Abstract
The critical role played by IgE in allergic asthma is well-documented and clinically precedented, but some patients in whom IgE neutralization may still offer clinical benefit are excluded from treatment with the existing anti-IgE therapy, omalizumab, due to high total IgE levels or body mass. In this study, we sought to generate a novel high affinity anti-IgE antibody (MEDI4212) with potential to treat a broad severe asthma patient population. Analysis of body mass, total and allergen-specific IgE levels in a cohort of severe asthmatics was used to support the rationale for development of a high affinity IgE-targeted antibody therapeutic. Phage display technology was used to generate a human IgG1 lead antibody, MEDI4212, which was characterized in vitro using binding, signaling and functional assay systems. Protein crystallography was used to determine the details of the interaction between MEDI4212 and IgE. MEDI4212 bound human IgE with an affinity of 1.95 pM and was shown to target critical residues in the IgE Cε3 domain critical for interaction with FcεRI. MEDI4212 potently inhibited responses through FcεRI and also prevented the binding of IgE to CD23. When used ex vivo at identical concentration, MEDI4212 depleted free-IgE from human sera to levels ~1 log lower than omalizumab. Our results thus indicate that MEDI4212 is a novel, high affinity antibody that binds specifically to IgE and prevents IgE binding to its receptors. MEDI4212 effectively depleted free-IgE from human sera ex vivo to a level (1 IU/mL) anticipated to provide optimal IgE suppression in severe asthma patients.
Introduction
Asthma is a chronic disorder characterized by airway inflammation, airways hyperresponsiveness and variable, reversible airway obstruction. Most patients have mild-to-moderate disease usually controlled by regular use of combined inhaled corticosteroids (ICS) and long-acting beta2 agonists (LABA) supplemented with short-acting beta2 agonists for symptomatic relief. However, asthma continues to be poorly controlled in a small subset of patients (~5%), who exhibit persistent symptoms, airflow obstruction or frequent exacerbations despite aggressive treatment including oral corticosteroids.Citation1 This has considerable effects on quality of life, disproportionate use of healthcare resources and adverse effects from regular systemic steroid use. Therefore, there is a substantial medical need for improved treatments in the poorly-controlled asthma patient population, including new biological agents.
The critical role played by IgE in type I hypersensitivity (allergic) responses is well documentedCitation2 and beneficial effects of targeting the IgE pathway in asthma are clinically validated.Citation3 Upon release from B lymphocytes IgE binds, via its Fc domain, to the high-affinity IgE receptor (FcεRI) present on mast cells and basophils. Cross-linking of receptor-bound IgE by allergen triggers cell activation and degranulation, resulting in release of histamine and other mediators of the allergic response. The role of the low affinity receptor, CD23, is complex, but consequences of CD23 interaction with IgE include regulation of IgE synthesis, allergen presentation,Citation4 allergen transportCitation5,Citation6 and cell-mediated effector functions.Citation7
Positive clinical experience with the IgE-specific antibody omalizumab (Xolair®, Genentech) in treatment of uncontrolled allergic asthma demonstrates the utility of targeting IgE. Omalizumab rapidly lowers serum free-IgE, preventing IgE binding to FcεRI and CD23. Omalizumab is indicated for adults and adolescents (≥ 12 y) with moderate-to-severe persistent asthma (US) or severe, persistent allergic asthma (EU) with positive skin test or in vitro reactivity to a perennial aeroallergen and symptoms inadequately controlled with ICS/LABA. Treatment of patients with omalizumab delivers both reduced exacerbations and reduced steroid requirements.Citation8-Citation10 In the EU, omalizumab is also approved for treating pediatric patients (6–12 y) with severe, persistent allergic asthma. In all patients, use of omalizumab is restricted by a complex dosing table incorporating baseline IgE and body mass.Citation11 In addition, the reported non-responder rate in eligible patients is 39%.Citation12 Thus, there remains need for optimized anti-IgE treatments that may overcome these disadvantages.
Here, we provide analysis of total and allergen-specific IgE levels in a severe asthma population to support rationale for a high affinity anti-IgE. We define the proportion of patients ineligible for omalizumab treatment, as well as possible causes of the sizable omalizumab-non-responding population. We also describe the generation and preclinical development of a novel high affinity anti-IgE antibody, MEDI4212, which has the potential to overcome these challenges and provide options for treatment of individuals with high body mass or high total/specific IgE.
Results
Analysis of omalizumab eligibility in severe asthma patients
To define the need for alternative anti-IgE therapies, investigation was performed in a cohort (n = 422) of severe asthmatics, i.e., the target patient population for a therapeutic anti-IgE molecule (), of which 67% (n = 283) were determined as atopic (Table S1). After taking into account additional exclusions based on IgE/bodyweight (US dosing table; Table S2) and non-response after 16 wk,Citation12 only 39% (26% of the entire severe asthmatic population) were potentially eligible to benefit from long-term omalizumab treatment. Thus, in the severe atopic asthmatic population, there appears to be substantial unmet need for therapies that can target a broader IgE/bodyweight range and improve efficacy.
