Abstract
Several novel anti-CD20 monoclonal antibodies are currently in development with the aim of improving the treatment of B cell malignancies. Mutagenesis and epitope mapping studies have revealed differences between the CD20 epitopes recognized by these antibodies. Recently, X-ray crystallography studies confirmed that the Type I CD20 antibody rituximab and the Type II CD20 antibody obinutuzumab (GA101) differ fundamentally in their interaction with CD20 despite recognizing a partially overlapping epitope on CD20. The Type I CD20 antibodies rituximab and ofatumumab are known to bind to different epitopes. The differences suggest that the biological properties of these antibodies are not solely determined by their core epitope sequences, but also depend on other factors, such as the elbow hinge angle, the orientation of the bound antibody and differential effects mediated by the Fc region of the antibody. Taken together, these factors may explain differences in the preclinical properties and clinical efficacy of anti-CD20 antibodies.
Introduction
CD20 is a transmembrane cellular protein that has been validated as a therapeutic target for treatment of B cell malignanciesCitation1 (). CD20 is highly expressed by over 95% of B cell lymphocytes throughout their development, from the pre-B cell stage until their final differentiation into plasma cells, but is absent on the hematopoietic stem cell.Citation2 Moreover, CD20 is believed to exist predominantly as a tetramer on the cell surface. It is also largely believed to be not usually shed or internalized upon antibody binding, meaning that therapeutic antibodies may be expected to recruit immune effectors cells and mediate sustained immunologic activity.Citation3 The physiological function of CD20 remains unclear,Citation1 although evidence suggested that it may be involved in calcium signaling downstream of B cell antigen receptor activation.Citation4
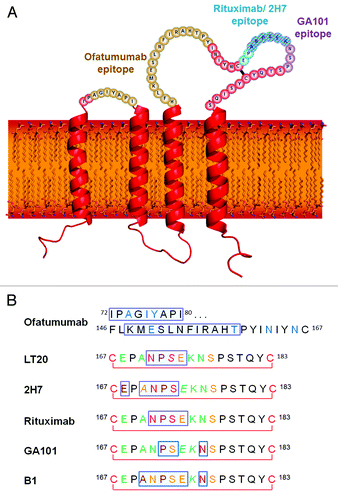
Figure 1. (A) The structure and topology of CD20 and the epitopes recognized by rituximab, ofatumumab and GA101. (B) Sequence alignment of CD20 epitopes recognized by CD20 antibodies based on published information. Core epitope residues are boxed in light blue. For 2F2 (ofatumumab), core epitope assignment is based on published work from Teeling et al. 46. For residues labeled in blue experimental evidence suggests a role in 2F2 binding. For the other antibodies, the following coloring scheme has been applied based on Pepscan results and FACS binding data of amino acid exchange mutants: green = almost any exchange tolerated at this position; brown = non-conservative exchange tested and not tolerated at this position; orange = conservative exchange tested and tolerated at this position; red = also conservative exchanges not tolerated at this position; black = position has not yet been evaluated. Italic font indicates that Pepscan and FACS binding results are discordant. Since the FACS binding results better reflect the native protein context, the coloring in such instants was based on the FACS binding data.
Rituximab (MabThera®; Rituxan®, Roche/Genentech/Biogen IDEC) was the first monoclonal antibody to be approved for the treatment of lymphoma, and it has changed the treatment of non-Hodgkin lymphoma (NHL) and chronic lymphocytic leukemia (CLL),Citation5 particularly in combination with chemotherapy where it has been shown to improve survival compared with chemotherapy alone.Citation6-Citation10 More recently, the use of rituximab in maintenance therapy has been shown to further improve outcomes in patients with follicular lymphoma (FL).Citation11-Citation16 This has established rituximab’s position as a standard-of-care therapy in the treatment of NHL and CLL.Citation17-Citation19 Other anti-CD20 antibodies have been introduced into use, including ofatumumab (Arzerra®; Genmab/GlaxoSmithKline), which is a human antibody approved for refractory CLL,Citation20,Citation21 and tositumomab (Bexxar®, GlaxoSmithKline) and ibritumomab tiuxetan (Zevalin®, Spectrum), which are murine antibodies used clinically as radioimmunoconjugates.Citation22 Ongoing research aims to develop novel anti-CD20 antibodies with improved properties and greater clinical efficacy. Critical to this process is a better understanding of the mechanisms by which anti-CD20 antibodies act and the relative contributions of different modes of action to clinical efficacy.
After binding to CD20-positive cells, antibodies are thought to trigger at least three different effector functions: (programmed) cell death (also termed as direct cell death or apoptosis), antibody-dependent cellular cytotoxicity (ADCC) or phagocytosis (ADCP) and complement-dependent cytotoxicity (CDC).Citation3,Citation23 Anti-CD20 antibodies are categorized as Type I or Type II according to their mode of CD20 binding and their primary mechanism for killing CD20-positive cellsCitation24-Citation29 ().
Table 1. Characteristics of Type I and II antibodies
This review article will focus on the application of anti-CD20 monoclonal antibodies to B cell malignancies; however, it should be noted that some of the antibodies discussed in this review have also been approvedCitation30 or are being investigatedCitation31 in the treatment of non-cancer indications (e.g., multiple sclerosis, rheumatoid arthritis, systemic lupus erythematosus).
Type I and Type II CD20 antibodies and their effector functions
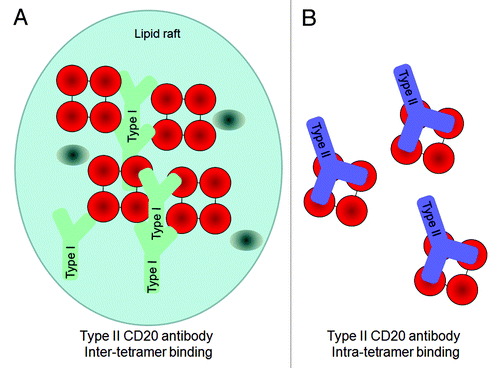
Most existing anti-CD20 antibodies, including rituximab, veltuzumab, ocrelizumab and ofatumumab, are categorized as Type I (). These antibodies are characterized by their ability to induce a translocation of CD20 into large lipid microdomains or ‘lipid rafts’ within the plasma membrane upon binding.Citation26,Citation32,Citation33 This clustering process enhances the recruitment and activation of complement, and hence Type I antibodies exert potent CDC.Citation25,Citation26 However, the contribution of complement activation to the depletion of B cells in vivo remains unclear.Citation3,Citation34 Another characteristic feature of Type I antibodies is that B cells can be bound by twice as many Type I antibodies compared with Type II antibodies,Citation27,Citation35 most likely due to different binding geometries. The biological significance of this is unknown, but it has been hypothesized that the 2:1 stochiometry could be explained by Type I antibodies binding between two CD20 tetramers, thereby crosslinking tetramers with two antibodies bound per tetramer, whereas Type II antibodies may bind within a tetramer, resulting in only one antibody bound per CD20 tetramerCitation29,Citation36 (). In line with this, the two known Type II anti-CD20 antibodies tositumomab (or B1) and obinutuzumab (GA101) (), do not induce accumulation of CD20 upon antibody binding in insoluble lipid rafts and show relatively little CDC activity.Citation25,Citation27 On the other hand, Type II antibodies are more potent than Type I antibodies in inducing homotypic adhesion and direct cell death.Citation24,Citation25,Citation27 Although this form of cell death was initially described as apoptosis, recent studies have demonstrated that it is a non-apoptotic form of direct cell death that follows an actin-dependent enhancement of cell-to-cell contact, the rupturing of lysosomes within the cytoplasmCitation28,Citation37,Citation38 and the generation of reactive oxygen species, but does not show the classical hallmarks of apoptosis such as DNA laddering or caspase dependence.Citation39
Table 2. Characteristics of selected anti-CD20 monoclonal antibodies
Figure 2. Hypothetical model for the 2:1 binding ratio of Type I and Type II CD20 antibodies binding to CD20 (tetramers, depicted in red). An explanation to explain the 2:1 binding stoichiometry between Type I and Type II CD20 antibodies is to assume that A) Type I antibodies bind between CD20 tetramer (inter-tetramer, depicted in red) resulting in accumulation in lipid rafts together with FcγRIIb (gray oval). In contrast, as Type II antibodies may bind within one tetramer (intra-tetramer).
