, Marinkovic-Petrovic A, Southwood S, Crimi C, Vang L, Walker L, Ishioka G, et al. Universal influenza DNA vaccine encoding conserved CD4+ T cell epitopes protects against lethal viral challenge in HLA-DR transgenic mice. Vaccine 2010; 28:664 - 72; http://dx.doi.org/10.1016/j.vaccine.2009.10.103; PMID: 19895924
- Richards KA, Topham D, Chaves FA, Sant AJ. Cutting edge: CD4 T cells generated from encounter with seasonal influenza viruses and vaccines have broad protein specificity and can directly recognize naturally generated epitopes derived from the live pandemic H1N1 virus. J Immunol 2010; 185:4998 - 5002; http://dx.doi.org/10.4049/jimmunol.1001395; PMID: 20889549
- Tu W, Mao H, Zheng J, Liu Y, Chiu SS, Qin G, Chan PL, Lam KT, Guan J, Zhang L, et al. Cytotoxic T lymphocytes established by seasonal human influenza cross-react against 2009 pandemic H1N1 influenza virus. J Virol 2010; 84:6527 - 35; http://dx.doi.org/10.1128/JVI.00519-10; PMID: 20410263
- Ge X, Tan V, Bollyky PL, Standifer NE, James EA, Kwok WW. Assessment of seasonal influenza A virus-specific CD4 T-cell responses to 2009 pandemic H1N1 swine-origin influenza A virus. J Virol 2010; 84:3312 - 9; http://dx.doi.org/10.1128/JVI.02226-09; PMID: 20071564
- Ellebedy AH, Ducatez MF, Duan S, Stigger-Rosser E, Rubrum AM, Govorkova EA, Webster RG, Webby RJ. Impact of prior seasonal influenza vaccination and infection on pandemic A (H1N1) influenza virus replication in ferrets. Vaccine 2011; 29:3335 - 9; http://dx.doi.org/10.1016/j.vaccine.2010.08.067; PMID: 20840835
- Krammer F, Palese P. Influenza virus hemagglutinin stalk-based antibodies and vaccines. Curr Opin Virol 2013;; pii:S1879 - 6257; PMID: 23978327
- Throsby M, van den Brink E, Jongeneelen M, Poon LL, Alard P, Cornelissen L, Bakker A, Cox F, van Deventer E, Guan Y, et al. Heterosubtypic neutralizing monoclonal antibodies cross-protective against H5N1 and H1N1 recovered from human IgM+ memory B cells. PLoS One 2008; 3:e3942; http://dx.doi.org/10.1371/journal.pone.0003942; PMID: 19079604
- Corti D, Voss J, Gamblin SJ, Codoni G, Macagno A, Jarrossay D, Vachieri SG, Pinna D, Minola A, Vanzetta F, et al. A neutralizing antibody selected from plasma cells that binds to group 1 and group 2 influenza A hemagglutinins. Science 2011; 333:850 - 6; http://dx.doi.org/10.1126/science.1205669; PMID: 21798894
- Schanen BC, De Groot AS, Moise L, Ardito M, McClaine E, Martin W, Wittman V, Warren WL, Drake DR 3rd. Coupling sensitive in vitro and in silico techniques to assess cross-reactive CD4(+) T cells against the swine-origin H1N1 influenza virus. Vaccine 2011; 29:3299 - 309; http://dx.doi.org/10.1016/j.vaccine.2011.02.019; PMID: 21349362
- Steere AC, Klitz W, Drouin EE, Falk BA, Kwok WW, Nepom GT, Baxter-Lowe LA. Antibiotic-refractory Lyme arthritis is associated with HLA-DR molecules that bind a Borrelia burgdorferi peptide. J Exp Med 2006; 203:961 - 71; http://dx.doi.org/10.1084/jem.20052471; PMID: 16585267
- Southwood S, Sidney J, Kondo A, del Guercio MF, Appella E, Hoffman S, Kubo RT, Chesnut RW, Grey HM, Sette A. Several common HLA-DR types share largely overlapping peptide binding repertoires. J Immunol 1998; 160:3363 - 73; PMID: 9531296
- De Groot AS, Marcon L, Bishop EA, Rivera D, Kutzler M, Weiner DB, Martin W. HIV vaccine development by computer assisted design: the GAIA vaccine. Vaccine 2005; 23:2136 - 48; http://dx.doi.org/10.1016/j.vaccine.2005.01.097; PMID: 15755584
- FDA Center for Biologics Evaluation and Research (CBER). Points to Consider on Plasmid DNA Vaccines for Preventive Infectious Diseases Indications. (Docket no. 96N-0400).
- Reed LJ, Muench HA. A simple method of estimating fifty percent endpoints. Am J Hyg 1938; 27:493 - 7