Abstract
The plasticity of natural immunoglobulin repertoires can be exploited for the generation of phage display libraries. Secondary lymphoid organs, such as the spleen and the lymph nodes, constitute interesting sources of diversity because they are rich in B cells, part of which can be affinity matured. These organs, however, differ in their anatomical structure, reflecting the different fluids they drain, which affects the B cell repertoires. The CDRH3 repertoires from these organs, extracted from naïve or immunized mice, were compared in the context of phage display libraries using human antibody framework families. Deep sequencing analysis revealed that all libraries displayed different CDRH3 repertoires, but the one derived from lymph nodes of naïve mice was the most diverse. Library performance was assessed by in vitro selection. For both organs, immunization increased substantially the frequency of molecules able to bind to the immunogen. The library derived from lymph nodes from naïve mice, however, was the most effective in generating diverse and high affinity candidates. These results illustrate that the use of a biased CDRH3 repertoire increases the performance of libraries, but reduces the clonal diversity, which may be detrimental for certain strategies.
Introduction
In vitro display and selection technologies involving phage,Citation1 bacteria,Citation2 yeastCitation3 or ribosomeCitation4 are commonly used to generate antibodies for research, diagnostic and therapeutic applications.Citation5 These approaches allow the isolation of antigen-specific antibody fragments from large libraries of immunoglobulin variable genes. The success of the selection process is dependent on the number, diversity and quality of the sequences present in the library. A direct correlation has been reported between the size of a library and the affinity of the candidates.Citation6-Citation8
Different strategies have been used for the diversification of antibody libraries. Oligonucleotides can be used to randomly diversify defined positions in the complementary-determining regions (CDR) of antibody genes.Citation9,Citation10 Alternatively, naturally rearranged variable genes isolated from humans or animals can be assembled to build antibody libraries.Citation11-Citation13,Citation18 Natural antibody repertoires are an attractive source of diversity because, during B cell maturation, productive gene rearrangements are controlled at several stages. This proofreading mechanism increases the frequency of genes encoding a functional variable domain that are incorporated into the library and, thus, its functionality. In contrast, artificial randomization of CDR sequences lead to a higher proportion of miss-folded and non-functional polypeptides. This limitation is particularly significant when considering the CDR3 of the heavy chain (CDRH3), which can be relatively long and is more difficult to diversify using oligonucleotides while maintaining proper protein folding. On the other hand, natural antibody repertoires contain variable domains that are less-represented or suboptimal from a stability and manufacturing standpoint, and are therefore less well-suited for industrial applications.
To overcome these limitations, we recently described a novel cloning strategy to recover CDRH3 sequences from human or other species. These repertoires are then integrated into human antibody frameworks selected for their representation in human repertoires and for their biochemical stability.Citation14 This approach allows for trapping of CDRH3 sequences from different sources into human antibody frameworks, expanding the diversity that can be exploited to generate human antibodies. For instance, CDRH3 sequences that have been biased against an antigen can be efficiently captured to generate target specific libraries and increase the frequency of isolating specific antibodies. This can be achieved by retrieving CDRH3 sequences from either immunized animals or pools of sequences that were enriched against an antigen using in vitro selection approaches such as phage display.Citation14
When B cells encounter an antigen recognized by their B cell receptor (BCR) and encounter T cell help, they undergo affinity maturation and a selection process to produce high affinity antibodies.Citation15 These events occur in the germinal centers of secondary lymphoid tissues such as lymph nodes and spleens. The proportion of B cells in these organs is 25% and 40%, respectively, which are further diversified by somatic hypermutation following immunization.Citation16 B cells from secondary lymphoid organs can be fused to myeloma cells to generate monoclonal antibodies via hybridoma technology.Citation17 As the lymph nodes and the spleen are anatomically different and drain antigens via different routes,Citation18,Citation19 differences in B cell populations and corresponding antibody repertoires have been reported.Citation20 These differences are further highlighted by the finding that hybridomas generated using B cells isolated from lymph nodes tend to provide higher affinity monoclonal antibodies.Citation21,Citation22
In this study, we explored spleen- and lymph node-derived immunoglobulin gene diversity as a source for antibody library construction. We used a CDRH3 diversity trapping approach to generate a panel of human single-chain variable fragment (scFv) libraries using CDRH3 sequences derived from spleens and lymph nodes isolated from the same group of naïve or immunized mice so that these repertoires could be directly compared. These human scFv libraries were characterized by next-generation sequencing (NGS) and their relative performance was evaluated according to their binding properties following phage display selection.
Results
Trapping murine CDRH3 repertoires into human scFv libraries
To create various CDRH3 repertoires, mice were immunized with two different antigens, i.e., human interferon gamma (hIFNγ) or chemokine ligand 5 (C-C motif) (hCCL5). After several booster immunizations, the sera were analyzed. They demonstrated an antigen-specific IgG response and the titers were similar between each animal (data not shown). Three days after a hyperboost, the spleen (S) and lymph nodes (LN) were obtained from the immunized mice and from a control group that did not receive any immunization, i.e., the naïve group. Cells from the spleen and the lymph nodes (4 or 5 mice per group) were separately isolated and mRNA extracted (). cDNA was synthesized and the murine CDRH3 sequences were then amplified using a set of primers covering 15 murine VH families followed by primers annealing at the boundaries of the CDRH3 and containing a FokI restriction site (). After amplification and purification, the DNA fragments were digested with FokI, generating cohesive ends compatible for ligation into the human acceptor scFv library, which was previously digested with BsmBI (). The acceptor library contained 7 VH and 7 VL stable human frameworks and synthetic diversity in the CDRL3. Using the spleen and lymph nodes from the three groups of animals, six independent phagemid libraries were generated by electroporation of E. coli cells. The size of each library ranged from 7.3 × 107 to 2.5 × 108 ().
