Abstract
Glycosylation is an important post-translational modification during protein production in eukaryotic cells, and it is essential for protein structure, stability, half-life, and biological functions. In this study, we produced various monoclonal antibody (mAb) glycoforms from Chinese hamster ovary (CHO) cells, including the natively glycosylated antibody, the enriched G0 form, the deglycosylated form, the afucosylated form, and the high mannose form, and we compared their intrinsic properties, side-by-side, through biophysical and biochemical approaches. Spectroscopic analysis indicates no measureable secondary or tertiary structural changes after in vitro or in vivo modification of the glycosylation pattern. Thermal unfolding experiments show that the high mannose and deglycosylated forms have reduced thermal stability of the CH2 domain compared with the other tested glycoforms. We also observed that the individual domain’s thermal stability could be pH dependent. Proteolysis analysis indicates that glycosylation plays an important role in stabilizing mAbs against proteases. The stability of antibody glycoforms at the storage condition (2–8 °C) and at accelerated conditions (30 and 40 °C) was evaluated, and the results indicate that glycosylation patterns do not substantially affect the storage stability of the antibody we studied.
Introduction
Post-translational modification is a common event during protein production from eukaryotic cells.Citation1,Citation2 It includes a wide variety of chemical modifications, such as acetylation,Citation3,Citation4 amidation,Citation5,Citation6 carboxylation,Citation7 glycosylation,Citation8 and phosphorylation.Citation9,Citation10 These important chemical modifications are essential for protein folding, stability, and function. Among various post-translational modifications, glycosylation is the most complex one from the perspective of chemical heterogeneity of modifications because it involves the covalent attachment of sugar moieties to proteins.Citation11,Citation12 Glycosylation pattern plays an important role in the biological activity of a protein, such as that of erythropoietinCitation13 and afucosylated monoclonal antibodies (mAbs).Citation14 There are two common types of glycosylation: O-linked glycosylation and N-linked glycosylation. In O-linked glycosylation, oligosaccharides are linked to amino acids with hydroxyl functional groups, such as serine (Ser), threonine (Thr), or tyrosine (Tyr). O-linked glycosylation is found in epidermal growth factor domains, urokinase, fetuin, and antifreeze glycoproteins.Citation11,Citation12 In N-linked glycosylation, oligosaccharides are linked to asparagine (Asn) in the conserved sequence of Asn-X-Ser/Thr, where X is any amino acid except proline (Pro).Citation15 N-linked glycosylation is found in plasma proteins, cell surface receptors, and lectins.Citation11
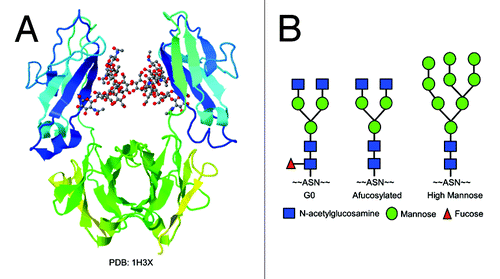
During therapeutic antibody production via mammalian cells, such as Chinese hamster ovary (CHO) cells, N-linked glycosylation is commonly observed. For the IgG1 type of antibodies, there is a conserved Asn297 glycosylation site in the heavy chain Fc region. During the post-translational modification, multiple sugar moieties (e.g., N-acetylglucosamine (GlcNAc), mannose, fucose, galactose) can be assembled with complex combinations to form a variety of glycosylation patterns. A crystal structure of the Fc region with glycan attachedCitation16 is shown in . The most common glycosylation pattern of recombinant IgG1 mAbs produced through CHO cells is the G0 form, which contains two core GlcNAc and one core fucose, three mannose, and terminal GlcNAc moieties (). Other glycosylation patterns include the afucosylated form, oligosaccharides that do not contain core fucose; and the high mannose form, an intermediate form before removal of extra mannose molecules by mannosidase and the following sugar moieties assembly (). The composition of oligosaccharides in the mAb Fc region plays a critical role in its biological activities, such as antibody-dependent cell-mediated cytotoxicity (ADCC) and complement dependent cytotoxicity (CDC).Citation17 It is reported that proteins without the core fucose increase ADCC activity in vivo due to increased affinity to FcγRIIIA.Citation14 The high mannose form also exhibits enhanced ADCC activities since it is an immature form and the core fucose is not attached.Citation18-Citation20 Complete removal of glycosylation from the Fc region of mAb results in loss of antibody’s effector functions through loss of the ability to bind to Fcγ receptors.Citation16
Figure 1. (A) Crystal structure of the Fc region of an IgG1 antibody with glycan attached (PDB: 1H3X). (B) Glycoform patterns of G0, afucosylated, and high mannose forms. Symbols: blue squares, N-acetylglucosamine (GlcNAc); green circles, mannose; red triangles, fucose.