The ADCC and ADCP activity of anti-CD20 antibodies is mediated by the interaction of their Fc region with FcγRIIIa and is not affected by the Type I or Type II character of the antibody. FcγRIIIa is expressed on various immune effector cells, most prominently macrophages/monocytes and natural killer cells. FcγRIIIa crosslinking by binding to CD20 on target cells stimulates release of lytic enzymes by the effector cells and induces cell killing or promotes the phagocytosis of the target CD20 positive cell.Citation3 Two variants of FcγRIIIa have been identified in humans: a predominant lower affinity form with a phenylalanine at position 158 (FcγRIIIa-158F) and a higher affinity form with valine at this position (FcγRIIIa-158V).Citation40-Citation42 The binding of the Fc region of antibodies to FcγRIIIa is dependent on interactions between the carbohydrate moieties of both the FcγRIIIa and antibody.Citation43 Notably, ADCC activity does not differ between Type I and Type II anti-CD20 antibodies,Citation3 but antibodies such as GA101 have been engineered for enhanced affinity for FcγRIIIa leading to an increased ability to bind and recruit effector cells and hence a higher ADCC level.Citation27,Citation44 The contribution of ADCC to the clinical activity of antibodies remains to be established. However, the expression of the higher affinity FcγRIIIa-158V genotype in lymphoma patients has been shown to be associated with an improved response to rituxumab (mono-) therapy,Citation40,Citation45 suggesting that enhanced FcγRIIIa affinity may confer a clinical advantage.
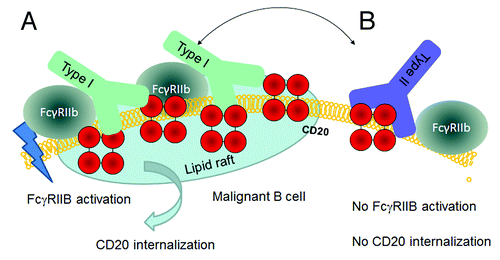
Recently, Beers and colleaguesCitation46 demonstrated an increased potency in depleting B cells from human CD20 transgenic mice of Type II antibodies compared with Type I antibodies. They attributed much of this disparity to the Type I antibody-mediated internalization of CD20 by B cells leading to reduced recruitment of macrophages (ADCP) and degradation of CD20/antibody complexes. The authors also noted that the type of disease affected the degree of internalization, with most cases of CLL and mantle cell lymphoma showing rapid CD20 internalization; this was in contrast to FL and DLBCL cells, which were more resistant to CD20 loss. The internalization process was promoted by the inhibitory FcγRIIb on target B cells and investigations have suggested that rituximab can crosslink CD20 and FcγRIIb on the same cell (in cis), whereas Type II antibodies do not appear to have this functionCitation47 ().
Figure 3. Hypothetical model for CD20 binding of Type I and Type II CD20 antibodies explaining the impact of FcγRIIb on internalization. A) Type I antibodies such as rituximab may bind to CD20 in a conformation that allows simultaneous binding to FcγRIIb and subsequent signaling followed by internalization in lipid rafts. B) Type II antibodies such as GA101 may bind in a conformation that does not allow simultaneous binding to FcγRIIb, thus resulting in reduced internalization.
Anti-CD20 antibodies possess complementarity-determining regions (CDR) that bind to a specific epitope on the antigen. Mutational analyses and peptide scanning studies have revealed differences between antibodies in their CD20 epitopes.Citation29,Citation48,Citation49 Recently, three-dimensional crystallographic representations of several antibodies in complex with CD20 confirmed fundamental differences in their interactions with CD20 () [rituximab,Citation50 C2H7 (ocrelizumab),Citation51 ofatumumab,Citation52 GA101].
Figure 4. Published crystal structures of CD20 antibodies. A) rituximab-CD20 complex,48 B) ofatumumab (no co-crystal structure is available),50 C) 2H7-CD20 complex,49 and D) GA101-CD20 complex.29 The heavy chain is colored in darker shades, the peptides derived from CD20 are colored in red where appropriate.
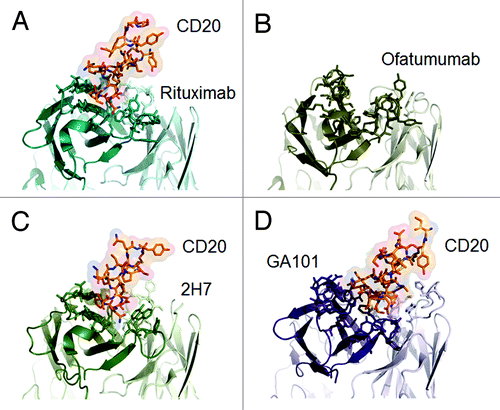
Structurally, CD20 comprises four hydrophobic membrane-spanning domains, two extracellular loops (one of approximately 44 amino acids and a smaller one of approximately seven amino acids), and intracellular N- and C-terminal regions (). The intracellular regions of CD20 can undergo phosphorylation upon antibody binding, thereby mediating cellular signaling.Citation1 Most of the epitopes involved in antibody recognition are located within the larger extracellular loop. Recently, Niederfellner and colleaguesCitation29 mapped the epitopes recognized by anti-CD20 antibodies. They showed that, despite recognizing an overlapping epitope on the large extracellular loop of CD20, Type II antibodies bind in a different orientation than Type I antibodies. For example, the core epitope of GA101 (a Type II antibody) is formed by residues 172–178, whereas the Type I antibody rituximab targets the more N-terminally comprising residues 168–175, with 170–173 contributing most essentially. For binding of Type II antibodies, asparagine 176 (N176) is a critical residue (), whereas this residue does not seem to make any contacts with CD20-bound Type I antibodies, as exemplified by the crystal structure of rituximab (). The crystal structure of the GA101–CD20 epitope peptide complex confirmed that the shift in the core epitope resulted in a fundamentally different orientation of GA101 with respect to CD20. Based upon the currently available data, we have generated a model of rituximab and GA101 bound to CD20 (). Ofatumumab, another Type I antibody, binds to both the large and small CD20 extracellular loops,Citation48,Citation52 as discussed below.
Figure 5. Comparison of A) rituximab (Type I) and B) GA101 (Type II) crystal structures in complex with CD20 peptide.29 While for rituximab N171 is deeply immersed and N176 has no contacts with the rituximab CDRs, N171 is not deeply immersed in the the GA101 CDRs and vice versa N176 makes contacts to residues F52/D57/D59 of GA101 supporting the C-terminal shift of the GA101 epitope.
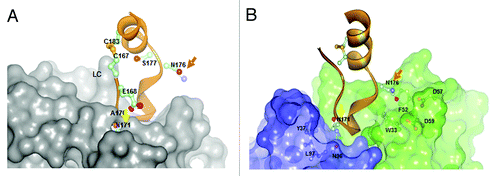
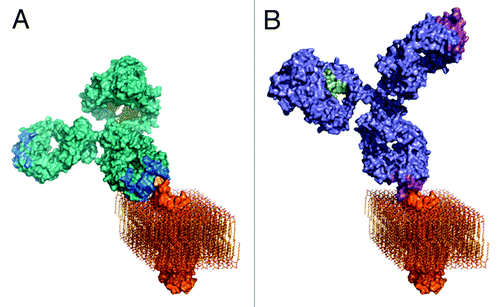
Figure 6. Three-dimensional models of A) rituximab and B) GA101. GA101 binds to the same binding epitope region of CD20 as rituximab, but in a different binding orientation. The molecular models were created by combining known structural data with the current knowledge and general understanding of antibody structure and membrane protein topology. The CD20 membrane protein model was created by combining the structural fragments of the crystallized CD20 antibody binding epitope and the transmembrane part of the HER2 receptor as a typical example of a membrane spanning molecule with known 3D information, and CD20 topology information.
Type I CD20 antibodies
Rituximab
Rituximab is a Type I chimeric (human–mouse) immunoglobulin (Ig)G1 anti-CD20 antibody. The CD20 epitope recognized by rituximab and other mouse-derived antibodies spans amino acid residues 168–175 of the CD20 protein, with the ANPS motif at residues 170–173 on the large extracellular loop appearing to be of critical importanceCitation29,Citation33,Citation48,Citation50,Citation53 (). These key residues have been shown to form a network of hydrogen bonds with residues of the surrounding CDR loops.Citation51 The particular importance of the alanine residue at position 170 (A170) and the proline residue at position 172 (P172) was shown by site-directed mutagenesis studies taking advantage of the fact that rituximab binds only human, but not mouse, CD20. Introducing the 170ANP172 motif into mouse CD20 conferred binding of rituximab. The importance of the 170ANPS173 region for rituximab binding in humans has also been established by the screening of libraries of phage-displayed peptides with different sequencesCitation53 where P172 was found to have a particular importance, since rituximab binds the human ANPS sequence but not the corresponding murine SNSS sequence.Citation53 Furthermore, mutation of the alanine and proline at positions 170 and 172 in human CD20 to serine was shown to abolish rituximab binding.Citation33,Citation48 Asparagine 171 (N171) was also found to be a key residue for rituximab binding as any amino acid replacement at this position, except histidine, resulted in a substantial loss of binding affinity to peptides representing the extracellular CD20 loop.Citation29
Phage-peptide screening also suggested that a second region of the epitope, 182YCYSI186, contributes to the binding of rituximab through conformational stabilization.Citation49 Furthermore, when Perosa and colleagues screened phage-display peptide libraries containing a repertoire of sequences of random 7- or 12-amino acid peptides they found that, while cyclic peptides mimicking the CD20 epitope were dependent on the 170ANPS173 motif, linear mimics that also bound rituximab required a different motif – WPxWLE – that does not correspond to any sequence present in CD20 itself.Citation53,Citation54 While the WPxWLE motif appears to share some rituximab contact points with 170ANPS173, these regions are conformationally different and have been proposed as distinct epitopes.Citation54 However, the functional role and significance of the WPxWLE sequence is unclear.