Figure 1. Capture of murine CDRH3 into a library of human scFv. (A) BalbC mice, divided in three groups, were either kept naïve or immunized with hIFNγ or hCCL5. (B) After sacrifice, the spleen and the lymph nodes were kept separated and VH repertoires were recovered by PCR. CDRH3 were then amplified from the VH pool by PCR, along with recognition sites for FokI, a type IIS restriction enzyme, were added to the inserts. CDR3 are represented in gray, CDR1 and 2 in white. (C) Murine inserts were then digested with FokI and (D) ligated to the human acceptor scFv library, itself digested with BsmBI. This second type IIS enzyme permitted the removal of a non-diversified stuffer sequence (“S”) at the location of CDRH3 and the generation of compatible cohesive ends for the incorporation of CDRH3. The acceptor library also contains synthetic diversity at the location of the CDRL3 (“L3”) and tags (“T,” a c-myc and a His tag) at the scFv C-terminal for purification purposes. Libraries featuring mouse CDRH3 were analyzed by NGS, which covered the CDRH3 and part of VH framework 3 (“NGS”). (E) The fusion to gIII allowed the expression of scFv at the surface of M13 phage for phage display selection.
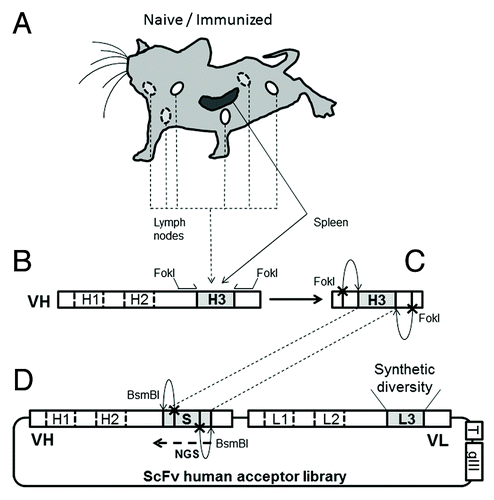
Table 1. Summary of libraries capturing murine CDRH3 and characterization by NGS
Characterization of the captured CDRH3 repertoires isolated from murine spleen and lymph nodes
The CDRH3 repertoires inserted in the six libraries were analyzed by NGS using the Illumina platform. Using a common sequencing primer annealing at the 3′ border of the CDRH3 (), the sequencing reads of 100 base pair (bp) allowed for the coverage of CDRH3 up to 32 amino acids. In most cases, enough sequence information in the framework 3 region could be obtained to determine the VH family in which the CDRH3 sequence had been inserted. For each library, between 3.5 × 106 and 4.9 × 106 sequences were obtained (data not shown). Of these, 1.0 × 106 were randomly selected from each library to homogenize the size of the data sample. Using the N2GSAb software, the number of unique CDRH3 sequences, their respective length, and the frequencies of the repeated sequences were determined (, and ).
Figure 2. Assessment of the murine nature of CDRH3 repertoires. (A) Length profile of CDRH3 represented as the percentage of unique CDRH3 in function of their length in amino acids. (B) Amino acid composition profile of unique CDRH3 of all lengths. The few stop codons found are likely due to cloning or sequencing errors. This analysis was performed by NGS.
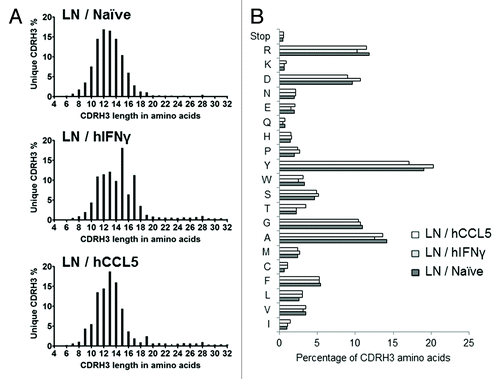
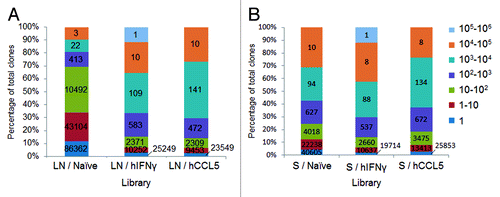
Figure 3. Evaluation of CDRH3 redundancy. Evaluation by NGS of CDRH3 repartition in the context of human scFv libraries. The percentage of total CDRH3 is represented in function of their frequency according to a color code. For each library, one million clones were analyzed. The values on the histograms are the number of unique CDRH3, i.e., the number of CDRH3 with a different amino acids sequence, corresponding to each section. (A) represents the level of redundancy of lymph nodes derived libraries while (B) represents the repartition of CDRH3 from spleens derived libraries.
The CDRH3 length distribution and amino acid (AA) composition found in the lymph nodes libraries () corresponded to the published characteristics of murine CDRH3 sequences.Citation23 The highest frequency and overall length of captured naive CDRH3 sequences (i.e., 12 AA) tended to be shorter than that reported for human CDRH3 (i.e., 14AA described for human) and contained a higher tyrosine content (17% to 20% vs. 14% for humanCitation23). These features remained present after immunization, albeit with a slight trend toward certain CDRH3 lengths (e.g., 15 and 17AA for the LN / hIFNγ library and 13 and 14 for the LN / hCCL5 library). Similar results were obtained with libraries capturing spleen derived CDRH3, as previously described.Citation14 These results demonstrate that mouse CDRH3 sequences were captured into human antibody frameworks. The distribution of sequences between the different VH families of the acceptor library was found to be equivalent (Fig. S1), indicating that no significant bias had been introduced during the cloning step.
