Abstract
We developed a rapid method to analyze Fc glycosylation of Fc fusion proteins, especially those with mutated Fc hinge regions. Fc fusion proteins were digested with IdeS, an IgG specific protease with exosites for substrate recognition and cleavage. The resultant fragments were directly analyzed through liquid chromatography mass spectrometry. The structures and relative quantities of Fc glycans were deduced from their masses and intensities. The separated substrate recognition and cleavage property of IdeS makes this method applicable to a broad range of Fc fusion proteins having either standard or non-canonical hinge regions.
Human Fc fusion has become an extremely popular protein expression/production platform for protein therapeutics. The Fc endows several advantages to the fusion partners, such as improved protein stability and solubility, simplified affinity purification and extended in vivo half-life.Citation1 Addition of Fc, however, introduces challenges to the analysis of the fusion proteins. Particularly, when glycosylated fusion partners are included, glycosylation characterization becomes fairly complicated. Glycosylation influences the physiochemical properties and biological functions of glycoproteins. The actual effects depend on both the glycan structures and locations. It has been shown that glycan structures at Asn297 in CH2 domain of Fc could modulate antibody binding to various cell surface receptors and affect the in vivo biological function of the fusion proteins. For example, an antibody produced in glyco-engineered Pichia with afucosylated glycans at Asn297 demonstrated stronger antibody-dependent cell-mediated cytotoxicity (ADCC) than the one produced in Chinese hamster ovary (CHO) cells with fucosylated glycans.Citation2
Like other glycoproteins, the glycosylation patterns of Fc fusion proteins, are largely decided by the production platforms, but other factors, such as fermentation conditions, purification steps and the local protein structure, can affect the glycan structures.Citation3 For antibodies and Fc fusion proteins, distinct glycosylation profiles were often detected in Fab/fusion partners and Fc region. For example, more sialylated glycans were observed on Fab fragment of antibodies or fusion partners of Fc fusion proteins than on Fc.Citation4
Given their unique influences on the in vitro and in vivo properties of fusion proteins, Fc glycans must be characterized specifically during Fc therapeutic development. Analytical characterization of antibody and Fc fusion therapeutics have been extensively reviewed.Citation5,Citation6 Conventionally, peptide mapping is the method of choice for comprehensive antibody glycosylation analysis. It frequently requires multiple sample preparation steps, lengthy high-performance liquid chromatography (HPLC) separation and time-consuming data analysis. As a result, it is not particularly attractive in high throughput screening of routine samples. A recent study using matrix-assisted laser desorption/ionization-time of flight mass spectrometry MALDI-TOF MS, instead of LC-MS, to analyze tryptic peptides showed improvement in throughput by eliminating the lengthy HPLC separation step. It helped improve the throughput of peptide mapping for antibody glycosylation analysis.Citation7 Moreover, an antibody-specific enzyme, papain, has been widely used to generate Fc and Fab fragments from full-length antibodies. The efficiency of papain digestion, however, varies substantially among different antibodies. Those with terminal N-acetyl glucosamine Fc glycans were found more resistant to papain digestion.Citation8 As such, certain glycan structures might be underrepresented in this approach.
IdeS (immunoglobulin-degrading enzyme of Streptococcus pyogenes), a recently identified cysteine protease, is highly efficient in digesting a large spectrum of IgGs, i.e., across different subclasses and species. Its application for the analysis of full-length IgGs has been reported.Citation9-Citation12 With its cleavage site located in the hinge region (—LLG/G—), IdeS demonstrated an exosite for its binding to Fc.Citation13 Many Fc fusion proteins with non-canonical hinge regions could be subject to IdeS digestion; therefore, its applications could be extended far beyond the standard full-length IgG. As described here, we demonstrated such utility by performing Fc glycosylation analysis of an Fc fusion protein with a mutated Fc hinge region.