Figure 1. Hypothetical successful treatment with omalizumab in a severe asthma cohort. 434 severe asthma patients were stratified for hypothetical eligibility for omalizumab treatments based on atopic status [at least one specific allergen result ≥ 0.35 kU/L from ALT. ALTERNATA, CAT DANDER, COCKROACH,, D. FARINAE, D. PTERONYSSINUS, DOG DANDER and additional panel of allergens dependent on country (Table S1)], IgE/bodyweight eligibility (based on US dosing table; Table S2) and responder status after 16 wk by physician’s global assessment (based on responder rate of 61% in Bousquet et al. 2007Citation12)
![Figure 1. Hypothetical successful treatment with omalizumab in a severe asthma cohort. 434 severe asthma patients were stratified for hypothetical eligibility for omalizumab treatments based on atopic status [at least one specific allergen result ≥ 0.35 kU/L from ALT. ALTERNATA, CAT DANDER, COCKROACH,, D. FARINAE, D. PTERONYSSINUS, DOG DANDER and additional panel of allergens dependent on country (Table S1)], IgE/bodyweight eligibility (based on US dosing table; Table S2) and responder status after 16 wk by physician’s global assessment (based on responder rate of 61% in Bousquet et al. 2007Citation12)](/cms/asset/c45c62e8-00ce-40f6-ba2e-488d05a5e51a/kmab_a_10928394_f0001.gif)
Antibody generation
Human IgE presented to a large phage library displaying human single-chain variable fragment (scFv)Citation13,Citation14 resulted in isolation of ENG085, an antibody specific for IgE that did not bind IgA, IgM, IgD or IgG. ENG085 demonstrated concentration-dependent inhibition of IgE binding to FcεRI (). The potency of this scFv in neutralizing human IgE bioactivity mediated through FcεRI was barely detectable, but, upon conversion to a human IgG molecule, a mean IC50 of 91 nM (n = 3) in an FcεRI-dependent calcium-signaling assay () was achieved. To improve affinity and generate a potential drug candidate, targeted mutagenesis of VH and VL CDR3 loops of the scFv was performed. This led to identification of a VH CDR3 able to pair with a variety of VL CDR3 sequences and achieve potent neutralization of FcεRI-mediated calcium signaling. The parent scFv clone, ENG085, was aligned to known human V gene germline sequences, with VH1-f_(DP-3) and Vλ1-e_(DPL8) being the closest match. Of 12 non-VernierCitation15 amino acid differences, 7 were reverted back to germline without detrimental effect, while 5 that could not be reverted without significant loss of activity were left unchanged. The CDR3 loops were replaced with the affinity optimized VH and VL CDR3s to generate the final lead human IgG1, MEDI4212.
Figure 2. In vitro characterization of MEDI4212. Neutralization of human IgE was assessed in FcεRI-dependent (A, B and C) or CD23-dependent (D and E) assays. (A) Human IgE-FcεRI binding assay; (B) RBL-ER51 calcium signaling; (C) LAD2 β-hexosaminidase release; (D) IM9 binding; (E) U937 phagocytosis of IGROV1 cells. Folate receptor α (FRα). Data points represent average of duplicate determinations from a representative experiment (A) or mean ± SEM of combined data from 3–11 experiments (B–E). All test compounds are IgG unless stated otherwise.
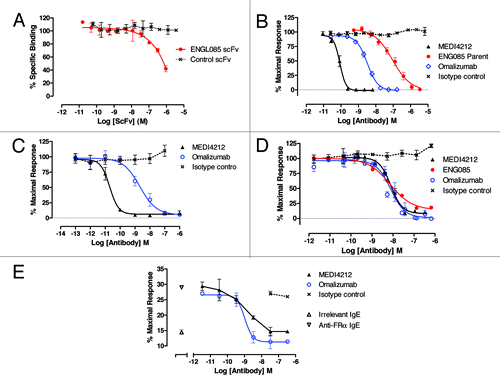
MEDI4212 inhibits IgE-mediated responses through FcεRI and CD23
MEDI4212 was characterized in a panel of in vitro assays reflecting the proposed mechanism of action. Omalizumab, an anti-IgE antibody approved for clinical use, was included as a comparator.
First, binding studies indicated an overall KD value for the MEDI4212-human IgE interaction of 1.95 pM, indicating 106-fold higher affinity than omalizumab ().
Table 1. Equilibrium dissociation constant (KD) measured by KinExA
Second, the ability of anti-IgE antibodies to neutralize human IgE bioactivity mediated through FcεRI was assessed. MEDI4212 inhibited human IgE-mediated calcium signaling in RBL-ER51 cells () and LAD2 degranulation () with IC50s of 84 pM and 20 pM, respectively, indicating ~1000-fold improved potency over parent antibody ENG085 and 30- to 100-fold higher potency than omalizumab ().
Table 2. MEDI4212 Inhibits IgE-mediated functional responses
Finally, the ability of MEDI4212 to inhibit IgE interaction with CD23 was also investigated. In a flow cytometry-based cell binding assay using IM9 B-lymphoblast cells expressing CD23 but not FcεRI on the cell surface, MEDI4212 inhibited IgE binding (IC50 8 nM; ). To measure functional responses through CD23, a cell-mediated killing assay was used.Citation16 IL-4 pre-treated monocytic cells (U937) were shown to mediate killing of anti-folate receptor α (FRα) IgE antibody-labeled IGROV1 target cells (expressing FRα) by CD23-mediated phagocytosis. MEDI4212 reduced phagocytosis (IC50 1.3 nM; ). These studies confirmed that MEDI4212 reduced IgE responses through CD23. In contrast to FcεRI-dependent assays, MEDI4212 demonstrated comparable potency to parent antibody ENG085 and omalizumab in blocking IgE binding and responses through CD23 ().
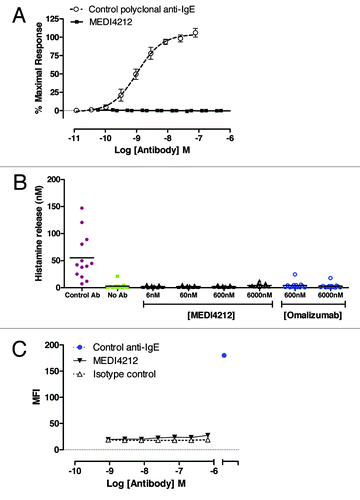
MEDI4212 does not trigger receptor-bound IgE
To avoid induction of inflammation, it was critical that MEDI4212 did not cross-link and activate IgE already bound to its receptors. To test this concept, we examined several different experimental systems that measured IgE-mediated activation of cells. The positive control anti-IgE polyclonal antibody, but not MEDI4212, triggered calcium signaling in IgE-primed RBL-ER51 cells () and histamine release in human whole blood (). These studies indicate that MEDI4212 does not cross-link or activate IgE bound to FcεRI. To determine if MEDI4212 could bind (and would therefore have potential to cross-link) IgE already bound to CD23, FITC-labeled test antibodies were assessed for binding to IgE-loaded RPMI-8866 B-lymphoblast cells by flow cytometry. MEDI4212 showed minimal binding to IgE-loaded cells relative to the commercially-available positive control ().