Although antibody glycosylation has been widely studied, most reports focus on biological activities and lack a thorough side-by-side comparison of molecular intrinsic biophysical and biochemical properties among various antibody glycoforms. These properties are important for efficacy, quality, and stability of therapeutic products. Recently, we published a reportCitation21 on the investigation of changes in structure and intrinsic molecular properties after complete removal of oligosaccharide from mAbs. The work described here focuses on the effect of the glycosylation pattern on antibody’s intrinsic properties. We produced a mAb with a variety of glycosylation patterns, including the enriched G0 form, which represents the primary glycoform during CHO cell production; the afucosylated form, which may exist in small quantities from the production using CHO cells and exhibits enhanced ADCC activities; the high mannose form, which is an immature glycoform with increased effector functions and is rarely observed from the production via CHO cells; and the deglycosylated form, which could be a good representative of aglycosyl antibody drugs, such as products from E. coli cell lines (). We used biophysical and biochemical methods to characterize how glycosylation patterns affect the overall secondary and tertiary structures and intrinsic properties of IgGs. Long-term stability of antibody glycoforms was also evaluated in our study. The stability results provide a good understanding of the correlation between the glycosylation pattern and shelf-life stability.
Through this detailed comparison, we hope to bridge glycoengineering work with product quality and stability. A thorough understanding of the effect of glycosylation pattern on the biophysical and biochemical properties and stability of mAbs will facilitate and accelerate the development of therapeutic antibodies.
Results
Antibody glycoform production
We used in vitro methods to produce antibodies with enriched G0 and complete deglycosylated glycoforms, and we used in vivo methods to produce afucosylated and high mannose antibody glycoforms. The monomeric purity of all antibodies was over 98% based on the size-exclusion chromatography (SEC) analysis (data not shown). To identify and confirm each in vitro and in vivo glycosylation modification, we compared the experimental mass of the heavy chain calculated from liquid chromatography–mass spectrometry (LC-MS) to its theoretical value. Heavy-chain masses of the modified glycoforms were found to correspond to the calculated theoretical masses (). We further estimated the homogeneity of glycoforms using capillary electrophoresis, which reached over 70% (data not shown).
Table 1. Mass of mAb heavy chains
Spectroscopic characterization of mAb structure
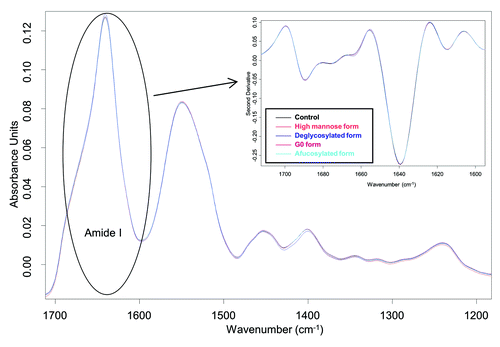
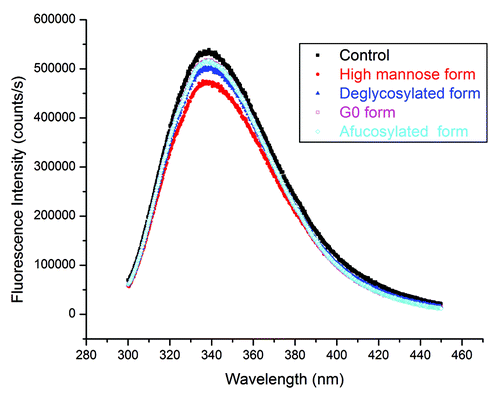