Mutagenesis studies can identify residues affecting antibody binding, but cannot define the contact sites between the CD20 epitope and the antibody. The structure of the rituximab:epitope complex has been determined by co-crystallizing a synthetic peptide mimic of the extracellular loop epitope of CD20 (residues 163–187) in complex with the antigen-binding fragment of rituximab.Citation50 The bound CD20 peptide forms a cyclic conformation owing to a disulfide bond between two cysteine residues, C167 and C183. This structure comprises a short N-terminal coil (residues 167–171), a 310 helix (residues 172–174), a small loop (residues 175–177) and a short C-terminal α-helix (residues 178 –184). The key 170ANPS173 motif is embedded in a cyclic, four-region pocket formed by the CDRs of the rituximab antibody (). Residues of the 170ANPS173 motif bind to CDR residues via numerous hydrogen bonds and van der Waals contacts. In accordance with evidence that P172 has a critical role in antibody binding, this residue is deeply buried in the CD20/Ab interface and forms additional hydrophobic and hydrophilic contacts with residues at the bottom of the CDR pocket that are likely to be important in maintaining the conformational stability of the epitope-antibody complex.Citation50 The 182YCYSI186 region at the C-terminus of the large extracellular loop of CD20 also appears to play a role in rituximab binding,Citation49 most likely through the formation of the disulfide bond that induces the cyclic conformation of the epitopeCitation50 loop necessary for the binding of CD20 to rituximab.Citation55 Abrogating the internal disulfide bridge (C167-C183) of the large extracellular loop seems to completely destabilize the CD20 protein, since expression of a CD20 variant with a C167S exchange is barely detectable by western blot analysis after transient transfection of HEK293 cells.Citation29
The knowledge of the CD20 epitope was used to design rituximab variants in which point mutations were inserted into the CDR to improve the binding characteristics of the antibody.Citation56 Rituximab variants that bound to CD20 with enhanced avidity, or with a reduced off-rate, did not show improved activity in terms of CDC, complement fixation or rafting. However, a variant with three mutational changes (H57DE/H102YK/L93NR/) was shown to mediate enhanced avidity-dependent ADCC and cell death.Citation56
In principle, genetic mutations in the rituximab epitope could reduce the binding and efficacy of the antibody, but clinical data in patients with DLBCL suggest that epitope mutations are very rare (0.4% of 264 patients at diagnosis and one of 15 patients at relapse) and are not an important cause of failure of treatment with rituximab in combination with conventional chemotherapy.Citation57
Veltuzumab
Veltuzumab (IMMU-106; hA20, Immunomodics, Nycomed) is a humanized IgG1κ Type I antibody in Phase 2 development for treatment of relapsed or refractory NHL and autoimmune diseasesCitation58 (). Veltuzumab has CDRs largely identical to those of rituximab with the exception of one residue, suggesting that it binds to the same epitope.Citation59 Veltuzumab competes for CD20 binding with rituximab and shows similar specificity, avidity and in vitro activity.Citation58,Citation59
AME-133v
AME-133v (Ocaratuzumab, LY2469298, MENTRIK) is a humanized IgG1 Type I antibody in Phase 2 development. AME-133v is an optimized version of rituximab with a Fab region engineered to improve CD20-binding affinity. AME-133v has a ca. 13- to 20-fold greater binding affinity for CD20 than rituximab.Citation60 The Fc region has been modified to improve affinity for FcγRIIIa-158F and -158V genotypes. As a result, AME-133v shows greater in vitro activation of natural killer cells and 5- to 7-fold more potent ADCC than rituximab.Citation44,Citation60 AME-133v recognizes the same epitope as rituximab.
Ibritumomab
Ibritumomab, a murine IgG1κ Type I antibody, is the antibody from which rituximab was derived and hence targets the same epitope as rituximab.Citation61 A radiolabeled form of the antibody, 90Y-ibritumomab tiuxetan (Zevalin, Spectrum), is used in the treatment of indolent NHLCitation62-Citation64 and as consolidation therapy following induction.Citation65,Citation66
Ocrelizumab
Ocrelizumab (PRO70769, Roche/Genentech) is a humanized anti-CD20 IgG1 Type I antibody that has been evaluated in a Phase 1/2 study in patients with relapsed/refractory FL and is currently in development for the treatment of multiple sclerosis.Citation67 Compared with rituximab, ocrelizumab shows lower CDC activity but greater ADCC activity and enhanced binding to the low-affinity FcγRIIIa variant.Citation67 Ocrelizumab is based on the murine Type I IgG2b antibody 2H7. The CDR loops of 2H7 are structurally similar to those of rituximab. Among the four CDR loops that interact with CD20, only one (H3) differs substantially from the rituximab counterpart in terms of residue sequence and conformation.Citation51 2H7 was first thought to recognize exactly the same epitope as rituximab. Early studies confirmed that residues A170 and P172 of CD20 are necessary for 2H7 binding, but suggested that they are not sufficient alone. Rather, the 162INxxN166 motif also appeared to be necessary for full binding of 2H7 in the presence of A170/P172, possibly because these residues may stabilize the conformation of the 2H7:CD20 complex. Mutation of the QTSK motif present in murine CD20 to 156RAHT159 (as present in human CD20) also improved the binding of 2H7, but was not necessary for full binding. In addition, 2H7 appears to only bind the oligomeric form of CD20 (e.g., tetramers).Citation33 Subsequent peptide scanning studies demonstrated that the core contact regions for 2H7 (CD20 positions 165–180) and rituximab (CD20 positions 165–182) are almost identical.Citation48 Crystallography has confirmed that the CDR loops of 2H7, like those of rituximab, form a deep pocket enclosing the critical 170ANPS173 epitope motif of CD20.Citation51 The P168 and P170 residues of 2H7 also form hydrogen bonds with CD20, while P175, which occurs in both 2H7 and rituximab, forms a hydrophilic interaction with CD20 that is oriented differently in the 2H7-CD20 and rituximab-CD20 complexes. As with rituximab, the cyclic conformation of the 2H7-CD20 complex is maintained by the disulfide bond of the peptide. The different structure of the H3 loop of 2H7, as compared with rituximab, alters the topology of the complex. These differences result in fewer binding interactions for 2H7, and hence a lower binding affinity, compared with rituximab.Citation51
PRO131921
PRO131921 (rhuMAb v114, Genentech) is a humanized IgG1 anti-CD20 antibody that was studied in two Phase 1 clinical trials, one for CLL and one for NHL. PRO131921 is derived from 2H7, but carries a modified Fc region with enhanced affinity for FcγRIIIa.Citation68 PRO131921 interacts with the same epitope as ocrelizumab.Citation69 Clinical development has been discontinued.Citation70
TRU-015
TRU-015 is a single-chain CD20-targeting protein that was derived from 2H7 and has a human IgG1 hinge that binds to the same epitope of 2H7.Citation51 TRU-015 was described to show reduced CDC activity but more in vitro and in vivo properties compared with rituximab.Citation71 Clinical development was discontinued.
Ofatumumab
Ofatumumab is a human IgG1 Type I antibody that is approved for the treatment of patients with CLL refractory to fludarabine and alemtuzumab.Citation20,Citation21 Ofatumumab is being studied in patients with lymphomas either as a single agent or in combination with chemotherapy.Citation58,Citation72-Citation74
Like rituximab, ofatumumab shows Type I anti-CD20 activity, including CD20 rafting and CDC activity,Citation35,Citation75 but binding studies suggest that ofatumumab recognizes an epitope different from that of rituximab. While the binding of rituximab is prevented by mutation of the A170/P172 residues, site-directed mutagenesis has shown that such mutations in the large extracellular loop of CD20 do not affect the binding of ofatumumab. Rather, the replacement of asparagine at position 163 (N163) or 166 (N166) with aspartic acid reduced ofatumumab binding by 50–75%. A triple mutant with mutations T159K, N163D and N166D did not bind ofatumumab at all.Citation48,Citation76 None of these single mutations affected rituximab binding, although the triple mutant showed slightly decreased binding. Peptide scanning analyses confirmed that ofatumumab (together with the four other human IgG1 or IgGM antibodies tested) does not recognize the A170/P172 motif. Instead, these human antibodies recognize a particular region in the large extracellular loop (146FLKMESLNFIRAHTP160) that is N-terminal to A170 and P172 ( and ). This region does not include the N163 and N166 residues shown by mutagenesis studies to be necessary for ofatumumab binding, suggesting that these residues indirectly contribute to the stability of the epitope rather than forming part of the binding site itself.Citation48
Peptide scanning and mutagenesis studies have revealed that the small extracellular loop of CD20 also contributes to the binding of ofatumumab. Binding of ofatumumab was almost completely prevented by the replacement of the entire small loop with an alternative sequence or by the insertion of three mutations (A74T, I76A and Y77S) in the loop. Neither the loop replacement nor these mutations affected the binding of rituximab.Citation76 These data confirm that ofatumumab recognizes an epitope distinct from that of rituximab, which comprises discontinuous sequences across both the large and small extracellular loops of CD20 ().