Figure 3. MEDI4212 does not trigger receptor-bound IgE. Cross-linking of FcεRI-bound IgE by MEDI4212, in comparison with a commercially available polyclonal anti-IgE, was measured by (A) calcium signaling in RBL-ER51 cells (B) human whole blood histamine release assay. (C) Binding to the IgE-loaded CD23 on RPMI-8866 cells was determined for test antibodies in comparison to a commercially available positive control anti-IgE. Data points represent mean ± SEM of combined data from 4–13 experiments (A and B) or average of duplicate determinations from a representative experiment (C).
Structural characterization of human IgE bound by MEDI4212
Protein crystallography was used to define the interaction between MEDI4212 and IgE. Because binding assays confirmed MEDI4212 bound to Cε3-Cε4 domain of the IgE Fc region with similar affinity to full-length protein, this domain was used to form a complex with MEDI4212 Fab and crystallized. The overall structure showed one Cε3-Cε4 bound two Fab molecules in close to symmetric fashion, and that each Fab engaged both IgE chains in binding (). The Cε3-Cε4 domain adopted an open conformation, similar to that capable of binding FcεRI.Citation17,Citation18
Figure 4. Structural definition of MEDI4212-IgE interaction. (A) Overview of the MEDI4212 Fab and IgE Cε3-Cε4 complex. The two identical IgE chains are colored dark red and salmon and the MEDI4212 fab domains blue, with the heavy chain dark blue and the light chain light blue. (B) A surface representation of MEDI4212 Fab illustrating the large fraction of framework residues in the paratope for IgE. MEDI4212 colored as previously, CDR- not engaged in IgE binding light pink, CDR engaged in binding magenta and framework/Vernier residues engaged in IgE binding in dark blue. (C) Framework residues in MEDI4212 (blue) involved in binding IgE (light red). Arg77 (right panel) forms a salt bridge directly with Glu472 in IgE. Ser75 (left panel) is involved in a stabilizing H-bonding network at the interface to IgE, positioning Asp72 in position to form a salt bridge with Arg342 of IgE. (D) Mode of action of MEDI4212 with respect to FcεRI binding. Overlay of the MEDI4212:IgE complex with the structure of FcεRI:IgE shows a direct overlap of the binding site of MEDI4212 and FcεRI. In addition a large volume, illustrated in orange, describes the steric clash between the two molecules trying to bind simultaneously. (E) Mode of action of MEDI4212 with respect to CD23. Overlay of the CD23:IgE complex structureCitation19 with the MEDI4212:IgE structure shows how the open conformation which IgE adopts in complex with MEDI4212 is incompatible with CD23 binding. The clash volume between CD23 and IgE is illustrated as an orange surface. For clarity, MEDI4212 and IgE of the CD23:IgE complex have been omitted in the picture.
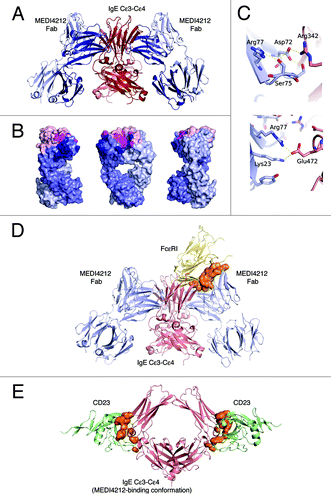
The structural epitope of IgE Cε3-Cε4 recognized by MEDI4212 was made up by amino acid residues Leu221, Arg223, Glu271 through to Asn275 inclusive, Ala309 to Thr315 inclusive, Thr317, Ser318 in Cε3, and Glu353 in Cε4. The sugar moieties GlcNAc1 and Man6 attached via Asn275 were in contact with the MEDI4212 heavy chain (data not shown). The total interaction area was 1150 Å2. Interestingly a large fraction, 57%, of the buried paratope surface constituted non-CDR residues (). It was clear that antibody framework residues contributed significantly to binding as full germlining of the VH resulted in loss of activity. shows how Ser75 and Arg77, two residues that could not be germlined without affecting potency, directly contributed to binding of IgE. It was also apparent that the VL CDR3 did not contribute to direct interaction with IgE, which may explain the ability of MEDI4212 VH to pair with multiple VL CDR3s without significant effects on potency.
Comparison of the MEDI4212 epitope with the FcεRI binding site on IgECitation17 indicated overlap within Cε3 at residues Arg274 and Asn275, suggesting MEDI4212 directly competes with FcεRI for IgE binding. In addition, when overlaying the two complex structures, there is clear spatial overlap between the MEDI4212 Fab and FcεRI ().
In contrast, comparison with the recently published structure of the CD23:IgE Cε3-Cε4 complexCitation19 revealed that, although MEDI4212 binds in close proximity to the CD23 binding site on IgE, it does not directly overlap this site. Instead, MEDI4212 prevents binding of CD23 by locking IgE in an open conformation incompatible with CD23 binding (). In addition, a minor steric clash between MEDI4212 and CD23 was identified (not shown).
Theoretical prediction of IgE suppression in humans
Having shown improved affinity and in vitro potency for MEDI4212 over omalizumab, it was important to understand how this might translate to potential benefits in the clinic.
It is reported that omalizumab may have altered affinity and form different antibody-antigen complexes in serum compared with PBS.Citation20 Thus, it was critical to confirm the high potency of MEDI4212 within this context. To investigate the ability of MEDI4212 to deplete free-IgE in human serum, we used an ex vivo IgE suppression assay with IgE levels spanning a broad range (25–1339 IU/mL). The omalizumab target free-IgE concentration (10 IU/mL, the level shown to relate to clinical efficacyCitation21) was achieved with only 2–3-fold molar excess of MEDI4212, whereas omalizumab required 100-fold excess (). Alternatively, for the same concentration of MEDI4212 as omalizumab, free-IgE levels could be suppressed ~10-fold lower. These data support concepts of greater coverage of high IgE patients and greater suppression of free-IgE with MEDI4212 compared with omalizumab.