The Fc fusion protein we chose was abatacept (Orencia®), which is a CHO cell-produced therapeutic protein with an Fc region of IgG1 fused to the extracellular domain of CTLA-4 (cytotoxic T-lymphocyte antigen).Citation14 The product is marketed for the treatment of rheumatoid arthritis. Different from typical IgG1 Fc, the hinge region of abatacept contains several mutations to accommodate the desired therapeutic profile. Among them,—CPPC—in the hinge region were mutated to—SPPS—, which abrogated the two disulfide bonds in the hinge region between the two heavy chains. Instead, a pair of Cys residues from the CTLA-4 domain formed a disulfide bond holding abatacept in its dimer configuration. Predicted from its amino acid sequence, abatacept has three N-linked glycosylation sites (Asn76, Asn108 in the CTLA-4 region and Asn207 in the Fc region). Additionally, O-linked glycosylation at Ser129 and Ser139 has also been identified through peptide mapping.Citation15
For IdeS digestion, 4 uL 25 mg/mL abatacept reconstituted from lyophilized powder was directly diluted in 96 uL 150 mM sodium chloride, 20 mM sodium phosphate pH 6.6 and incubated with 1 uL IdeS (Bulldog Bio, Portsmouth, NH) at 37°C for 30 min. Because the reported O-linked glycosylation and sialylation might complicate the assignment and quantitation of N-linked glycan structures, we treated 50 ug and 10 ug of IdeS digested abatacept with 1 uL PNGase F (New England BioLabs) and 1 uL neuraminidase (New England BioLabs), respectively, at 37°C for 30 min. The digested samples were then directly loaded onto an Agilent Q-TOF 6520 mass spectrometer coupled with Agilent 1200 HPLC (Agilent). An in-line MassPREP Micro desalt cartridge (Waters, Milford, MA) was used to remove salts in the samples prior to directing the flow to the mass spectrometer. The proteins were eluted off the cartridge in a one-step gradient: 100% buffer A (0.1% formic acid in water) flowed at 1 mL per min for the first two minutes, then the flow was set for 100% buffer B (0.1% formic acid, 10% water and 90% acetonitrile) at 0.4 mL per min from 2.01 to 7 min. At the end of the gradient, a three minute post-run of 100% buffer A was performed to equilibrate the cartridge. On the mass spectrometer, data from 300 to 3000 m/z were collected from 2 to 7 min. The dual ESI ion source was set as follows: gas temp at 350°C; drying gas at 13 L/min; nebulizer at 45 psig; fragmentor at 150 V; skimmer at 65 V; Oct1 RF VPP at 750 V; Vcap at 3500 V. The MS spectra were analyzed using MassHunter software and the deconvolution was done using maximum entropy with deconvoluted mass range set from 10 to 100 KDa.
While abatacept has a reported mass of ~92 kDa according to MALDI-TOF analysis, LC-MS analysis of intact abatacept produced a basically non-deconvolutable spectrum (data not shown), which was probably due to the heterogeneity of glycosylation. The results of LC-MS analysis of IdeS-treated abatacept are presented in . The deconvoluted spectrum of IdeS- and PNGase F-treated abatacept is shown in . The peak of 23746.90 Da was the aglycosylated Fc fragment. It was in a good agreement with the theoretically calculated mass of 23745.80 Da (the intra-chain disulfide bonds were fully oxidized and C-terminal Lys was completely processed). The other three peaks, 24071.08 Da, 24395.38 Da and 24719.63 Da, demonstrated sequential 324 Da mass additions to the aglycosylated Fc fragment.
Figure 1. Deconvoluted mass spectra of abatacept digested with IdeS and PNGase F (A), IdeS (B), IdeS and neuraminidase (C). Protein sequence of abatacept is showed. Amino acid mutations, N-linked glycosylation sites and potential O-linked glycosylation sites are highlighted in red, blue and underlined, respectively.
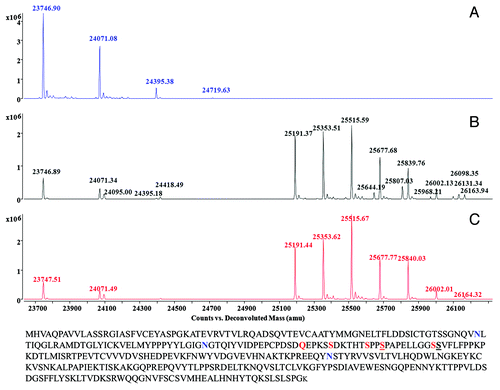
The repeating, uniform mass addition of 324 Da, however, was not due to O-linked glycosylation, in which the glycans are attached to either a serine or threonine on the protein through N-acetylgalactosamine. Instead, they were most likely from the non-enzymatic maltose glycation through the primary amine group on the lysine residues. Abatacept has two distinct formulations for clinic administrations, a lyophilized powder containing 50 mg maltose per 25 mg protein and a liquid solution without maltose. Maltose glycation on proteins through lysine has been observed.Citation16 One maltose added 324 Da to the glycated protein, which was the exact mass addition we observed here. It was reasonable for us to believe that the observed 324 Da modification was a maltose glycation that resulted from the storage of reconstituted abatacept powder formulated with maltose. A comparison of abatacept in both formulations would be helpful to further confirm the glycation modifications, but such experiments were not performed because we did not have access to both the liquid formulation and the un-dissolved lyophilized form.