The analysis of the diversity of CDRH3 sequences in the libraries took into account only those in the correct reading frame (). The CDRH3 sequences captured from the naïve lymph nodes were more diverse compared with the naïve spleen, i.e., unique CDRH3 sequences comprised 19% vs. 13%, respectively, of the totals (). The difference in diversity could also be observed by analyzing the frequency of CDRH3 fragments. In the LN / Naïve library, 69% of CDRH3 sequences were represented less than a 100 times while only 31% were more abundant (). In contrast in the S / Naïve library, 25% of CDRH3 were sequenced less than 100 times while 75% were highly repeated (). After immunization (), the diversity of unique CDRH3 sequences dropped from 19% to 4% in the LN libraries, and from 13% to 5% and 9% in the S libraries, indicating that the CDRH3 repertoires were biased in vivo. In line with this, CDRH3 sequences, represented more than 100 times, reached 85% and 86% for LN libraries and 86% and 82% for S libraries (i.e., immunization with hIFNγ and hCCL5, respectively) (). Furthermore, as the CDRH3 sequences are inserted in different VH frameworks and combined with different VL, the scFv sequence diversity is not limited to the CDRH3, and is actually considerably higher, as was confirmed by sequencing a panel of randomly selected clones from each library using the Sanger method (data not shown).
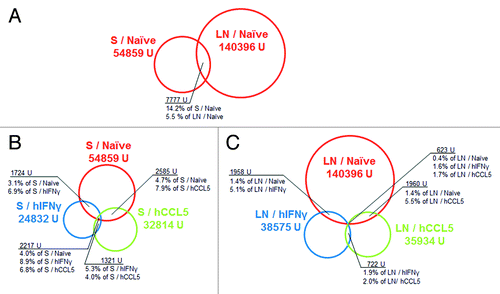
To further analyze the captured diversity, the overlap of the CDRH3 unique sequences between libraries was assessed (, Table S1). For the two naïve CDRH3 repertoires, the common sequences represented 5.5% of the LN library and 14.2% for the S library, highlighting that most of the CDRH3 sequences in these organs differed although they had been obtained from the same animals (). Following immunization, CDRH3 repertoires showed limited overlap with the naïve repertoire (, Table S1), probably resulting from the affinity maturation when generating high affinity antigen-specific antibodies. The overlap observed in the naïve and immunized spleen libraries reached 15.8% (the sum of 6.9 + 8.9%; , Table S1) with hIFNγ immunization and 14.7% (the sum of 7.9 + 6.8%: , Table S1) with hCCL5 immunization. The overlap between the LN / Naïve and each LN / immunized libraries was 6.7% (5.1 + 1.6) of the LN / hIFNγ sequences and 7.2% (5.5 + 1.7) of the LN / hCCL5 sequences (, Table S1). These results, which demonstrate a larger overlap in the S libraries, suggest that the immunization induced a stronger bias in the repertoires of lymph nodes compared with the spleen.
Figure 4. Overlap between CDRH3 repertoires. Evaluation of the unique CDRH3 sequences in common between libraries. The analysis included one million clones for each library (NGS). Diversity of unique CDRH3 (i.e., all CDRH3 with a different amino acids sequence) is represented by circles which size is proportional to the size of the sample. Naïve libraries are represented in red, libraries biased against hIFNγ in blue and libraries biased against hCCL5 in green. For each library, the number of unique CDRH3 is described with the same color code, U refers to “unique sequences.” Unique sequences in common between libraries are symbolized by the overlap between circles. Numbers in black represent the number of unique sequences corresponding to each section determined by a black line. (A) represents the overlap between both naïve libraries, (B) represents the overlap between the spleen derived libraries, and (C) represents the overlap between the lymph nodes derived libraries.
Overall, the NGS characterization revealed that the CDRH3 repertoire captured from naïve lymph nodes was the most diverse and that the repertoires isolated from immunized animals had been biased in vivo. This bias was particularly apparent for the repertoires derived from lymph nodes, while only a trend toward a higher redundancy was observed for the spleen repertoires (). Notably, the CDRH3 repertoire from naïve spleens had a profile closer to immune repertoires ().
Relative performance of libraries incorporating CDRH3 repertoires from spleen and lymph nodes
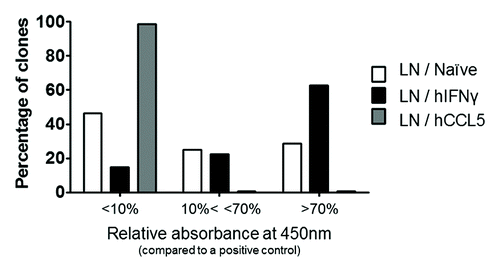
After characterizing the CDRH3 repertoires, the libraries were used in phage display selection against hIFNγ to assess their performance. This was evaluated by their ability to generate specific and potent binders against hIFNγ. Three rounds of selection were performed to enrich scFv repertoires for clones specific for hIFNγ. For each LN library, 528 random clones from the output of selection were tested as soluble scFv by ELISA for specific binding to hIFNγ. The results () showed an increased frequency of scFv binding to hIFNγ for the LN / hIFNγ library (85%) compared with the LN / Naïve library (54%), while the LN / hCCL5 library was found to perform very poorly against hIFNγ (1% of hits). In addition, the frequency of scFv producing an absorbance of > 70% of the positive control was higher in the candidates obtained from the LN / hIFNγ library (63%) compared with those from the LN / Naïve library (29%) (). The same relative performance of the libraries was observed when testing the individual clones by ELISA using scFv expressed at the surface of M13 bacteriophage, which generally leads to an increased signal due to avidity and signal amplification (Fig. S2). Taken together, these results indicate that immune and naïve features of the lymph node derived repertoires were transferred to the libraries via capture of the CDRH3. The same evaluation has been performed with the S libraries and similar results were obtained.