According to crystallography, the region of the ofatumumab molecule that binds with CD20 comprises six CDR loops, which form a deep pocket. Around the periphery of the pocket are hydrophobic residues (Y32, W94, W53, I58, Y60, Y102 and Y105) and at the bottom of the pocket is a positively charged residue (R91).Citation52 It should be noted that the crystal structure of the Fab fragment of ofatumumab was determined in the absence of CD20Citation52 (). The hydrophobic pocket formed by the CDRs of ofatumumab is thought to interact with hydrophobic residues on both the large and small extracellular loops of CD20, and possibly with the cell membrane itself. The negatively charged N-terminal E150 residue of the large extracellular loop of CD20 is thought to interact with the positively charged R91 residue at the bottom of the CDR pocket of ofatumumab.
The binding of ofatumumab to the large and small extracellular loop of CD20 was hypothesized to position ofatumumab closer to the surface of the CD20 cell membrane than antibodies binding the large loop. This could be expected to facilitate the deposition of activated complement on the cell surface and hence the amplification of the complement response.Citation77 However, the impact of this is unclear as the CD20 extracellular loop is very small compared with the size of an antibody so that the antibody-binding domain of CD20 is already membrane-proximal. In addition to the difference in binding sites between ofatumumab and rituximab, studies have suggested that ofatumumab dissociates more slowly from the cell surface than rituximabCitation35 and exhibits greater CDC activity than rituximab in various B cell lines.Citation35,Citation48,Citation75,Citation77 Furthermore, CDC by ofatumumab was found to be less dependent on the cell-surface density of CD20 than CDC by rituximab.Citation48 The differential action of ofatumumab on the complement has been supported by direct visualization of complement-mediated cell killing obtained using spinning-disk confocal microscopy.Citation77 Compared with rituximab, ofatumumab has been shown to be more active in both the deposition of complement and in causing morphologic effects induced by the membrane attack complexes of complement, namely blebbing (the formation of bulges in the cell membrane) and the creation of long, thin ‘streamer’ structures that extend from the cell membrane. Other data, however, have suggested that the preclinical activity of ofatumumab and rituximab are similar, demonstrating comparable levels of CDC, ADCC, whole blood B cell depletion and antitumor activity in preclinical assays and models.Citation78
Hu8E4
Hu8E4 is a humanized Type I antibody incorporating CDRs from the mouse IgG2 anti-CD20 antibody, 8E4, grafted onto human light and heavy framework chains. Compared with rituximab, hu8E4 showed similar levels of ADCC and direct cell death against human lymphoma cells in vitro, but greater CDC and greater antitumor activity in lymphoma models in mice.Citation79 The epitope recognized by Hu8E4 is not currently known.
Ublituximab
Ublituximab (LFB-R603, LFP) is a chimeric glyco-engineered anti-CD20 antibody with enhanced FcγRIII affinity (as compared with rituximab) that acts via enhanced induction of ADCC. The CD20 epitope of ublituximab is unknown. Preclinical studies imply that ublituximab can disrupt NF-κB/Snail/RKIP/PTEN/AKT signaling in B cell NHL cell lines that are resistant to chemotherapy and immunochemotherapy.Citation80 Ublituximab is currently in a Phase 1/2 clinical study in NHL.
Type II CD20 antibodies
Tositumomab
Radiolabeled 131I-tositumomab (Bexxar, GlaxoSmithKline), a murine IgG2aλ antibody, known as B1 in the scientific literature, is used clinically in extensively pretreated patients with NHL.Citation81 The activity of tositumomab is mainly achieved through its radioisotope rather than its antibody type.Citation82 The non-radioactive parental antibody B1, however, is the prototypic Type II CD20 antibody that displays all typical features of a Type II anti-CD20 activity, i.e., it binds B cells at approximately half the density of Type I antibodies and induces homotypic aggregation and cell death, but not rafting.Citation33,Citation35,Citation37 In transgenic mice expressing human CD20, tositumomab depleted normal B cells (both circulating and within lymphoid tissues) for significantly longer than rituximab,Citation34 although there was no difference in the CD20 binding affinities or biological half-lives of the antibody. Mutational studies showed that the 170ANP172 epitope motif of CD20 is critical to full binding of tositumomab, just as for rituximab.Citation33 Peptide scanning studies have confirmed that tositumomab shares most of the core contact region (positions 170–182) used by rituximab.Citation48 Importantly, both Type II antibodies, B1 and GA101, do not tolerate well substitutions of N176, while all Type I antibodies tested do ().
GA101
GA101 (obinutuzumab, Roche) is a Type II, glyco-engineered, humanized IgG1κ anti-CD20 antibody derived from the murine antibody Bly-1Citation27 (). GA101 is in Phase 2 and 3 clinical trials for the treatment of patients with NHL and CLL.
GA101 shows biological activity characteristic of a Type II anti-CD20 antibody. It binds to the surface of the CD20 cell at a lower density than rituximab, and unlike Type I antibodies, GA101 does not induce rafting of CD20 and shows low CDC activity. GA101 triggers pronounced homotypic adhesion of lymphoma cells and high levels of direct cell killing activity that is superior to that of rituximab and tositumomab.Citation27 GA101 was significantly more effective than rituximab in depleting B cells in whole blood samples from healthy donors (n = 10) and from an individual with CLL.Citation27 GA101 also showed greater inhibition of tumor growth than rituximab, including complete tumor remission in xenograft models of human DLBCL and improved survival in a model of advanced, disseminated mantle cell lymphoma. GA101 and rituximab showed similar activity in depleting B cells from peripheral blood in cynomolgus monkeys, but GA101 was more effective in depleting B cells in spleen and lymph nodes.Citation27
In addition to the antibody type, these characteristics also result from two unique, engineered features of the GA101 molecule, namely a non-fucosylated Fc portion and a modified elbow hinge region.Citation27,Citation29 GA101 has been glyco-engineered to produce a non-fucosylated Fc region that substantially enhances the affinity of this antibody for both the FcγRIIIa-158F and FcγRIIIa-158V variants. This modification leads to an increased ability to bind and recruit effector cells and hence to an increased ADCC activity against lymphoma cells compared with rituximab.Citation27 The elbow hinge region of GA101 between the variable region and the first constant domain was modified during the humanization process. A valine residue present in the parental murine B-ly1 antibody at Kabat position 11 was replaced by leucine present in B-lyl. This mutation widens the elbow angle for GA101 by almost 30° compared with rituximab and 2H7 as determined by X-ray structure analysis.Citation29 Mutagenesis experiments indicate that this mutation enhances its Type II antibody characteristics, including the increased direct cell death induction.Citation27 By mutating the Kabat 11 position, direct cell death induction can be switched on and off, although the CDRs of the antibody remain unchanged and binding to CD20 per se is retained.Citation27
Positional mapping has confirmed that the epitopes of GA101 and rituximab overlap;Citation29 however, the GA101 epitope is shifted toward the C-terminus of CD20, with N176 contributing to binding of Type II but not of Type I antibodies (). The core of the GA101 epitope consists of an extended region, 170ANPSEKNSP178, rather than the 170ANPS173 motif that is critical to rituximab binding.Citation29
The relative roles of these residues in GA101 binding has been confirmed by crystallography ( and ). N171 forms hydrogen bonds with GA101 but is not essential for binding. P172 and S173 both contribute to the binding of GA101, while residues at position 174–176 (174EKN176) form an extensive network of hydrogen bonds with the CDR of GA101.Citation29 Unlike ofatumumab,Citation48,Citation52 GA101 does not appear to directly interact with the small extracellular loop of CD20 or the region preceding the larger loop.Citation29 However, Pepscan analyses indicate that residues from positions 142–160 affect GA101 binding, suggesting that they might indirectly stabilize the epitope conformation (unpublished observations).