Figure 5. IgE suppression in humans. (A) Suppression of free-IgE in ex vivo human sera from 7 donors with baseline IgE levels 25–1339 IU/mL. Data from each donor at each antibody concentration are plotted as separate data points. (B) Mechanistic PK/PD model. Simulated serum free IgE or specific IgE per basophil profiles following administrations of MEDI4212 and omalizumab in (C) typical asthma patient; (D) High IgE / bodyweight; (E) Low total IgE with high IgE specificity.
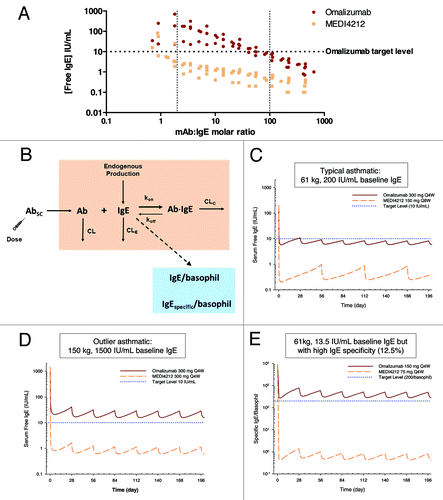
A mechanistic pharmacokinetics/pharmacodynamics (PK/PD) model () was then constructed and translational simulations performed to predict the IgE response in asthma patients following anti-IgE treatment. Simulated serum free-IgE levels following repeated omalizumab and MEDI4212 dosing in a typical asthma patient (61 kg, 200 IU/mL baseline IgE) are shown in . The improvement in binding affinity is anticipated to translate to more substantial free-IgE suppression by MEDI4212.
Further simulations were performed to evaluate potential use of MEDI4212 in asthma patients ineligible for omalizumab treatment. MEDI4212 is predicted to suppress free serum IgE to the 10 IU/mL target level even in patients with extremely high baseline IgE (1500 IU/mL) and high body weight (150 kg) (), as well as eliminate antigen-specific IgE on basophils in patients with low baseline IgE (13.5 IU/mL) but high allergen specificity (12.5%) (). The target level (200 specific IgE per basophil) corresponded to IC90 value determined from in vitro histamine release experiments.Citation22
Analysis of allergen-specific IgE in severe asthma patients
As in vitro and in silico data showed the capacity for MEDI4212 to suppress free-IgE considerably lower than omalizumab, we investigated the potential value of this feature in severe allergic asthma patients. Serum levels of specific IgE against a panel of perennial allergens were measured ( and Table S1). Atopic status was determined by a test result ≥ 0.35 IU/mL of any single allergen-specific IgE and hypothetical suppression of total and specific IgE was investigated. At the omalizumab-defined target free-IgE level of 10 IU/mL,Citation21 individuals with > 3.5% specific IgE did not fall below the 0.35kU/L threshold, resulting in a high proportion of patients (57%; n = 160/283) that would still test positive for atopic status. Omalizumab has a reported non-responder rate of 39%,Citation12 surprisingly close to the 57% with > 3.5% specific IgE in this cohort. We therefore speculate that a significant proportion of omalizumab non-responders may be due to insufficient IgE suppression.
Figure 6. Allergen-specific IgE analysis in severe atopic asthma patients. Total vs. allergen-specific IgE levels in severe atopic asthma. Atopic was defined as least one specific allergen result ≥ 0.35 kU/L from ALT. ALTERNATA, CAT DANDER, COCKROACH, D. FARINAE, D. PTERONYSSINUS, DOG DANDER and additional panel of allergens dependent on country (Table S1).
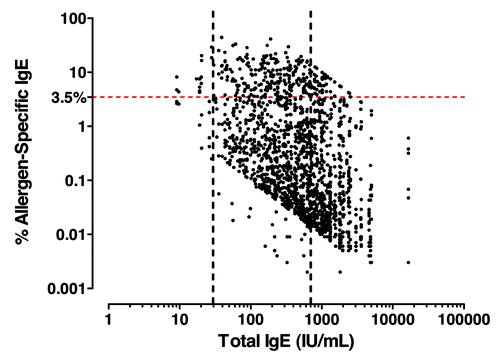
In contrast, a free-IgE level of 1 IU/mL captured > 99% of patients in our cohort below the allergen-specific IgE atopy threshold, suggesting this is the optimal ‘target’ free-IgE in a severe asthma population. MEDI4212 reduced serum free-IgE levels 10-fold lower than omalizumab (), reaching the new 1 IU/mL target. We propose that by fully suppressing IgE, MEDI4212 may provide improved efficacy compared with omalizumab in up to 55% of severe allergic asthma patients.
Discussion
IgE is a critical mediator of allergic responses.Citation2 Powerful actions through FcεRI, triggering degranulation and release of inflammatory mediators from mast cells and basophils, are well defined. Furthermore, clinical experience with IgE-specific antibody omalizumab demonstrates benefits of suppressing IgE in treatment of atopic asthma.Citation8-Citation10 However, the modest affinity of omalizumab for IgE prevents its use in patients with high IgE levels.Citation11 Even within the eligible population, there is evidence that IgE responses may not be completely eliminated in many patients.Citation23,Citation24 Published studies of total and specific IgE levels in a severe asthma population to investigate these issues are few.Citation25 Here, we include such an analysis, which defines numbers of severe asthma patients ineligible for treatment with omalizumab and may also shed light on the sizable non-responding population. In addition, we describe a high affinity anti-IgE antibody, MEDI4212, with potential to enable treatment of individuals with higher body mass or total/specific serum IgE than currently possible with omalizumab.