Figure 5. Screening of selection outputs from lymph nodes libraries in the scFv format. For each library, 528 random clones from the selection round 3 were independently cultured and tested by scFv ELISA against hIFNγ. Revelation was performed via HRP and read at 450nm. The frequency of clones is provided according to their level of absorbance at 450nm. Clones were classified according to their level of absorbance relative to a positive control, i.e., displaying absorbance values above 70% of the signal of a positive control scFv (absorbance ~1.6), between 10% and 70% (absorbance 0.2 and 1.6), and below 10%.
To assess the diversity of binders, a selection of scFv specific to hIFNγ was sequenced from each library by the Sanger method, allowing for complete coverage of the VH and VL sequences. With the exception of LN / hCCL5, > 50 clones were analyzed for each library. Only 5 clones of LN / hCCL5 were sequenced because of the very low hit rate obtained with this library. A larger diversity of scFv was obtained from the LN / Naïve library compared with the immunized LN libraries (), and these candidates were different from those retrieved from the spleen derived libraries following the same procedure.Citation14
Table 2. Frequency and potency of binders derived from lymph nodes diversity
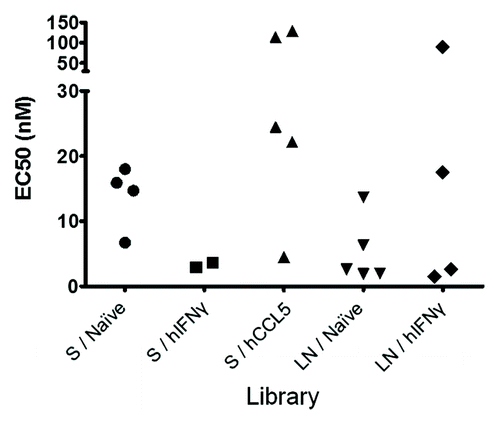
The candidates identified by screening and defined by sequencing were produced, purified as soluble scFv and quantified. They were then tested at the same concentrations in a dose-response ELISA for binding to hIFNγ to evaluate their apparent affinity (EC50) for the target hIFNγ (). The frequency of the CDRH3 sequences corresponding to each candidate was also retrieved by NGS before selection and after the third round of selection (). The ranking before and after selection are different, in particular for the immune libraries. This indicates that the most frequent sequences that were integrated in the unselected libraries did not necessarily lead to the generation of a functional and antigen specific binding site when inserted in the novel context of the human antibody frameworks used in this study. The EC50 for clones representing more than 1% of the total sequences after the selection round three are presented in . The EC50 values indicate that potent (i.e., < 10 nM) binders of hIFNγ were isolated from the S / hIFNγ and the LN / hIFNγ libraries. In contrast, libraries biased against another target due to the immunization with hCCL5, mainly provided low affinity binders of hIFNγ (20‒150 nM). These data indicate that the immune features of murine antibody repertoires were transferred to the human acceptor library. The LN / Naïve library, which initially contained the most diverse CDRH3 repertoire, also allowed for the identification of the largest panel of potent (i.e., <10 nM) candidates (, ).
Figure 6. Relative performance of most amplified binders. Representation of all the scFv defined as binders to hIFNγ from each library and with a frequency defined by deep sequencing above 1% at the selection round 3. Clones are grouped according to their library of origin and represented by their EC50 in nM (defined by ELISA). None of the binders found from the library LN / hCCL5 having a frequency above 1%, the library is not represented in this graph.
Specificity of scFv isolated from different repertoires
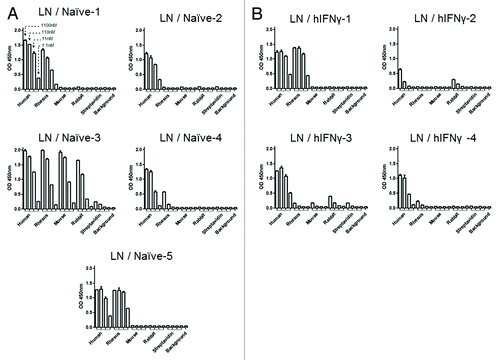
To assess whether scFv isolated from naïve and immunized CDRH3 repertoires targeted similar epitopes, we compared their binding capacity against IFNγ from human, rhesus, mouse and rabbit using purified scFv in a dose-response ELISA. Only scFv with a frequency over 1% at the selection round three (as defined by NGS) were tested. Cross-reactivity profiles from the LN- and the S-derived libraries are presented in and in Figure S3, respectively. The results indicate that several patterns of cross-reactivity with different species-derived IFNγ could be identified. ScFv derived from the LN / Naïve library covered three patterns, i.e., LN / Naïve-2 (clone ID) was not cross-reactive, LN / Naïve-1 4 and 5 were cross-reactive with rhesus only and LN / Naïve-3 was cross-reactive with rhesus, mouse and rabbit (). ScFv capturing LN / hIFNγ CDRH3 displayed two cross-reactivity patterns, i.e., LN / hIFNγ-1 and 4 were cross-reactive with rhesus only and LN / hIFNγ-2 and 3 were cross-reactive with rabbit only (). For the spleen derived libraries, the S / Naïve and the S / hIFNγ scFv presented three and two cross-reactivity patterns, respectively, while the library biased for hCCL5 produced scFv all specific to hIFNγ only. These data indicate that the clones tested bound to different epitopes on hIFNγ, some being conserved between species and others not. The scFv were also tested against streptavidin (Figs. Seven and S3) and other control targets to ensure their specificity (data not shown). These results demonstrate that the diversity of scFv was translated by a diversity of epitopes covered for the naïve and the hIFNγ biased libraries, but not for the S / hCCL5 library biased against another target. Notably, some cross-reactivity patterns were only found in the naïve or the hIFNγ biased libraries, which suggests that these libraries could be complementary for covering a maximum of hIFNγ epitopes.