The extended binding site sequence of GA101 may explain its high binding affinity for CD20. Moreover, GA101 binds CD20 with a different topology compared with other antibodies owing both to its unique epitope and elbow angle ( and ). Rituximab and 2H7 bind to CD20 in positions oriented toward the core of the epitope. In comparison, the bound GA101 is rotated 90° clockwise around its middle axis and tilted about 70° toward the C-terminus of the peptide.Citation29 This topologic difference may explain several differences observed between the arrangement and conformation of rituximab–CD20 and GA101–CD20 complexes. According to protein tomography analysis, GA101 often binds monovalently to CD20, whereas rituximab binds the peptide mostly bivalently. This may favor intra- rather than inter-tetramer bindingCitation36 (). Electron densities observed in protein tomography suggest that GA101 appears to bind to CD20 tetramers, while rituximab also binds to large CD20 complexes consisting of network-like structures of unidentified proteins.Citation29 The latter might represent higher order signaling complexes assembled in lipid rafts, e.g., the tetraspanin network. It is thought that the different geometry of the antibody–CD20 complexes may, in part, explain the differences in preclinical and clinical activity.
We believe that the differences in CD20 internalization and FcγRIIb dependence reported by other groupsCitation45,Citation82 might be related to differences in the orientation of the antibodies after binding to CD20. Recent work with TNFR agonistic antibodies including CD40 and DR5 antibodies has shown that binding to CD40 and FcγRIIb in cis is required to mediate potent CD40 or DR5 activation.Citation83-Citation85 We propose that Type I CD20 antibodies bind to CD20 on B cells in a conformation that allows simultaneous binding to FcγRIIb on the same cell (in cis) resulting in crosslinking, FcγRIIb co-activation and CD20 co-internalization upon binding potentially in lipid rafts. Vice versa, the biological effects could be explained by the different binding conformation of Type II CD20 antibodies that might prevent simultaneous binding in cis to FcγRIIB, which precludes FcγRIIb crosslinking and CD20 co-internalization ().
Other antibodies
hOUM3 and hOUbM6
hOUBM3 and hOUBM6 are humanized versions of the murine antibodies 1k1782 and 1k1791 that were previously identified as having properties and epitope specificities different from rituximab and ibritumomab.Citation83 In preclinical studies, variants of hOUBM6 showed higher CDC levels, similar or higher ADCC levels and similar depletion of leukemia and lymphoma cells compared with rituximab.Citation75
Residues A170 and P172 of CD20 are not essential for binding of hOUBM3 and hOUBM6, suggesting that the epitope for these antibodies indeed differs from that of rituximab. According to the limited available data, the epitope for hOUBM6 includes the motifs 287ES288, 156RAHT159 and 162INIYN166.Citation75 Researchers reporting preclinical studies of a series of hOUBM3 and hOUBM6 variants recently proposed a classification scheme based on the affinity (measured by the dissociation constant) and the epitope of antibodies, rather than biological effects as used to categorize Type I and II anti-CD20 antibodies.Citation75 The affinity was correlated with potential to induce direct cell death, allowing antibodies to be defined into Group A and Group B antibodies. Group A antibodies (hOUBM3, hOUBM6 clones with lower Kd, and ofatumumab) exhibited high affinity and did not induce direct cell death in lymphoma cells. Group B antibodies (i.e., rituximab and hOUBM6 clones with high Kd) had lower affinity and induced apoptosis. The researchers proposed that antibodies with lower affinity might induce direct cell death more efficiently by binding simultaneously to two CD20 dimers, cross-linking them and bringing them into close proximity with each other. The authors further subcategorized antibodies according to the similarity with ibritumomab, the murine version of rituximab. Thus, antibodies with a non-ibritumomab-like epitope profile included hOUBM3, hOUBM6 and ofatumumab, and those with an ibritumomab-like profile were rituximab and 2H7. The relationship between these affinity/Kd and epitope categories and the conventional Type I and II categories of anti-CD20 antibody remains to be established.
Conclusions
Characterization of anti-CD20 antibodies epitope specificity has revealed variations that may contribute to differences in the effects caused by these molecules. The relationship between the epitope and the biological effect is not always clear and there is no apparent link between epitope and antibody type. For example, ofatumumab and rituximab are both classified as Type I antibodies and yet they recognize different CD20 epitopes. Conversely, tositumomab shows Type II activity but targets an epitope similar to one recognized by rituximab, so subtle differences in the interaction of anti-CD20 antibodies with their target can profoundly change the biological outcome.
These differences may affect the orientation of the antibodies in complex with their respective CD20 peptides, but other factors like the elbow-hinge angle and Fc effects also play a role. GA101 and rituximab, for example, bind CD20 in different orientations, even though their epitopes are largely shared. This appears to result in different overall conformations of bivalently bound CD20 complexes. The relative contribution of these factors to preclinical and clinical efficacy remains to be established. In general, it is not advisable to select therapeutic antibody candidates solely based on binding affinity and epitope binning data without testing them also in a functional biological assay, as demonstrated by the substantially different biological effects of rituximab and GA101 with only subtle differences in their epitopes.
Further studies are required to determine whether differences in molecular and preclinical pharmacology translate into differences in clinical outcomes. Phase 3 head-to-head trials comparing GA101 or ofatumumab, with rituximab are currently recruiting and should help in optimization of existing antibody use and development of future treatments.
| Abbreviations: | ||
| ADCC | = | antibody-dependent cellular cytotoxicity |
| ADCP | = | antibody-dependent cellular phagocytosis |
| CDC | = | complement-dependent cytotoxicity |
| CDR | = | complementarity-determining region |
| CLL | = | chronic lymphocytic leukemia |
| DLBCL | = | diffuse large B cell lymphoma |
| FcɣR | = | Fcɣ receptor |
| FL | = | follicular lymphoma |
| Ig | = | immunoglobulin |
| NHL | = | non-Hodgkin lymphoma |
| MS | = | multiple sclerosis |
Acknowledgments
We thank all members and contributors in the GA101 preclinical team and the GA101 global life cycle team.
Potential Conflicts of interest
C.K., E.M. and P.U. are employees of Roche Glycart A.G., W.S., G.G., M.S., G.N. are employees of Roche Diagnostics GmbH, all other authors do not have a conflict of interest to declare. Writing support was provided by Zoe Crossman, Health Interactions, UK and Rachel Edwards, Prism Ideas, UK.
References
- Cragg MS, Walshe CA, Ivanov AO, Glennie MJ. The biology of CD20 and its potential as a target for mAb therapy. Curr Dir Autoimmun 2005; 8:140 - 74; http://dx.doi.org/10.1159/000082102; PMID: 15564720
- Stashenko P, Nadler LM, Hardy R, Schlossman SF. Characterization of a human B lymphocyte-specific antigen. J Immunol 1980; 125:1678 - 85; PMID: 6157744
- Glennie MJ, French RR, Cragg MS, Taylor RP. Mechanisms of killing by anti-CD20 monoclonal antibodies. Mol Immunol 2007; 44:3823 - 37; http://dx.doi.org/10.1016/j.molimm.2007.06.151; PMID: 17768100
- Polyak MJ, Li H, Shariat N, Deans JP. CD20 homo-oligomers physically associate with the B cell antigen receptor. Dissociation upon receptor engagement and recruitment of phosphoproteins and calmodulin-binding proteins. J Biol Chem 2008; 283:18545 - 52; http://dx.doi.org/10.1074/jbc.M800784200; PMID: 18474602