The extent of maltose glycation in the sample was quantitated according to the abundance of each species in the spectra. The results were showed in , and they were used in the quantitative analysis of various N-linked glycans detected in , which shows the deconvoluted spectrum of abatacept treated with IdeS only. There were two clusters of peaks. The first cluster comprised Fc fragments with no or partial N-linked glycosylation. The mass of 24095.00 Da matched the theoretical mass of the Fc fragment containing an N-acetyl-glucosamine with the core fucose. The 24418.49 Da peak represented its glycated form. In the second cluster, 25191.37 Da and 25353.51 Da were the Fc fragments with typical G0F, G1F structures, which are commonly observed on the Fc region of antibodies produced in CHO cells; nomenclature and structures of detected N-linked glycans are listed in . The 25515.59 Da peak, however, could be the Fc fragment with either a G2F glycan or a G0F glycan with a single maltose glycation event. Likewise, 25677.68 Da, 25839.76 Da, 26002.13 Da and 26163.94 Da could be the Fc fragments with only N-linked glycans or N-linked glycans containing additional maltose glycation or a mixture of both.
Table 1. Detected glycosylation on Fc fragments and the relative compositions. Glycation contents were determined based on . N-linked glycan quantitation was calculated based on
To determine the actual composition of each peak, we assessed the contribution of glycated species based on the glycation efficiency calculated from (also showed in ) with the assumption that the occurrences of glycation and N-linked glycosylation were independent to each other. Based on our calculations, 25515.59 Da, 25677.68 Da, 25839.76 Da, 26002.13 Da and 26163.94 Da peaks contained both glycated and non-glycated Fc fragments. When calculating the relative ratios of the Fc fragments with various N-linked glycans, we took only the peaks without glycation into account and the contribution from the glycated species was subtracted based on the glycation efficiency obtained from .
The sialylated glycans were confirmed by treating the sample with neuraminidase. The masses present in were not observed in (with neuraminidase treatment), indicating they were sialylated. The Fc fragment with the mass of 25644.19 Da in contained the glycan structure of A1F-Gal; 25807.03 Da was the mass of the Fc fragment with A1F glycan; 25968.21 Da indicated the A1F+Gal structure, 26131.34 Da indicated the same N-linked glycan with a maltose glycation; 26098.35 Da was the Fc fragment with an A2F structure. The observed extra galactose addition to G2F and A1F structures suggested α-gal structure on abatacept, which was previously reported.Citation17 The relative composition of glycation and various N-linked glycans were calculated separately and summarized in .
In conclusion, we demonstrated here a simple procedure combining IdeS digestion with LC-MS for Fc glycosylation analysis of Fc fusion proteins with non-canonical hinge regions and glycosylated fusion partners. In this method, the structures and relative composition of each N-linked Fc glycans were assigned and assessed based on the masses and relative abundances of Fc fragments. As examined in this study, the Fc region of abatacept contains uncommon α-galactosylation, in addition to the typical G0F, G1F, G2F and sialylated structures. Moreover, the maltose glycation presumably caused by reconstituting and storing lyophilized abatacept in a buffer containing the reducing sugar of maltose was also detected. As shown, this rapid procedure was very effective for the analysis of Fc fusion proteins with mutated Fc hinge regions and extended the utility of IdeS digestion beyond the analysis of the standard full-length IgG.
Disclosure of Potential Conflicts of Interest
No potential conflict of interest was disclosed.