Figure 7. Evaluation of the cross specificity of clones. A selection of clones from each LN library was tested for their cross reactivity on IFNγ. Purified scFv were tested at four concentrations (1100, 110, 11 and 1.1 nM, n = 2) for their specificity to a panel of IFNγ from different species, i.e., human, rhesus monkey, mouse and rabbit, and streptavidin as a negative control. Revelation was performed via HRP and read at 450 nm. All clones above 1% frequency at the selection round 3 were tested.
Discussion
Our aim was to investigate novel diversification strategies when building human immunoglobulin libraries by exploiting CDRH3 combinatorial designs. To achieve this, we explored natural repertoires extracted from different organ types to define the most appropriate source for the generation of such libraries. Secondary lymphoid tissues contain a high percentage of B cells and, thus represent good sources of naturally rearranged immunoglobulin genes. As the spleen and the lymph nodes drain different body fluids, i.e., blood and lymph, respectively, B cells present in these organs are preferentially exposed to different types of pathogens,Citation18,Citation19 potentially biasing the repertoire. The spleen is specialized in clearing bacteria from the blood and driving the humoral response against bacteria capsular polysaccharides. Indeed, splenectomy results in increased susceptibility to infection by this class of pathogens.Citation24-Citation26 In addition, the spleen and the lymph nodes are characterized by different anatomical structures that may further affect the B cell repertoires. Thus, we compared the performance of human antibody libraries featuring CDRH3 repertoires isolated from murine spleen and lymph nodes with, and prior to, immunization.
Qualitative analysis by NGS of the generated libraries highlighted major differences between the CDRH3 repertoires. The CDRH3 sequence diversity recovered from naïve lymph nodes was superior compared with naïve spleen, i.e., more unique sequences were obtained and less redundant motifs. Furthermore, when used in selection, the performance of the naïve lymph node library correlated with its higher diversity, leading to the identification of diverse and potent binders compared with the other libraries. Thus, the lymph nodes from naïve animals appear to represent a better source of sequences to build a general-purpose library, in principle suitable for the identification of antibodies against any target.
We also observed a polarization of the CDRH3 repertoire isolated from spleen and lymph nodes from immunized animals, with an increase of highly repeated sequences and a decrease in single and low copy sequences. These differences between naïve and immune repertoires were evident in the case of lymph nodes, but the naïve spleen repertoire appeared already relatively polarized. This result may be due to the fact that, although mice were kept in specific pathogen-free conditions, the animals are still exposed to the same environment and maintain an intestinal bacteria flora, thereby allowing the repertoire of the spleen to be biased. Taken together, anatomical and functional differences between spleen and lymph nodes appear to shape the B cell and immunoglobulin repertoires, which in this study were recovered independently as a source of CDRH3 sequences to build human antibody libraries.
The relative performance of the lymph node-derived libraries during selections against hIFNγ further confirms our previous findings that the immune characteristics of the immunoglobulin repertoire from an immunized animal can be transferred via CDRH3 sequences to an antibody library.Citation14 Indeed, screening of the output of selection against hIFNγ using the S / hIFNγ and LN / hIFNγ libraries allowed the identification of binders at higher frequency compared with the other libraries. In contrast, the CDRH3 repertoire that was biased by immunization with hCCL5, led to the very poor performance of the S / hCCL5 and LN / hCCL5 libraries for selections against hIFNγ.
The six libraries tested in this study yielded antibodies with different sequences and different binding profiles against IFNγ from various species, indicating that they interact with these proteins differently. In addition, the scFv isolated from each library were unique and were not found in another library. In many instances, and in particular for therapeutic and diagnostic applications, obtaining a large panel of diverse antibodies is important for identification of a candidate with the desired characteristics. Thus, the CDRH3 capturing approach described here represents a way to probe and exploit different repertoires that can be derived from natural sources to widen the spectrum of human antibody candidates.
Overall, our results indicate that the lymph nodes of naïve mice provided the most diverse and best source of CDRH3 sequences to generate general purpose, or so-called universal, libraries. Libraries based on CDRH3 sequences biased against an antigen isolated from spleen or lymph nodes performed equally, facilitating the identification of candidates by increasing the hit rate and yielding additional candidates that could not be retrieved from the naïve libraries.
Materials and Methods
Construction of the acceptor library
A library of phagemid vectors was generated encoding human scFv sequences. This library included 7 human VH and 7 human VL framework sequences of which CDR1 and 2 were kept germline while synthetic diversity was added at the level of the CDRL3. At the location of the CDRH3, a non-diversified “stuffer” sequence that could be digested off via type IIS restriction enzyme, BsmBI, was added, thereby allowing the integration of diversity in only one cloning step. This phagemid library was electroporated into TG1 E. coli cells, generating a diversity of 2.2 x109 transformants. The library with the ability to accept CDRH3 sequences after BsmBI digestion was referred as the acceptor library.
Immunization
As described earlier,Citation14 BalbC mice were either immunized or kept naïve to allow comparison of murine CDRH3 repertoires, focused or not, in the context of human scFv sequences. Five mice were kept naïve, and 2 groups of 4 mice each were immunized with hIFNγ or hCCL5 administered in RIBI via intraperitoneal (IP) and subcutaneous (SC) injection (mice were immunized both IP and IV along each boost); both targets were produced at NovImmune SA. After sacrifice, the spleen and lymph nodes (popliteal, inguinal, brachio, axillary, cervical) were recovered and kept separated for RNA extraction.