- Keating GM. Rituximab: a review of its use in chronic lymphocytic leukaemia, low-grade or follicular lymphoma and diffuse large B-cell lymphoma. Drugs 2010; 70:1445 - 76; http://dx.doi.org/10.2165/11201110-000000000-00000; PMID: 20614951
- Marcus R, Imrie K, Belch A, Cunningham D, Flores E, Catalano J, et al. CVP chemotherapy plus rituximab compared with CVP as first-line treatment for advanced follicular lymphoma. Blood 2005; 105:1417 - 23; http://dx.doi.org/10.1182/blood-2004-08-3175; PMID: 15494430
- Forstpointner R, Dreyling M, Repp R, Hermann S, Hänel A, Metzner B, et al, German Low-Grade Lymphoma Study Group. The addition of rituximab to a combination of fludarabine, cyclophosphamide, mitoxantrone (FCM) significantly increases the response rate and prolongs survival as compared with FCM alone in patients with relapsed and refractory follicular and mantle cell lymphomas: results of a prospective randomized study of the German Low-Grade Lymphoma Study Group. Blood 2004; 104:3064 - 71; http://dx.doi.org/10.1182/blood-2004-04-1323; PMID: 15284112
- Hiddemann W, Kneba M, Dreyling M, Schmitz N, Lengfelder E, Schmits R, et al. Frontline therapy with rituximab added to the combination of cyclophosphamide, doxorubicin, vincristine, and prednisone (CHOP) significantly improves the outcome for patients with advanced-stage follicular lymphoma compared with therapy with CHOP alone: results of a prospective randomized study of the German Low-Grade Lymphoma Study Group. Blood 2005; 106:3725 - 32; http://dx.doi.org/10.1182/blood-2005-01-0016; PMID: 16123223
- van Oers MH, Klasa R, Marcus RE, Wolf M, Kimby E, Gascoyne RD, et al. Rituximab maintenance improves clinical outcome of relapsed/resistant follicular non-Hodgkin lymphoma in patients both with and without rituximab during induction: results of a prospective randomized phase 3 intergroup trial. Blood 2006; 108:3295 - 301; http://dx.doi.org/10.1182/blood-2006-05-021113; PMID: 16873669
- Herold M, Pasold R, Srock S, Neser S, Niederwieser D, Neubauer A, et al. Results of a prospective randomised open label phase III study comparing rituximab plus mitoxantrone, chlorambucile, prednisolone chemotherapy (R-MCP) versus MCP alone in untreated advanced indolent non-Hodgkin's lymphoma (NHL) and mantle-cell-lymphoma (MCL). Blood 2004; 104:584
- Fowler NH. Role of maintenance rituximab (rituxan) therapy in the treatment of follicular lymphoma. P.T. 2011; 36:590 - 8
- Salles G, Seymour JF, Offner F, López-Guillermo A, Belada D, Xerri L, et al. Rituximab maintenance for 2 years in patients with high tumour burden follicular lymphoma responding to rituximab plus chemotherapy (PRIMA): a phase 3, randomised controlled trial. Lancet 2011; 377:42 - 51; http://dx.doi.org/10.1016/S0140-6736(10)62175-7; PMID: 21176949
- van Oers MH, Van Glabbeke M, Giurgea L, Klasa R, Marcus RE, Wolf M, et al. Rituximab maintenance treatment of relapsed/resistant follicular non-Hodgkin’s lymphoma: long-term outcome of the EORTC 20981 phase III randomized intergroup study. J Clin Oncol 2010; 28:2853 - 8; http://dx.doi.org/10.1200/JCO.2009.26.5827; PMID: 20439641
- Forstpointner R, Unterhalt M, Dreyling M, Böck HP, Repp R, Wandt H, et al, German Low Grade Lymphoma Study Group (GLSG). Maintenance therapy with rituximab leads to a significant prolongation of response duration after salvage therapy with a combination of rituximab, fludarabine, cyclophosphamide, and mitoxantrone (R-FCM) in patients with recurring and refractory follicular and mantle cell lymphomas: Results of a prospective randomized study of the German Low Grade Lymphoma Study Group (GLSG). Blood 2006; 108:4003 - 8; http://dx.doi.org/10.1182/blood-2006-04-016725; PMID: 16946304
- Martinelli G, Schmitz SF, Utiger U, Cerny T, Hess U, Bassi S, et al. Long-term follow-up of patients with follicular lymphoma receiving single-agent rituximab at two different schedules in trial SAKK 35/98. J Clin Oncol 2010; 28:4480 - 4; http://dx.doi.org/10.1200/JCO.2010.28.4786; PMID: 20697092
- Vidal L, Gafter-Gvili A, Leibovici L, Shpilberg O. Rituximab as maintenance therapy for patients with follicular lymphoma. Cochrane Database Syst Rev 2009; CD006552; PMID: 19370640
- Eichhorst B, Dreyling M, Robak T, Montserrat E, Hallek M, ESMO Guidelines Working Group. Chronic lymphocytic leukemia: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol 2011; 22:Suppl 6 vi50 - 4; http://dx.doi.org/10.1093/annonc/mdr377; PMID: 21908504
- Tilly H, Dreyling M, ESMO Guidelines Working Group. Diffuse large B-cell non-Hodgkin’s lymphoma: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol 2010; 21:Suppl 5 v172 - 4; http://dx.doi.org/10.1093/annonc/mdq203; PMID: 20555073
- Dreyling M, Ghielmini M, Marcus R, Salles G, Vitolo U, ESMO Guidelines Working Group. Newly diagnosed and relapsed follicular lymphoma: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol 2011; 22:Suppl 6 vi59 - 63; http://dx.doi.org/10.1093/annonc/mdr388; PMID: 21908506
- Lemery SJ, Zhang JZ, Rothmann MD, Yang J, Earp JC, Zhao H, et al. U.S. Food and Drug Administration approval: ofatumumab for the treatment of patients with chronic lymphocytic leukemia refractory to fludarabine and alemtuzumab. Clin Cancer Res 2010; 16:4331 - 8; http://dx.doi.org/10.1158/1078-0432.CCR-10-0570; PMID: 20601446
- Wierda WG, Kipps TJ, Mayer J, Stilgenbauer S, Williams CD, Hellmann A, et al, Hx-CD20-406 Study Investigators. Ofatumumab as single-agent CD20 immunotherapy in fludarabine-refractory chronic lymphocytic leukemia. J Clin Oncol 2010; 28:1749 - 55; http://dx.doi.org/10.1200/JCO.2009.25.3187; PMID: 20194866
- Illidge T, Morschhauser F. Radioimmunotherapy in follicular lymphoma. Best Pract Res Clin Haematol 2011; 24:279 - 93; http://dx.doi.org/10.1016/j.beha.2011.03.005; PMID: 21658624
- Lim SH, Beers SA, French RR, Johnson PW, Glennie MJ, Cragg MS. Anti-CD20 monoclonal antibodies: historical and future perspectives. Haematologica 2010; 95:135 - 43; http://dx.doi.org/10.3324/haematol.2008.001628; PMID: 19773256
- Chan HT, Hughes D, French RR, Tutt AL, Walshe CA, Teeling JL, et al. CD20-induced lymphoma cell death is independent of both caspases and its redistribution into triton X-100 insoluble membrane rafts. Cancer Res 2003; 63:5480 - 9; PMID: 14500384
- Cragg MS, Glennie MJ. Antibody specificity controls in vivo effector mechanisms of anti-CD20 reagents. Blood 2004; 103:2738 - 43; http://dx.doi.org/10.1182/blood-2003-06-2031; PMID: 14551143
- Cragg MS, Morgan SM, Chan HT, Morgan BP, Filatov AV, Johnson PW, et al. Complement-mediated lysis by anti-CD20 mAb correlates with segregation into lipid rafts. Blood 2003; 101:1045 - 52; http://dx.doi.org/10.1182/blood-2002-06-1761; PMID: 12393541
- Mössner E, Brünker P, Moser S, Püntener U, Schmidt C, Herter S, et al. Increasing the efficacy of CD20 antibody therapy through the engineering of a new type II anti-CD20 antibody with enhanced direct and immune effector cell-mediated B-cell cytotoxicity. Blood 2010; 115:4393 - 402; http://dx.doi.org/10.1182/blood-2009-06-225979; PMID: 20194898
- Alduaij W, Ivanov A, Honeychurch J, Cheadle EJ, Potluri S, Lim SH, et al. Novel type II anti-CD20 monoclonal antibody (GA101) evokes homotypic adhesion and actin-dependent, lysosome-mediated cell death in B-cell malignancies. Blood 2011; 117:4519 - 29; http://dx.doi.org/10.1182/blood-2010-07-296913; PMID: 21378274
- Niederfellner G, Lammens A, Mundigl O, Georges GJ, Schaefer W, Schwaiger M, et al. Epitope characterization and crystal structure of GA101 provide insights into the molecular basis for type I/II distinction of CD20 antibodies. Blood 2011; 118:358 - 67; http://dx.doi.org/10.1182/blood-2010-09-305847; PMID: 21444918
- MabThera SmPC. June 2012. Available at: http://www.ema.europa.eu/docs/en_GB/document_library/EPAR_-_Product_Information/human/000165/WC500025821.pdf. Accessed October 2012.
- Clinical trials information is available at: http://www.clinicaltrials.gov. Accessed October 2012.