References
- Beck A, Reichert JM. Therapeutic Fc-fusion proteins and peptides as successful alternatives to antibodies. MAbs 2011; 3:415 - 6; http://dx.doi.org/10.4161/mabs.3.5.17334; PMID: 21785279
- Li H, Sethuraman N, Stadheim TA, Zha D, Prinz B, Ballew N, et al. Optimization of humanized IgGs in glycoengineered Pichia pastoris.. Nat Biotechnol 2006; 24:210 - 5; http://dx.doi.org/10.1038/nbt1178; PMID: 16429149
- Hossler P, Khattak SF, Li ZJ. Optimal and consistent protein glycosylation in mammalian cell culture. Glycobiology 2009; 19:936 - 49; http://dx.doi.org/10.1093/glycob/cwp079; PMID: 19494347
- Mimura Y, Ashton PR, Takahashi N, Harvey DJ, Jefferis R. Contrasting glycosylation profiles between Fab and Fc of a human IgG protein studied by electrospray ionization mass spectrometry. J Immunol Methods 2007; 326:116 - 26; http://dx.doi.org/10.1016/j.jim.2007.07.014; PMID: 17714731
- Beck A, Wagner-Rousset E, Ayoub D, Van Dorsselaer A, Sanglier-Cianférani S. Characterization of therapeutic antibodies and related products. Anal Chem 2013; 85:715 - 36; http://dx.doi.org/10.1021/ac3032355; PMID: 23134362
- Beck A, Diemer H, Ayoub D, Debaene F, Wagner-Rousset E, Carapito C, et al. Analytical characterization of biosimilar antibodies and Fc-fusion proteins. Trends Anal Chem 2013; http://dx.doi.org/10.1016/j.trac.2013.02.014.
- Baković MP, Selman MHJ, Hoffmann M, Rudan I, Campbell H, Deelder AM, et al. High-throughput IgG Fc N-glycosylation profiling by mass spectrometry of glycopeptides. J Proteome Res 2013; 12:821 - 31; http://dx.doi.org/10.1021/pr300887z; PMID: 23298168
- Raju TS, Scallon B. Fc glycans terminated with N-acetylglucosamine residues increase antibody resistance to papain. Biotechnol Prog 2007; 23:964 - 71; PMID: 17571902
- Chevreux G, Tilly N, Bihoreau N. Fast analysis of recombinant monoclonal antibodies using IdeS proteolytic digestion and electrospray mass spectrometry. Anal Biochem 2011; 415:212 - 4; http://dx.doi.org/10.1016/j.ab.2011.04.030; PMID: 21596014
- Goetze AM, Zhang Z, Liu L, Jacobsen FW, Flynn GC. Rapid LC-MS screening for IgG Fc modifications and allelic variants in blood. Mol Immunol 2011; 49:338 - 52; http://dx.doi.org/10.1016/j.molimm.2011.09.002; PMID: 21945018
- Janin-Bussat MC, Tonini L, Huillet C, Colas O, Klinguer-Hamour C, Corvaïa N, et al. Cetuximab Fab and Fc N-glycan fast characterization using IdeS digestion and liquid chromatography coupled to electrospray ionization mass spectrometry. Methods Mol Biol 2013; 988:93 - 113; http://dx.doi.org/10.1007/978-1-62703-327-5_7; PMID: 23475716
- Wang B, Gucinski AC, Keire DA, Buhse LF, Boyne MT 2nd. Structural comparison of two anti-CD20 monoclonal antibody drug products using middle-down mass spectrometry. Analyst 2013; 138:3058 - 65; http://dx.doi.org/10.1039/c3an36524g; PMID: 23579346
- Vincents B, von Pawel-Rammingen U, Björck L, Abrahamson M. Enzymatic characterization of the streptococcal endopeptidase, IdeS, reveals that it is a cysteine protease with strict specificity for IgG cleavage due to exosite binding. Biochemistry 2004; 43:15540 - 9; http://dx.doi.org/10.1021/bi048284d; PMID: 15581366
- Linsley PS, Ledbetter JA, Damle NK, Brady W. CTLA4Ig fusion proteins. United States Patent 1998; Nr. 5844095.
- Bongers J, Devincentis J, Fu J, Huang P, Kirkley DH, Leister K, et al. Characterization of glycosylation sites for a recombinant IgG1 monoclonal antibody and a CTLA4-Ig fusion protein by liquid chromatography-mass spectrometry peptide mapping. J Chromatogr A 2011; 1218:8140 - 9; http://dx.doi.org/10.1016/j.chroma.2011.08.089; PMID: 21978954
- Montgomery H, Tanaka K, Belgacem O. Glycation pattern of peptides condensed with maltose, lactose and glucose determined by ultraviolet matrix-assisted laser desorption/ionization tandem mass spectrometry. Rapid Commun Mass Spectrom 2010; 24:841 - 8; http://dx.doi.org/10.1002/rcm.4455; PMID: 20187122
- Bosques CJ, Collins BE, Meador JW 3rd, Sarvaiya H, Murphy JL, Dellorusso G, et al. Chinese hamster ovary cells can produce galactose-α-1,3-galactose antigens on proteins. Nat Biotechnol 2010; 28:1153 - 6; http://dx.doi.org/10.1038/nbt1110-1153; PMID: 21057479