Extraction and capture of murine CDRH3
Cells from the spleens and the lymph nodes were harvested on the day of sacrifice by cutting organs into pieces, in the presence of DNase (DN25 Sigma) and collagenase (Type 4 from Invitrogen), and kept at -80°C in RNAlater (Ambion). Spleens were also treated with ACK (NH4Cl 0.15M, KHCO3 1M, Na2EDTA 0.1mM) to remove red blood cells. An mRNA extraction (RNAqueous kit, Ambion) was then performed on the cell preparations, followed by reverse transcription (Ready-to-Go You Prime beads, GE Healthcare) and nested PCR for the recovery of the CDRH3 sequences. In order to avoid cross contamination between the repertoires, mRNA isolation was performed on different days for the different groups of animals. In addition, rigorous cleaning of the equipment was performed between extractions and only disposable precast gels (Invitrogen) were used for DNA purification. During the last amplification, recognition sites for FokI type IIS restriction enzymes were also added using biotinylated primers. The acceptor library and the CDRH3 inserts were digested with BsmBI and FokI (both from New England Biolabs), respectively, which generated compatible cohesive ends, and purified with Chroma Spin T1000 columns (Clontech) and Dynabeads (M280 Streptavidin from Invitrogen), respectively. CDRH3 were then ligated into the acceptor library pool with Rapid DNA Ligation Kit (Roche) and electroporated into TG1 E. coli cells. Each library reached around 108 transformants ().
Sequencing
A selection of clones was sequenced with the Sanger method (service provided by Fasteris SA). TG1 colonies were separately cultured overnight at 37°C in 2mL of LB AG and vectors were purified with the Qiaprep Spin Miniprep kit (Qiagen).
Libraries and selection rounds 3 were also analyzed by NGS via the HiSeq Illumina platform (service provided by Fasteris SA) and 87% to 92% of the bases had a Phred score ≥ 30. Sequencing primers, designed in VH J region, could cover CDRH3 and part of VH framework 3, allowing identification of families using DNA signatures. Analysis of large data sets was performed with the N2GSAb software developed at NovImmune.Citation27 For each library, the analysis was performed from a pool of one million random clones to ensure samples were of comparable size.
Selection against hIFNγ
Phage particles displaying scFv fragments were produced and purified. Libraries electroporated into TG1 cells were cultured at 37°C and 240 rpm in 2xTY AG until OD600 0.5. Cells were then infected with M13K07 helper phage (MOI of 10) 1 h at 37°C and 100 rpm. The medium was replaced by 2xTY AK to select cells that incorporated a helper phage vector. Cells were incubated overnight at 30°C with shaking at 280 rpm for robust production of phage. Viral particles were purified from supernatants by two consecutive PEG precipitations (using 1/3 v/v of 20% PEG-8000/2.5 M NaCl). Purified phage were used to infect fresh TG1 cells, which were then titrated. Phage preparations were thus evaluated from 5 × 1013 to 1 × 1014 pfu/mL.
Phage from each library were used in three rounds of phage display selection against the target hIFNγ (biotinylated, produced at NovImmune SA). For each library, 1010 viral particles were blocked in skimmed milk 3% (w/v) and deselected twice via incubation for 1 h on Dynabeads (M-280 Invitrogen) coated with streptavidin. Phage remaining in the supernatant were panned on Dynabeads precoated with biotinylated hIFNγ (30 min at a concentration of 100 nM). The mix was incubated for 2 h at room temperature so that phage could specifically bind to hIFNγ. Beads were washed five times with PBS/0.1% Tween 20 (Sigma) and twice with PBS to remove non-specific phage. Phage remaining bound to the beads surface were eluted with 10 mM triethylamine TEA (Sigma) for 20 min and the solution was neutralized 10 min with 1 M TRIS-HCl pH 7.4 (Sigma). The mix was used to infect 10 mL of fresh TG1 cells at OD600 0.5 for 1 h at 37°C and 90 rpm. Infected cells were spread on 2xTY AG bioassay plates and incubated at 30°C overnight. Colonies were scraped off the plate surface and kept at -80°C after addition of glycerol (Sigma, 17% final). For the following rounds of selection, 20 μL of the output of the preceding round was cultured at 37°C and 240 rpm in 20 mL 2xTY AG 4%. When the culture reached an OD600 0.5, cells were infected with helper phage (MOI of 10) for 1 h at 37°C and 90 rpm, the medium was changed to 2xTY AK and the cells incubated overnight at 30°C and 280 rpm. Phage particles excreted in the supernatant were then used as the input of the next selection round.
ELISA screening
Individual TG1 clones from the output of the third selection rounds were cultured separately in 96-wells plates, in 2xTY AG overnight at 30°C and 150rpm. These master plates were supplemented with glycerol to 17% for storage at -80°C and used as a source for the various screenings. Clones were tested in ELISA for their specificity for hIFNγ after being produced as soluble scFv or as scFv displayed on phage.
Screening ELISA was first performed in the scFv format (6 plates tested per library, i.e., 528 individual clones). Clones were cultured in 2xTY AG for 6 h at 37°C, followed by IPTG addition (1 mM final) and incubated overnight at 30°C and 150 rpm for scFv production. A positive control clone encoding a scFv with high potency against hIFNγ was also included in each plate. Streptavidin plates (Greiner Bio-one) were coated with 50 μL of biotinylated hIFNγ (1 μg/mL) for 1 h at 2 μg/mL. After washing, 50 μL of cell culture supernatants were added to plates that were incubated for 2 h at room temperature. After washing, remaining scFv were revealed via a mouse anti-c-myc (produced at NovImmune SA) and then a goat anti-mouse Fcγ HRP (Jackson). Then, TMB (Sigma) was added, followed by H2SO4 (2N) for blocking. In the final step, absorbance was read at 450 nm (Synergy HT, BioTeK).