MEDI4212 is a human IgG1λ monoclonal antibody generated by phage-display technology that selectively binds to the Cε3-Cε4 domain of human IgE. The initial lead antibody, ENG085, isolated from a naïve antibody library had relatively low starting potency (91 nM). A combinatorial mutagenesis strategy taking advantage of CDR loop synergyCitation26 was successfully employed and improved affinity of the final IgG by > 45,000 fold over the starting parent antibody. Investigation of the paratope through protein crystallography suggested this gain was achieved through optimization of VH CDR3 alone. Interestingly, besides VH CDR3, the MEDI4212 VH framework regions also appeared to play a significant role in the interaction with IgE, explaining why attempts to revert certain framework residues to germline significantly affected activity. Framework regions are commonly present in antigen recognition to a variable degree and can comprise up to 15% of the buried surface area of an antibody-antigen complex.Citation27 The presence of 57% buried surface area of non-CDR residues in the MEDI4212-IgE interface represents a novel observation and suggests that mutagenesis of these non-CDR regions could contribute to further affinity improvements for this antibody.
MEDI4212 selectively binds human IgE with affinity of 1.95 pM, more than 100-fold higher than the affinity of omalizumab for IgE, and was shown by crystallography studies to target critical residues in the IgE Cε3 domain critical for interaction with FcεRI.Citation17,Citation28 Consistent with this, MEDI4212 is more efficient at inhibiting IgE binding to, and subsequent activation of, FcεRI than omalizumab.
Omalizumab is licensed for use in atopic asthma, which in our cohort accounts for ~two-thirds of severe asthmatics. Analysis of this population shows that 38% of allergic asthma patients are ineligible for omalizumab treatment due to low IgE < 30 IU/mL (3.3%) or high IgE/ bodyweight (34.3%). Thus, a significant potential for expansion of the patient population exists beyond those served by omalizumab. In silico translational simulations using a mechanistic PK/PD model suggested that MEDI4212, based on free-IgE levels reported to correlate with efficacy,Citation21 could achieve adequate suppression in individuals with high (700–1500 IU/mL) baseline IgE. In support of these assumptions, ex vivo treatment of human sera with MEDI4212 resulted in potent suppression of endogenous IgE levels at concentrations of MEDI4212 50-fold lower than omalizumab.
In addition to expanding the patient population beyond the current IgE/bodyweight limits of omalizumab, a crucial question is whether there is potential for better efficacy with MEDI4212. Patients in our severe asthma cohort with < 76 IU/mL serum IgE (corresponding to a subgroup with lower omalizumab efficacy described by Bousquet et al.Citation12) had higher proportions of allergen specific IgE (67%, n = 29/43) with > 3.5% specific IgE for at least 1 allergen) compared with 55% (n = 131/240) in the subgroup with serum IgE ≥ 76 IU/mL. As our study as well as othersCitation24,Citation29-Citation31 suggest the inability of omalizumab recommended doses to fully suppress IgE activity in patients with > 3–4% of specific IgE to a single allergen, these data highlight the possibility that reduced efficacy with omalizumab in this case may be due to incomplete target suppression. With adequate IgE suppression, there may also be no need to exclude atopic patients with low total IgE (< 30 IU/mL) that often have a high fraction of specific IgECitation29-Citation31 (87.5%, n = 7/8 in our cohort).
The ability to treat low baseline IgE (greater probability of high specific IgE) patients may be even more important if potential treatment of non-atopic asthmatics is considered. Here a much higher proportion (51%) have low total IgE levels (< 76 IU/mL). Although omalizumab has never been fully evaluated and is not currently licensed in this patient subgroup, emerging dataCitation32-Citation36 suggests that IgE may play a pathogenic role.
Most importantly, analysis of total and specific IgE in severe asthmatics suggests that at 5–10 IU/mL of free serum IgE, the lowest levels reached in omalizumab clinical studies,Citation37 38–57% of patients would not have their free allergen-specific IgE suppressed below the ‘atopy’ threshold of 0.35 IU/mL for every allergen. It is therefore interesting to note that reported non-responder rates following omalizumab treatment are 30–39%,Citation12,Citation38 a very similar number. There is also a spectrum of efficacy level reportedCitation38 and perhaps some patients classified as responders might not have achieved maximal response. Our data suggest that a target free-IgE level of 1 IU/ml appears optimal to achieve maximal efficacy in severe asthma. Furthermore, in vitro and in silico analysis highlight that MEDI4212 should achieve this 1 IU/mL target. We speculate that MEDI4212 may lead to an increase in responder rate or efficacy compared with omalizumab by improving IgE suppression in the significant number of severe asthma patients with > 3–4% allergen-specific IgE.
In conclusion, we used an integrated approach to drug discoveryCitation26 that led to the generation of MEDI4212, a novel anti-IgE antibody that has been evaluated in a Phase 1 clinical study (NCT01544348). The data presented here suggest that MEDI4212 may reduce free-IgE to a level (1 IU/mL) anticipated to provide optimal IgE suppression in severe asthma patients.
Materials and Methods
Subjects
Subjects (18–75 y, body mass index 16–40kg/m2) with physician-diagnosed asthma, asthma controller regimen consistent with Step 4 or 5 of the Global Initiative for Asthma guidelines (GINA Report, Global Strategy for Asthma Management and Prevention, updated December 2009. Available from: www.ginasthma.org) for at least 6 mo and 2–6 documented asthma exacerbations in the past year were included in the analysis. Key exclusion criteria were additional respiratory pathology, cigarette smoking ≥ 10 pack-yrs, recent infection or treatment with immunosuppressive medication (> 10 mg oral prednisone or equivalent per day) or any biologic agent.
The entire cohort included 452 severe asthmatics. Only subjects with complete data for atopic status, total and specific IgE, and bodyweight (n = 422) were included in the analysis reported here. Sensitivity analysis revealed that the missing values had little effect on the results presented here and will not invalidate the results.
This study was conducted in accordance with principles of the Declaration of Helsinki and International Conference on Harmonisation Guidance for Good Clinical Practice. Independent ethics committee approval was obtained. All subjects provided written informed consent.
IgE assays
Total, allergen-specific and free-IgE levels were determined by ImmunoCAPTM (Phadia AB) according to manufacturer’s instructions.