After confirming the glycosylation modifications, we used Fourier transform infrared (FTIR) spectroscopy to evaluate the secondary structure and intrinsic fluorescence to characterize the tertiary structure of antibody glycoforms. shows the original infrared (IR) absorbance spectra. In order to better understand the secondary structure, we calculated the second derivatives of the amide I region of antibody glycoforms using OPUS 6.5 software (Bruker Corp.), and these data are shown in the insert of . The amide I region is between 1700–1600 cm−1. Its signal mainly comes from the C=O bond stretching vibration, and it contains important information regarding protein secondary structures.Citation22,Citation23 From , neither the original spectra nor the second derivatives show notable alteration of the secondary structure as a function of the glycoforms present. Next, we characterized the tertiary structure of antibody glycoforms using intrinsic fluorescence primarily from tryptophan (Trp) residues. From the X-ray crystal structure of the Fc region (PDB: 1H3X), it was shown that the distance to the Trp closest to the glycosylation site is ~10 Å (1 Å = 0.1 nm). Thus, any changes in fluorescence would be induced by tertiary structural changes of mAbs instead of the direct interaction with glycans. Trp emission spectra in revealed that the fluorescence emission peaks of all antibody glycoforms are between 337–339 nm. Considering the variability of the fluorescence method, it suggests that there is no overall tertiary structural alteration after either in vitro or in vivo glycosylation modification.
Thermal unfolding
The effect of the glycosylation pattern on the secondary or tertiary structure of the antibody appears to be unmeasurable using FTIR spectroscopy or intrinsic fluorescence analysis, but glycosylation could play an important role in the thermal stability of mAbs.Citation24 To explore this hypothesis, we used differential scanning calorimetry (DSC) to measure thermal unfolding transitions of antibody glycoforms at pH 5.5 (). Assignments of the thermal transition of the individual domain of IgG1 antibodies were reported previously.Citation25 Normally the first thermal transition peak is from the contribution of the CH2 domain and the following two peaks represent Fab and CH3 regions’ unfolding. Sometimes the transitions of Fab and CH3 regions are very close and merge into a single peak, which is what was observed in this study. There is a single peak between 75–85 °C among all antibody glycoforms, which indicates that there is no clear distinction of the thermal unfolding between Fab and CH3 regions for the antibody we studied. Furthermore, the single peak between 75–85 °C has similar transition temperatures for all glycoforms, suggesting that there is no effect on the thermal stability of Fab and CH3 regions from oligosaccharides.
Figure 4. Thermal unfolding of glycoforms from DSC measurements. Thermogram line colors: black, control; red, high mannose form; blue, deglycosylated form; magenta, G0 form; cyan, afucosylated form.
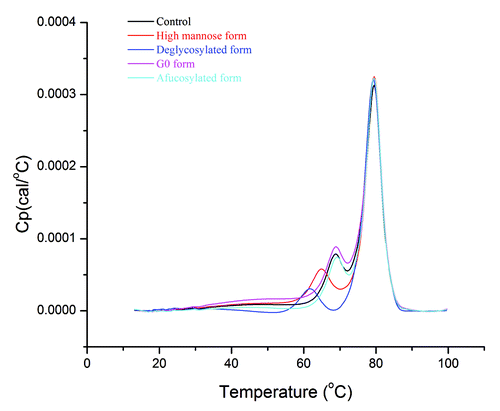
Nevertheless, the transition peak of the CH2 domain is well-resolved most of the time, which provides an opportunity to elucidate the effect of oligosaccharides on its nearby regions. As shown in , there is no substantial difference among the control, G0, and afucosylated forms. However, the high mannose form exhibits lower unfolding temperatures than the control, G0, or afucosylated forms, and the deglycosylated form has the lowest thermal transition among all. Thermal unfolding data indicate that oligosaccharides in the CH2 domain play an important role in stabilizing its local region.