The screening ELISA was also performed using phage particles displaying the scFv at their surface (1 plate tested per library, i.e., 88 individual clones). Clones were cultured in 2xTY AG for 6 h at 37°C and then infected with the helper phage (MOI of 10) for 1 h. The medium was replaced by 2xTY AK. Plates were incubated overnight at 30°C and 150 rpm for phage production. The ELISA procedure was similar to the one described for scFv, except that the first detection antibody was a mouse anti-M13 P8 (Abcam).
Dose-response ELISA using purified scFv
A selection of clones were produced as soluble scFv and purified via their His tag with Ni-NTA agarose chromatography (Qiagen). Purified scFv were quantified using a Nanodrop ND 1000 spectrophotometer (Witec AG). Serial dilutions of scFv were applied in duplicates to streptavidin plates (Grener Bio-one) coated with biotinylated hIFNγ (coating with 50 µL of hIFNγ at 1 µg/mL) for 2 h at room temperature (as described for the screening ELISA). After washing, specific scFv were detected using a mouse anti-c-myc antibody and a goat anti-mouse Fcγ HRP antibody, followed by the addition of TMB and H2SO4 (2 N). Absorbance was then read at 450 nm (Synergy HT from Bio TeK).
Cross-specific ELISA
A selection of clones, including those most amplified from each library, were tested in dose-response ELISA against a panel of IFNγ from different species (i.e., human, rhesus, mouse and rabbit) in duplicates. Targets (produced at NovImmune SA, biotinylated and with a His tag) were coated for 30 min at 1 µg/mL in 50 µL of PBS BSA 1% on microplates precoated with streptavidin (Greiner Bio-one). Purified scFv were tested at 1100 nM, 110 nM, 11 nM and 1.1 nM in 50 µL PBS BSA 1%. Plates were revealed using a mouse anti-c-myc and goat anti-mouse Fcγ HRP as described in the dose-response ELISA section.
| Abbreviations: | ||
| AA | = | amino acid |
| BCR | = | B cell receptor |
| CDR | = | complementary determining region |
| CDRH3 | = | CDR3 of the heavy chain of an antibody |
| ELISA | = | enzyme-linked immunosorbent assay |
| hCCL5 | = | chemokine ligand 5 (C-C motif) |
| hIFNγ | = | human interferon gamma |
| IFNγ | = | interferon gamma |
| IP | = | intraperitoneal |
| LN | = | lymph node |
| NGS | = | next generation sequencing |
| S | = | spleen |
| SC | = | subcutaneous |
Additional material
Download Zip (207.2 KB)Acknowledgments
The authors wish to thank Ulla Ravn and Gérard Didelot for expert assistance for next generation sequencing analysis and Vanessa Buatois for help with animal immunization.
Submitted
05/23/2013
Revised
06/28/2013
Accepted
06/28/2013
Disclosure of Potential Conflicts of Interest
No potential conflict of interest was disclosed.
References
- McCafferty J, Griffiths AD, Winter G, Chiswell DJ. Phage antibodies: filamentous phage displaying antibody variable domains. Nature 1990; 348:552 - 4; http://dx.doi.org/10.1038/348552a0; PMID: 2247164
- Mazor Y, Van Blarcom T, Mabry R, Iverson BL, Georgiou G. Isolation of engineered, full-length antibodies from libraries expressed in Escherichia coli. Nat Biotechnol 2007; 25:563 - 5; http://dx.doi.org/10.1038/nbt1296; PMID: 17435747
- Feldhaus MJ, Siegel RW, Opresko LK, Coleman JR, Feldhaus JMW, Yeung YA, et al. Flow-cytometric isolation of human antibodies from a nonimmune Saccharomyces cerevisiae surface display library. Nat Biotechnol 2003; 21:163 - 70; http://dx.doi.org/10.1038/nbt785; PMID: 12536217
- Hanes J, Plückthun A. In vitro selection and evolution of functional proteins by using ribosome display. Proc Natl Acad Sci U S A 1997; 94:4937 - 42; http://dx.doi.org/10.1073/pnas.94.10.4937; PMID: 9144168
- Thie H, Meyer T, Schirrmann T, Hust M, Dübel S. Phage display derived therapeutic antibodies. Curr Pharm Biotechnol 2008; 9:439 - 46; http://dx.doi.org/10.2174/138920108786786349; PMID: 19075684
- Marks JD, Hoogenboom HR, Bonnert TP, McCafferty J, Griffiths AD, Winter G. By-passing immunization. Human antibodies from V-gene libraries displayed on phage. J Mol Biol 1991; 222:581 - 97; http://dx.doi.org/10.1016/0022-2836(91)90498-U; PMID: 1748994
- Sheets MD, Amersdorfer P, Finnern R, Sargent P, Lindquist E, Schier R, et al. Efficient construction of a large nonimmune phage antibody library: the production of high-affinity human single-chain antibodies to protein antigens. Proc Natl Acad Sci U S A 1998; 95:6157 - 62; http://dx.doi.org/10.1073/pnas.95.11.6157; PMID: 9600934
- Vaughan TJ, Williams AJ, Pritchard K, Osbourn JK, Pope AR, Earnshaw JC, et al. Human antibodies with sub-nanomolar affinities isolated from a large non-immunized phage display library. Nat Biotechnol 1996; 14:309 - 14; http://dx.doi.org/10.1038/nbt0396-309; PMID: 9630891