Recombinant proteins
IgE was purified from U266 cells.Citation39 Cε3-Cε4 expression and purification is provided in supplementary methods. IgGCitation40 and scFvCitation13 were expressed and purified as previously described. Omalizumab was sourced from a pharmacy, reconstituted according to the package insert and further diluted as required in PBS.
MEDI4212 generation and characterization assays
Generation of MEDI4212, affinity measurements, RBL-ER51 calcium signaling, LAD2 degranulation, IM9 and RPMI-8866 IgE binding, and basophil histamine release assays are described in supplementary methods. Data from cell-based assays were analyzed using Microsoft Excel and Graphpad Prism software. Data are expressed as geometric mean (95% confidence intervals) unless stated otherwise.
Epitope mapping
X-ray crystallography was used to visualize the MEDI4212-IgE interaction. Monomerized Fab was mixed with IgE domain Cε3-Cε4, resultant complex purified by size exclusion chromatography and crystals obtained belonging to the trigonal space group P3221. Diffraction data to 2.85Å resolution were collected at the European Synchrotron Radiation Facility. The structure was solved by Molecular Replacement.Citation41 Details are provided in supplementary methods.
In silico translational simulations
A model was constructed to describe the PK of MEDI4212, binding of MEDI4212 to endogenous IgE and disposition of MEDI4212-IgE complex based on a study of omalizumab in asthma patients.Citation42 Model parameters were adjusted to incorporate KinExA-determined affinity values for MEDI4212 and omalizumab (). MEDI4212 affinity was adjusted upward by 6.7 pM, a theoretical component of in vivo affinity associated with the elimination kinetics of IgG-IgE immunocomplex calculated as the ratio of reported elimination rate of omalizumab-IgE immunocomplex.Citation42 and association rate constant to IgE.Citation43 All other PK and PD parameters were assumed identical for omalizumab and MEDI4212. The model was extended to simulate amount of specific IgE on basophils, with assumptions that MEDI4212 does not alter the fraction of antigen-specific IgE, and, at low serum IgE levels, specific IgE/basophil correlates with serum IgE.
| Abbreviations: | ||
| IgE | = | Immunoglobulin E |
| IgG | = | Immunoglobulin G |
| mAb | = | monoclonal antibody |
| CDR | = | complementarity-determining region |
| Fab | = | antibody binding fragment |
| Fc | = | Fragment crystallizable |
| IC50 | = | half maximal inhibitory concentration |
| IC90 | = | 90% inhibitory concentration |
| Kd | = | equilibrium dissociation constant |
| scFv | = | single chain variable fragment |
| VH | = | variable heavy |
| VL | = | variable light |
| HC | = | heavy chain |
| LC | = | light chain |
| ICS | = | inhaled corticosteroids |
| LABA | = | Long acting β2 agonists |
Additional material
Download Zip (323.4 KB)Disclosure of Potential Conflicts of Interest
This work was funded by AstraZeneca. Support is acknowledged by Cancer Research UK (C30122/A11527); CR UK/EPSRC/MRC/NIHR KCL/UCL Comprehensive Cancer Imaging Centre (C1519/A10331); the National Institute for Health Research (NIHR) Biomedical Research Centre based at Guy's and St Thomas' NHS Foundation Trust and King's College London. The views expressed are those of the author(s) and not necessarily those of the NHS, the NIHR or the Department of Health
Acknowledgments
We thank Ed Piper for access to details of the patient cohort, Athula Herath for statistical support, Steven Coats and Matthew Sleeman for helpful review of the manuscript, Isabelle de Mendez and Duncan Cochrane for support during the project, Martin Strain and Peter Cariuk for technical input, and Faisal Uddin, Ulf Sivars, Elke Lullau, Mark Farmery, Aleksei Rozkov, MedImmune DNA chemistry and IgG teams for reagent generation.
References
- Peters SP, Ferguson G, Deniz Y, Reisner C. Uncontrolled asthma: a review of the prevalence, disease burden and options for treatment. Respir Med 2006; 100:1139 - 51; http://dx.doi.org/10.1016/j.rmed.2006.03.031; PMID: 16713224
- Gould HJ, Sutton BJ, Beavil AJ, Beavil RL, McCloskey N, Coker HA, Fear D, Smurthwaite L. The biology of IGE and the basis of allergic disease. Annu Rev Immunol 2003; 21:579 - 628; http://dx.doi.org/10.1146/annurev.immunol.21.120601.141103; PMID: 12500981
- Chang TW, Wu PC, Hsu CL, Hung AF. Anti-IgE antibodies for the treatment of IgE-mediated allergic diseases. Adv Immunol 2007; 93:63 - 119; http://dx.doi.org/10.1016/S0065-2776(06)93002-8; PMID: 17383539
- Wilcock LK, Francis JN, Durham SR. IgE-facilitated antigen presentation: Role in allergy and the influence of allergen immunotherapy. Immunol Allergy Clin North Am 2006; 26:333,47, viii-ix.