- Deans JP, Robbins SM, Polyak MJ, Savage JA. Rapid redistribution of CD20 to a low density detergent-insoluble membrane compartment. J Biol Chem 1998; 273:344 - 8; http://dx.doi.org/10.1074/jbc.273.1.344; PMID: 9417086
- Polyak MJ, Deans JP. Alanine-170 and proline-172 are critical determinants for extracellular CD20 epitopes; heterogeneity in the fine specificity of CD20 monoclonal antibodies is defined by additional requirements imposed by both amino acid sequence and quaternary structure. Blood 2002; 99:3256 - 62; http://dx.doi.org/10.1182/blood.V99.9.3256; PMID: 11964291
- Beers SA, Chan CH, James S, French RR, Attfield KE, Brennan CM, et al. Type II (tositumomab) anti-CD20 monoclonal antibody out performs type I (rituximab-like) reagents in B-cell depletion regardless of complement activation. Blood 2008; 112:4170 - 7; http://dx.doi.org/10.1182/blood-2008-04-149161; PMID: 18583569
- Teeling JL, French RR, Cragg MS, van den Brakel J, Pluyter M, Huang H, et al. Characterization of new human CD20 monoclonal antibodies with potent cytolytic activity against non-Hodgkin lymphomas. Blood 2004; 104:1793 - 800; http://dx.doi.org/10.1182/blood-2004-01-0039; PMID: 15172969
- Cragg MS. CD20 antibodies: doing the time warp. Blood 2011; 118:219 - 20; http://dx.doi.org/10.1182/blood-2011-04-346700; PMID: 21757627
- Ivanov A, Beers SA, Walshe CA, Honeychurch J, Alduaij W, Cox KL, et al. Monoclonal antibodies directed to CD20 and HLA-DR can elicit homotypic adhesion followed by lysosome-mediated cell death in human lymphoma and leukemia cells. J Clin Invest 2009; 119:2143 - 59; PMID: 19620786
- Jak M, van Bochove G, Klein C, Umana P, Eldering E, Van Oers MHJ. CD40 stimulation sensitizes CLL cells to CD20-triggered cell death by rituximab and GA101 via a different mechanism. Blood 2011; 118:5178 - 88
- Honeychurch J, Alduaij W, Azizyan M, Cheadle EJ, Pelicano H, Ivanov A, et al. Antibody-induced nonapoptotic cell death in human lymphoma and leukemia cells is mediated through a novel reactive oxygen species-dependent pathway. Blood 2012; 119:3523 - 33; http://dx.doi.org/10.1182/blood-2011-12-395541; PMID: 22354003
- Cartron G, Dacheux L, Salles G, Solal-Celigny P, Bardos P, Colombat P, et al. Therapeutic activity of humanized anti-CD20 monoclonal antibody and polymorphism in IgG Fc receptor FcgammaRIIIa gene. Blood 2002; 99:754 - 8; http://dx.doi.org/10.1182/blood.V99.3.754; PMID: 11806974
- Cartron G, Watier H, Golay J, Solal-Celigny P. From the bench to the bedside: ways to improve rituximab efficacy. Blood 2004; 104:2635 - 42; http://dx.doi.org/10.1182/blood-2004-03-1110; PMID: 15226177
- Koene HR, Kleijer M, Algra J, Roos D, von dem Borne AE, de Haas M. Fc gammaRIIIa-158V/F polymorphism influences the binding of IgG by natural killer cell Fc gammaRIIIa, independently of the Fc gammaRIIIa-48L/R/H phenotype. Blood 1997; 90:1109 - 14; PMID: 9242542
- Ferrara C, Stuart F, Sondermann P, Brünker P, Umaña P. The carbohydrate at FcgammaRIIIa Asn-162. An element required for high affinity binding to non-fucosylated IgG glycoforms. J Biol Chem 2006; 281:5032 - 6; http://dx.doi.org/10.1074/jbc.M510171200; PMID: 16330541
- Bowles JA, Wang SY, Link BK, Allan B, Beuerlein G, Campbell MA, et al. Anti-CD20 monoclonal antibody with enhanced affinity for CD16 activates NK cells at lower concentrations and more effectively than rituximab. Blood 2006; 108:2648 - 54; http://dx.doi.org/10.1182/blood-2006-04-020057; PMID: 16825493
- Weng WK, Levy R. Two immunoglobulin G fragment C receptor polymorphisms independently predict response to rituximab in patients with follicular lymphoma. J Clin Oncol 2003; 21:3940 - 7; http://dx.doi.org/10.1200/JCO.2003.05.013; PMID: 12975461
- Beers SA, French RR, Chan HT, Lim SH, Jarrett TC, Vidal RM, et al. Antigenic modulation limits the efficacy of anti-CD20 antibodies: implications for antibody selection. Blood 2010; 115:5191 - 201; http://dx.doi.org/10.1182/blood-2010-01-263533; PMID: 20223920
- Lim SH, Vaughan AT, Ashton-Key M, Williams EL, Dixon SV, Chan HT, et al. Fc gamma receptor IIb on target B cells promotes rituximab internalization and reduces clinical efficacy. Blood 2011; 118:2530 - 40; http://dx.doi.org/10.1182/blood-2011-01-330357; PMID: 21768293
- Teeling JL, Mackus WJ, Wiegman LJ, van den Brakel JH, Beers SA, French RR, et al. The biological activity of human CD20 monoclonal antibodies is linked to unique epitopes on CD20. J Immunol 2006; 177:362 - 71; PMID: 16785532
- Binder M, Otto F, Mertelsmann R, Veelken H, Trepel M. The epitope recognized by rituximab. Blood 2006; 108:1975 - 8; http://dx.doi.org/10.1182/blood-2006-04-014639; PMID: 16705086
- Du J, Wang H, Zhong C, Peng B, Zhang M, Li B, et al. Structural basis for recognition of CD20 by therapeutic antibody Rituximab. J Biol Chem 2007; 282:15073 - 80; http://dx.doi.org/10.1074/jbc.M701654200; PMID: 17395584
- Du J, Wang H, Zhong C, Peng B, Zhang M, Li B, et al. Crystal structure of chimeric antibody C2H7 Fab in complex with a CD20 peptide. Mol Immunol 2008; 45:2861 - 8; http://dx.doi.org/10.1016/j.molimm.2008.01.034; PMID: 18346788
- Du J, Yang H, Guo Y, Ding J. Structure of the Fab fragment of therapeutic antibody Ofatumumab provides insights into the recognition mechanism with CD20. Mol Immunol 2009; 46:2419 - 23; http://dx.doi.org/10.1016/j.molimm.2009.04.009; PMID: 19427037
- Perosa F, Favoino E, Caragnano MA, Dammacco F. Generation of biologically active linear and cyclic peptides has revealed a unique fine specificity of rituximab and its possible cross-reactivity with acid sphingomyelinase-like phosphodiesterase 3b precursor. Blood 2006; 107:1070 - 7; http://dx.doi.org/10.1182/blood-2005-04-1769; PMID: 16223774
- Perosa F, Favoino E, Vicenti C, Guarnera A, Racanelli V, De Pinto V, et al. Two structurally different rituximab-specific CD20 mimotope peptides reveal that rituximab recognizes two different CD20-associated epitopes. J Immunol 2009; 182:416 - 23; PMID: 19109173
- Ernst JA, Li H, Kim HS, Nakamura GR, Yansura DG, Vandlen RL. Isolation and characterization of the B-cell marker CD20. Biochemistry 2005; 44:15150 - 8; http://dx.doi.org/10.1021/bi0511078; PMID: 16285718
- Li B, Zhao L, Guo H, Wang C, Zhang X, Wu L, et al. Characterization of a rituximab variant with potent antitumor activity against rituximab-resistant B-cell lymphoma. Blood 2009; 114:5007 - 15; http://dx.doi.org/10.1182/blood-2009-06-225474; PMID: 19828699
- Johnson NA, Leach S, Woolcock B, deLeeuw RJ, Bashashati A, Sehn LH, et al. CD20 mutations involving the rituximab epitope are rare in diffuse large B-cell lymphomas and are not a significant cause of R-CHOP failure. Haematologica 2009; 94:423 - 7; http://dx.doi.org/10.3324/haematol.2008.001024; PMID: 19211644
- Morschhauser F, Leonard JP, Fayad L, Coiffier B, Petillon MO, Coleman M, et al. Humanized anti-CD20 antibody, veltuzumab, in refractory/recurrent non-Hodgkin’s lymphoma: phase I/II results. J Clin Oncol 2009; 27:3346 - 53; http://dx.doi.org/10.1200/JCO.2008.19.9117; PMID: 19451441
- Goldenberg DM, Rossi EA, Stein R, Cardillo TM, Czuczman MS, Hernandez-Ilizaliturri FJ, et al. Properties and structure-function relationships of veltuzumab (hA20), a humanized anti-CD20 monoclonal antibody. Blood 2009; 113:1062 - 70; http://dx.doi.org/10.1182/blood-2008-07-168146; PMID: 18941114
- Forero-Torres A, de Vos S, Pohlman BL, Pashkevich M, Cronier DM, Dang NH, et al. Results of a phase 1 study of AME-133v (LY2469298), an Fc-engineered humanized monoclonal anti-CD20 antibody, in FcγRIIIa-genotyped patients with previously treated follicular lymphoma. Clin Cancer Res 2012; 18:1395 - 403; http://dx.doi.org/10.1158/1078-0432.CCR-11-0850; PMID: 22223529