- Griffiths AD, Williams SC, Hartley O, Tomlinson IM, Waterhouse P, Crosby WL, et al. Isolation of high affinity human antibodies directly from large synthetic repertoires. EMBO J 1994; 13:3245 - 60; PMID: 8045255
- Hoogenboom HR, Winter G. By-passing immunisation. Human antibodies from synthetic repertoires of germline VH gene segments rearranged in vitro. J Mol Biol 1992; 227:381 - 8; http://dx.doi.org/10.1016/0022-2836(92)90894-P; PMID: 1404359
- Amersdorfer P, Wong C, Chen S, Smith T, Deshpande S, Sheridan R, et al. Molecular characterization of murine humoral immune response to botulinum neurotoxin type A binding domain as assessed by using phage antibody libraries. Infect Immun 1997; 65:3743 - 52; PMID: 9284147
- Burton DR, Barbas CF 3rd, Persson MA, Koenig S, Chanock RM, Lerner RA. A large array of human monoclonal antibodies to type 1 human immunodeficiency virus from combinatorial libraries of asymptomatic seropositive individuals. Proc Natl Acad Sci U S A 1991; 88:10134 - 7; http://dx.doi.org/10.1073/pnas.88.22.10134; PMID: 1719545
- Pelat T, Thullier P. Non-human primate immune libraries combined with germline humanization: an (almost) new, and powerful approach for the isolation of therapeutic antibodies. MAbs 2009; 1:377 - 81; http://dx.doi.org/10.4161/mabs.1.4.8635; PMID: 20068407
- Venet S, Ravn U, Buatois V, Gueneau F, Calloud S, Kosco-Vilbois M, et al. Transferring the characteristics of naturally occurring and biased antibody repertoires to human antibody libraries by trapping CDRH3 sequences. PLoS One 2012; 7:e43471 - 43471; http://dx.doi.org/10.1371/journal.pone.0043471; PMID: 22937053
- Mebius RE, Kraal G. Structure and function of the spleen. Nat Rev Immunol 2005; 5:606 - 16; http://dx.doi.org/10.1038/nri1669; PMID: 16056254
- Mak TW and Saunders ME. The immune response, basic and clinical principles. Waltham (MA): Elsevier Academic Press; 2006.
- Köhler G, Milstein C. Continuous cultures of fused cells secreting antibody of predefined specificity. Nature 1975; 256:495 - 7; http://dx.doi.org/10.1038/256495a0; PMID: 1172191
- Gretz JE, Norbury CC, Anderson AO, Proudfoot AE, Shaw S. Lymph-borne chemokines and other low molecular weight molecules reach high endothelial venules via specialized conduits while a functional barrier limits access to the lymphocyte microenvironments in lymph node cortex. J Exp Med 2000; 192:1425 - 40; http://dx.doi.org/10.1084/jem.192.10.1425; PMID: 11085745
- Nolte MA, Beliën JA, Schadee-Eestermans I, Jansen W, Unger WW, van Rooijen N, et al. A conduit system distributes chemokines and small blood-borne molecules through the splenic white pulp. J Exp Med 2003; 198:505 - 12; http://dx.doi.org/10.1084/jem.20021801; PMID: 12900524
- Carey JB, Moffatt-Blue CS, Watson LC, Gavin AL, Feeney AJ. Repertoire-based selection into the marginal zone compartment during B cell development. J Exp Med 2008; 205:2043 - 52; http://dx.doi.org/10.1084/jem.20080559; PMID: 18710933
- Başalp A, Yücel F. Development of mouse hybridomas by fusion of myeloma cells with lymphocytes derived from spleen, lymph node, and bone marrow. Hybrid Hybridomics 2003; 22:329 - 31; http://dx.doi.org/10.1089/153685903322538863; PMID: 14678651
- Mirza IH, Wilkin TJ, Cantarini M, Moore K. A comparison of spleen and lymph node cells as fusion partners for the raising of monoclonal antibodies after different routes of immunisation. J Immunol Methods 1987; 105:235 - 43; http://dx.doi.org/10.1016/0022-1759(87)90271-7; PMID: 3320207
- Zemlin M, Klinger M, Link J, Zemlin C, Bauer K, Engler JA, et al. Expressed murine and human CDR-H3 intervals of equal length exhibit distinct repertoires that differ in their amino acid composition and predicted range of structures. J Mol Biol 2003; 334:733 - 49; http://dx.doi.org/10.1016/j.jmb.2003.10.007; PMID: 14636599
- Amlot PL, Hayes AE. Impaired human antibody response to the thymus-independent antigen, DNP-Ficoll, after splenectomy. Implications for post-splenectomy infections. Lancet 1985; 1:1008 - 11; http://dx.doi.org/10.1016/S0140-6736(85)91613-7; PMID: 2859463
- Kraal G. Cells in the marginal zone of the spleen. Int Rev Cytol 1992; 132:31 - 74; http://dx.doi.org/10.1016/S0074-7696(08)62453-5; PMID: 1555921
- Likhite VV. Immunological impairment and susceptibility to infection after splenectomy. JAMA 1976; 236:1376 - 7; http://dx.doi.org/10.1001/jama.1976.03270130038024; PMID: 989094
- Ravn U, Didelot G, Venet S, Ng KT, Gueneau F, Rousseau F, et al. Deep sequencing of phage display libraries to support antibody discovery. Methods 2013; 60:99 - 110; http://dx.doi.org/10.1016/j.ymeth.2013.03.001; PMID: 23500657