- Li H, Nowak-Wegrzyn A, Charlop-Powers Z, Shreffler W, Chehade M, Thomas S, Roda G, Dahan S, Sperber K, Berin MC. Transcytosis of IgE-antigen complexes by CD23a in human intestinal epithelial cells and its role in food allergy. Gastroenterology 2006; 131:47 - 58; http://dx.doi.org/10.1053/j.gastro.2006.03.044; PMID: 16831589
- Palaniyandi S, Tomei E, Li Z, Conrad DH, Zhu X. CD23-dependent transcytosis of IgE and immune complex across the polarized human respiratory epithelial cells. J Immunol 2011; 186:3484 - 96; http://dx.doi.org/10.4049/jimmunol.1002146; PMID: 21307287
- Karagiannis SN, Bracher MG, Hunt J, McCloskey N, Beavil RL, Beavil AJ, Fear DJ, Thompson RG, East N, Burke F, et al. IgE-antibody-dependent immunotherapy of solid tumors: cytotoxic and phagocytic mechanisms of eradication of ovarian cancer cells. J Immunol 2007; 179:2832 - 43; PMID: 17709497
- Ayres JG, Higgins B, Chilvers ER, Ayre G, Blogg M, Fox H. Efficacy and tolerability of anti-immunoglobulin E therapy with omalizumab in patients with poorly controlled (moderate-to-severe) allergic asthma. Allergy 2004; 59:701 - 8; http://dx.doi.org/10.1111/j.1398-9995.2004.00533.x; PMID: 15180756
- Busse W, Corren J, Lanier BQ, McAlary M, Fowler-Taylor A, Cioppa GD, van As A, Gupta N. Omalizumab, anti-IgE recombinant humanized monoclonal antibody, for the treatment of severe allergic asthma. J Allergy Clin Immunol 2001; 108:184 - 90; http://dx.doi.org/10.1067/mai.2001.117880; PMID: 11496232
- Solèr M, Matz J, Townley R, Buhl R, O’Brien J, Fox H, Thirlwell J, Gupta N, Della Cioppa G. The anti-IgE antibody omalizumab reduces exacerbations and steroid requirement in allergic asthmatics. Eur Respir J 2001; 18:254 - 61; http://dx.doi.org/10.1183/09031936.01.00092101; PMID: 11529281
- Lanier B. Unanswered clinical questions and speculation about the role of anti-immunoglobulin E in atopic and nonatopic disease. Allergy Asthma Proc 2006; 27:Suppl 1 S37 - 42; PMID: 16722331
- Bousquet J, Rabe K, Humbert M, Chung KF, Berger W, Fox H, Ayre G, Chen H, Thomas K, Blogg M, et al. Predicting and evaluating response to omalizumab in patients with severe allergic asthma. Respir Med 2007; 101:1483 - 92; http://dx.doi.org/10.1016/j.rmed.2007.01.011; PMID: 17339107
- Vaughan TJ, Williams AJ, Pritchard K, Osbourn JK, Pope AR, Earnshaw JC, McCafferty J, Hodits RA, Wilton J, Johnson KS. Human antibodies with sub-nanomolar affinities isolated from a large non-immunized phage display library. Nat Biotechnol 1996; 14:309 - 14; http://dx.doi.org/10.1038/nbt0396-309; PMID: 9630891
- Lloyd C, Lowe D, Edwards B, Welsh F, Dilks T, Hardman C, Vaughan T. Modelling the human immune response: performance of a 1011 human antibody repertoire against a broad panel of therapeutically relevant antigens. Protein Eng Des Sel 2009; 22:159 - 68; http://dx.doi.org/10.1093/protein/gzn058; PMID: 18974080
- Foote J, Winter G. Antibody framework residues affecting the conformation of the hypervariable loops. J Mol Biol 1992; 224:487 - 99; http://dx.doi.org/10.1016/0022-2836(92)91010-M; PMID: 1560463
- Bracher M, Gould HJ, Sutton BJ, Dombrowicz D, Karagiannis SN. Three-colour flow cytometric method to measure antibody-dependent tumour cell killing by cytotoxicity and phagocytosis. J Immunol Methods 2007; 323:160 - 71; http://dx.doi.org/10.1016/j.jim.2007.04.009; PMID: 17531261
- Garman SC, Wurzburg BA, Tarchevskaya SS, Kinet JP, Jardetzky TS. Structure of the Fc fragment of human IgE bound to its high-affinity receptor Fc epsilonRI alpha. Nature 2000; 406:259 - 66; http://dx.doi.org/10.1038/35018500; PMID: 10917520
- Holdom MD, Davies AM, Nettleship JE, Bagby SC, Dhaliwal B, Girardi E, Hunt J, Gould HJ, Beavil AJ, McDonnell JM, et al. Conformational changes in IgE contribute to its uniquely slow dissociation rate from receptor FcɛRI. Nat Struct Mol Biol 2011; 18:571 - 6; http://dx.doi.org/10.1038/nsmb.2044; PMID: 21516097
- Dhaliwal B, Yuan D, Pang MO, Henry AJ, Cain K, Oxbrow A, Fabiane SM, Beavil AJ, McDonnell JM, Gould HJ, et al. Crystal structure of IgE bound to its B-cell receptor CD23 reveals a mechanism of reciprocal allosteric inhibition with high affinity receptor FcεRI. Proc Natl Acad Sci U S A 2012; 109:12686 - 91; http://dx.doi.org/10.1073/pnas.1207278109; PMID: 22802656
- Demeule B, Shire SJ, Liu J. A therapeutic antibody and its antigen form different complexes in serum than in phosphate-buffered saline: a study by analytical ultracentrifugation. Anal Biochem 2009; 388:279 - 87; http://dx.doi.org/10.1016/j.ab.2009.03.012; PMID: 19289095
- Hochhaus G, Brookman L, Fox H, Johnson C, Matthews J, Ren S, Deniz Y. Pharmacodynamics of omalizumab: implications for optimised dosing strategies and clinical efficacy in the treatment of allergic asthma. Curr Med Res Opin 2003; 19:491 - 8; http://dx.doi.org/10.1185/030079903125002171; PMID: 14594521
- MacGlashan DW Jr., W. MD. Releasability of human basophils: cellular sensitivity and maximal histamine release are independent variables. J Allergy Clin Immunol 1993; 91:605 - 15; http://dx.doi.org/10.1016/0091-6749(93)90266-I; PMID: 7679683
- Corren J, Shapiro G, Reimann J, Deniz Y, Wong D, Adelman D, Togias A. Allergen skin tests and free IgE levels during reduction and cessation of omalizumab therapy. J Allergy Clin Immunol 2008; 121:506 - 11; http://dx.doi.org/10.1016/j.jaci.2007.11.026; PMID: 18269927
- Johansson SG, Nopp A, Oman H, Ankerst J, Cardell LO, Grönneberg R, Matsols H, Rudblad S, Strand V, Stålenheim G. The size of the disease relevant IgE antibody fraction in relation to ‘total-IgE’ predicts the efficacy of anti-IgE (Xolair) treatment. Allergy 2009; 64:1472 - 7; http://dx.doi.org/10.1111/j.1398-9995.2009.02051.x; PMID: 19393000
- Wahn U, Martin C, Freeman P, Blogg M, Jimenez P. Relationship between pretreatment specific IgE and the response to omalizumab therapy. Allergy 2009; 64:1780 - 7; http://dx.doi.org/10.1111/j.1398-9995.2009.02119.x; PMID: 19627273
- Minter RR, Cohen ES, Wang B, Liang M, Vainshtein I, Rees G, et al. Protein engineering and preclinical development of a GM-CSF receptor antibody for the treatment of rheumatoid arthritis. Br J Pharmacol 2012; In press PMID: 22913645
- Sundberg EJ. 2. structural basis of antibody-antigen interactions. In: Reineke U, Schutkowski M, eds. Epitope Mapping Protocols. 2nd ed.: Humana Press, 2009.