- Gordon LI, Molina A, Witzig T, Emmanouilides C, Raubtischek A, Darif M, et al. Durable responses after ibritumomab tiuxetan radioimmunotherapy for CD20+ B-cell lymphoma: long-term follow-up of a phase 1/2 study. Blood 2004; 103:4429 - 31; http://dx.doi.org/10.1182/blood-2003-11-3883; PMID: 15016644
- Witzig TE, White CA, Wiseman GA, Gordon LI, Emmanouilides C, Raubitschek A, et al. Phase I/II trial of IDEC-Y2B8 radioimmunotherapy for treatment of relapsed or refractory CD20(+) B-cell non-Hodgkin’s lymphoma. J Clin Oncol 1999; 17:3793 - 803; PMID: 10577851
- Witzig TE, Gordon LI, Cabanillas F, Czuczman MS, Emmanouilides C, Joyce R, et al. Randomized controlled trial of yttrium-90-labeled ibritumomab tiuxetan radioimmunotherapy versus rituximab immunotherapy for patients with relapsed or refractory low-grade, follicular, or transformed B-cell non-Hodgkin’s lymphoma. J Clin Oncol 2002; 20:2453 - 63; http://dx.doi.org/10.1200/JCO.2002.11.076; PMID: 12011122
- Wiseman GA, Gordon LI, Multani PS, Witzig TE, Spies S, Bartlett NL, et al. Ibritumomab tiuxetan radioimmunotherapy for patients with relapsed or refractory non-Hodgkin lymphoma and mild thrombocytopenia: a phase II multicenter trial. Blood 2002; 99:4336 - 42; http://dx.doi.org/10.1182/blood.V99.12.4336; PMID: 12036859
- Morschhauser F, Radford J, Van Hoof A, Vitolo U, Soubeyran P, Tilly H, et al. Phase III trial of consolidation therapy with yttrium-90-ibritumomab tiuxetan compared with no additional therapy after first remission in advanced follicular lymphoma. J Clin Oncol 2008; 26:5156 - 64; http://dx.doi.org/10.1200/JCO.2008.17.2015; PMID: 18854568
- Hainsworth JD, Spigel DR, Markus TM, Shipley D, Thompson D, Rotman R, et al. Rituximab plus short-duration chemotherapy followed by Yttrium-90 Ibritumomab tiuxetan as first-line treatment for patients with follicular non-Hodgkin lymphoma: a phase II trial of the Sarah Cannon Oncology Research Consortium. Clin Lymphoma Myeloma 2009; 9:223 - 8; http://dx.doi.org/10.3816/CLM.2009.n.044; PMID: 19525191
- Morschhauser F, Marlton P, Vitolo U, Lindén O, Seymour JF, Crump M, et al. Results of a phase I/II study of ocrelizumab, a fully humanized anti-CD20 mAb, in patients with relapsed/refractory follicular lymphoma. Ann Oncol 2010; 21:1870 - 6; http://dx.doi.org/10.1093/annonc/mdq027; PMID: 20157180
- Sikder MA, Friedberg JW. Beyond rituximab: The future of monoclonal antibodies in B-cell non-Hodgkin lymphoma. Curr Hematol Malig Rep 2008; 3:187 - 93; http://dx.doi.org/10.1007/s11899-008-0027-5; PMID: 20425465
- Shields RL, Namenuk AK, Hong K, Meng YG, Rae J, Briggs J, et al. High resolution mapping of the binding site on human IgG1 for Fc gamma RI, Fc gamma RII, Fc gamma RIII, and FcRn and design of IgG1 variants with improved binding to the Fc gamma R. J Biol Chem 2001; 276:6591 - 604; http://dx.doi.org/10.1074/jbc.M009483200; PMID: 11096108
- Friedberg JW, Vose JM, Kahl BS, Brunvand MW, Goy A, Kasamon YL, et al. A phase I study of PRO131921, a novel anti-CD20 monoclonal antibody in patients with relapsed/refractory CD20+ indolent NHL: Correlation between clinical responses and AUC pharmacokinetics. Blood 2009; 114:3742
- Hayden-Ledbetter MS, Cerveny CG, Espling E, Brady WA, Grosmaire LS, Tan P, et al. CD20-directed small modular immunopharmaceutical, TRU-015, depletes normal and malignant B cells. Clin Cancer Res 2009; 15:2739 - 46; http://dx.doi.org/10.1158/1078-0432.CCR-08-1694; PMID: 19351771
- Czuczman MS, Fayad L, Delwail V, Cartron G, Jacobsen E, Kuliczkowski K, et al, 405 Study Investigators. Ofatumumab monotherapy in rituximab-refractory follicular lymphoma: results from a multicenter study. Blood 2012; 119:3698 - 704; http://dx.doi.org/10.1182/blood-2011-09-378323; PMID: 22389254
- Czuczman MS, Hess G, Gadeberg OV, Pedersen LM, Goldstein N, Gupta I, et al, 409 Study Investigators. Chemoimmunotherapy with ofatumumab in combination with CHOP in previously untreated follicular lymphoma. Br J Haematol 2012; 157:438 - 45; http://dx.doi.org/10.1111/j.1365-2141.2012.09086.x; PMID: 22409295
- Hagenbeek A, Plesner T, Johnson P, Pedersen L, Walewski J, Hellman A, et al. HuMax-CD20, a novel fully human anti-CD20 monoclonal antibody: Results of a phase I/II trial in relapsed or refractory follicular non-Hodgkins's lymphoma. Blood 2005; 106:Abstract 4760; PMID: 16174759
- Uchiyama S, Suzuki Y, Otake K, Yokoyama M, Ohta M, Aikawa S, et al. Development of novel humanized anti-CD20 antibodies based on affinity constant and epitope. Cancer Sci 2010; 101:201 - 9; http://dx.doi.org/10.1111/j.1349-7006.2009.01392.x; PMID: 19930155
- Engelberts P, Beurskens F, Mackus W, Bakker J, Vink T, Tiebout A, et al. Ofatumuab targets a conformational membrane-proximal epitope which contains amino acids located in the small and large loops of CD20. Haematologica 2010; 95:46
- Beum PV, Lindorfer MA, Beurskens F, Stukenberg PT, Lokhorst HM, Pawluczkowycz AW, et al. Complement activation on B lymphocytes opsonized with rituximab or ofatumumab produces substantial changes in membrane structure preceding cell lysis. J Immunol 2008; 181:822 - 32; PMID: 18566448
- Herter S, Waldhauer I, Otz T, Herting F, Lang S, Nicolini V, et al. . Superior Efficacy of the Novel Type II, Glycoengineered CD20 Antibody GA101vs. the Type I CD20 Antibodies Rituximab and Ofatumumab. Blood2010; 116:3925.
- Wu L, Wang C, Zhang D, Zhang X, Qian W, Zhao L, et al. Characterization of a humanized anti-CD20 antibody with potent antitumor activity against B-cell lymphoma. Cancer Lett 2010; 292:208 - 14; http://dx.doi.org/10.1016/j.canlet.2009.12.004; PMID: 20056316
- Baritaki S, Militello L, Malaponte G, Spandidos DA, Salcedo M, Bonavida B. The anti-CD20 mAb LFB-R603 interrupts the dysregulated NF-κB/Snail/RKIP/PTEN resistance loop in B-NHL cells: role in sensitization to TRAIL apoptosis. Int J Oncol 2011; 38:1683 - 94; PMID: 21455568
- Kaminski MS, Zelenetz AD, Press OW, Saleh M, Leonard J, Fehrenbacher L, et al. Pivotal study of iodine I 131 tositumomab for chemotherapy-refractory low-grade or transformed low-grade B-cell non-Hodgkin’s lymphomas. J Clin Oncol 2001; 19:3918 - 28; PMID: 11579112
- Davis TA, Kaminski MS, Leonard JP, Hsu FJ, Wilkinson M, Zelenetz A, et al. The radioisotope contributes significantly to the activity of radioimmunotherapy. Clin Cancer Res 2004; 10:7792 - 8; http://dx.doi.org/10.1158/1078-0432.CCR-04-0756; PMID: 15585610
- Nishida M, Usuda S, Okabe M, Miyakoda H, Komatsu M, Hanaoka H, et al. Characterization of novel murine anti-CD20 monoclonal antibodies and their comparison to 2B8 and c2B8 (rituximab). Int J Oncol 2007; 31:29 - 40; PMID: 17549402
- Stein R, Qu Z, Chen S, Rosario A, Shi V, Hayes M, et al. Characterization of a new humanized anti-CD20 monoclonal antibody, IMMU-106, and Its use in combination with the humanized anti-CD22 antibody, epratuzumab, for the therapy of non-Hodgkin’s lymphoma. Clin Cancer Res 2004; 10:2868 - 78; http://dx.doi.org/10.1158/1078-0432.CCR-03-0493; PMID: 15102696
- Forero-Torres A, de Vos S, Pohlman BL, Pashkevich M, Cronier DM, Dang NH, et al. Results of a phase 1 study of AME-133v (LY2469298), an Fc-engineered humanized monoclonal anti-CD20 antibody, in FcγRIIIa-genotyped patients with previously treated follicular lymphoma. Clin Cancer Res 2012; 18:1395 - 403; http://dx.doi.org/10.1158/1078-0432.CCR-11-0850; PMID: 22223529