- Sayers I, Cain SA, Swan JR, Pickett MA, Watt PJ, Holgate ST, Padlan EA, Schuck P, Helm BA. Amino acid residues that influence Fc epsilon RI-mediated effector functions of human immunoglobulin E. Biochemistry 1998; 37:16152 - 64; http://dx.doi.org/10.1021/bi981456k; PMID: 9819207
- Johansson SG, Oman H, Nopp A, Pettersson S. The importance of IgE antibody levels in anti-IgE treatment. Allergy 2006; 61:1216 - 9; http://dx.doi.org/10.1111/j.1398-9995.2006.01172.x; PMID: 16942572
- Ankerst J, Nopp A, Johansson SG, Adédoyin J, Oman H. Xolair is effective in allergics with a low serum IgE level. Int Arch Allergy Immunol 2010; 152:71 - 4; http://dx.doi.org/10.1159/000260086; PMID: 19940508
- Hamilton RG, MacGlashan DW Jr., Saini SS. IgE antibody-specific activity in human allergic disease. Immunol Res 2010; 47:273 - 84; http://dx.doi.org/10.1007/s12026-009-8160-3; PMID: 20066506
- Bachert C, van Steen K, Zhang N, Holtappels G, Cattaert T, Maus B, et al. Specific IgE against staphylococcus aureus enterotoxins: An independent risk factor for asthma. J Allergy Clin Immunol 2012; 130:376,381 e8.
- de Llano LP, Vennera MdelC, Álvarez FJ, Medina JF, Borderías L, Pellicer C, González H, Gullón JA, Martínez-Moragón E, Sabadell C, et al, Spanish Registry. Effects of omalizumab in non-atopic asthma: results from a Spanish multicenter registry. J Asthma 2013; 50:296 - 301; http://dx.doi.org/10.3109/02770903.2012.757780; PMID: 23350994
- Menzella F, Piro R, Facciolongo N, Castagnetti C, Simonazzi A, Zucchi L. Long-term benefits of omalizumab in a patient with severe non-allergic asthma. Allergy Asthma Clin Immunol 2011; 7:9; http://dx.doi.org/10.1186/1710-1492-7-9; PMID: 21609447
- Takhar P, Corrigan CJ, Smurthwaite L, O’Connor BJ, Durham SR, Lee TH, Gould HJ. Class switch recombination to IgE in the bronchial mucosa of atopic and nonatopic patients with asthma. J Allergy Clin Immunol 2007; 119:213 - 8; http://dx.doi.org/10.1016/j.jaci.2006.09.045; PMID: 17208604
- van den Berge M, Pauw RG, de Monchy JG, van Minnen CA, Postma DS, Kerstjens HA. Beneficial effects of treatment with anti-IgE antibodies (Omalizumab) in a patient with severe asthma and negative skin-prick test results. Chest 2011; 139:190 - 3; http://dx.doi.org/10.1378/chest.10-0128; PMID: 21208879
- Lowe PJ, Tannenbaum S, Gautier A, Jimenez P. Relationship between omalizumab pharmacokinetics, IgE pharmacodynamics and symptoms in patients with severe persistent allergic (IgE-mediated) asthma. Br J Clin Pharmacol 2009; 68:61 - 76; http://dx.doi.org/10.1111/j.1365-2125.2009.03401.x; PMID: 19660004
- Braunstahl GJ, Chen CW, Maykut R, Georgiou P, Peachey G, Bruce J. The eXpeRience registry: the ‘real-world’ effectiveness of omalizumab in allergic asthma. Respir Med 2013; 107:1141 - 51; http://dx.doi.org/10.1016/j.rmed.2013.04.017; PMID: 23721684
- Ikeyama S, Nakagawa S, Arakawa M, Sugino H, Kakinuma A. Purification and characterization of IgE produced by human myeloma cell line, U266. Mol Immunol 1986; 23:159 - 67; http://dx.doi.org/10.1016/0161-5890(86)90038-6; PMID: 3702874
- Persic L, Roberts A, Wilton J, Cattaneo A, Bradbury A, Hoogenboom HR. An integrated vector system for the eukaryotic expression of antibodies or their fragments after selection from phage display libraries. Gene 1997; 187:9 - 18; http://dx.doi.org/10.1016/S0378-1119(96)00628-2; PMID: 9073061
- Rossmann MG. The Molecular Replacement Method. New York: Gordon & Breach, 1972.
- Hayashi N, Tsukamoto Y, Sallas WM, Lowe PJ. A mechanism-based binding model for the population pharmacokinetics and pharmacodynamics of omalizumab. Br J Clin Pharmacol 2007; 63:548 - 61; http://dx.doi.org/10.1111/j.1365-2125.2006.02803.x; PMID: 17096680
- Putnam WS, Li J, Haggstrom J, Ng C, Kadkhodayan-Fischer S, Cheu M, Deniz Y, Lowman H, Fielder P, Visich J, et al. Use of quantitative pharmacology in the development of HAE1, a high-affinity anti-IgE monoclonal antibody. AAPS J 2008; 10:425 - 30; http://dx.doi.org/10.1208/s12248-008-9045-4; PMID: 18686041