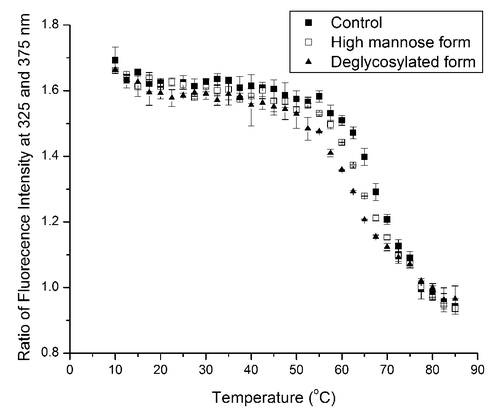
Because the high mannose and deglycosylated forms show notable thermal transition shifts of the CH2 domain compared with that of the control form in DSC, we further confirmed the thermal stability trend using intrinsic fluorescence, which serves as an orthogonal method to the DSC measurement. To reduce the noise of fluorescence data, we monitored the fluorescence intensity ratio at 325 and 375 nm at each temperature interval between 10–85 °C and plotted it vs. temperature (). Between 50–75 °C, thermal transitions of glycoforms exhibit the same stability ranking as that from DSC data: deglycosylated form < high mannose form < control. Our data also indicate that although intrinsic fluorescence cannot detect any overall tertiary structural change of mAbs after glycosylation pattern modifications (), it can be a useful approach to characterize protein thermal unfolding.
Figure 5. Thermal unfolding of control (filled squares), high mannose (open squares), and deglycosylated (filled triangles) forms from fluorescence measurements. Ratios of fluorescence intensities at 325 and 375 nm are plotted vs. temperature.
During production, purification, storage, and administration, therapeutic mAbs would be exposed to a wide range of pH conditions. Thus, it is important to understand how the glycosylation pattern affects antibody properties under various pH conditions. Since the DSC measurement is a sensitive approach to detect the oligosaccharide impact on antibodies, we evaluated the thermal stability of antibody glycoforms at various pH conditions. Because there is no substantial thermal stability alteration for G0 and afucosylated glycoforms compared with the control form (), we focused on control, high mannose, and deglycosylated forms in the pH dependent study to simplify the comparison. We tested a pH range of 4.0–7.5. Data are shown in −E and . We found that the three tested forms show similar trends: from pH 4.0–5.5, the first transition temperature (Tm1) increases about 14–17 °C and the main transition temperature (Tm2) increases about 6–7 °C. From pH 5.5–6.5, Tm1 increases about 4 °C while Tm2 has a slight decrease (≤ 1 °C). Tm1 of the control form becomes undetectable as it merges into the main transition. On the other hand, a new shoulder peak around 82.5 °C (Tm3) emerged for all three glycoforms. For all glycoforms from pH 6.5–7.5, there is a slight increase in Tm1, a slight decrease in Tm2, and lack of the trend in Tm3.
Figure 6. Thermal unfolding of control (solid line), high mannose (dashed line), and deglycosylated (dotted line) forms from DSC measurements. Thermograms: (A) at pH 4.0, (B) at pH 5.5, (C) at pH 6.5, (D) at pH 7.0, (E) at pH 7.5.
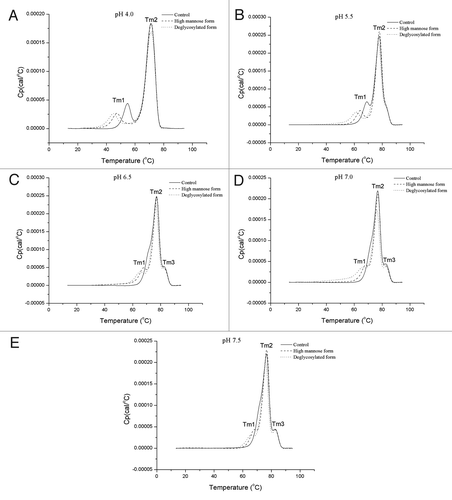
Table 2. Thermal transition temperatures (°C) measured by DSC at various pH conditions
The disappearance of Tm1 in the control form appears to be due to the proximity of its first transition to the main peak (unfolding of Fab and CH3 regions) and it merges into the main transition peak as its transition temperature increases at high pH conditions. On the other hand, because Tm1 values of high mannose and deglycosylated forms are much lower compared with the control form due to the reduced thermal stability of the CH2 domain, they remain distinct from the main peak even at high pH conditions. As a result, three individual peaks are observed for thermograms of high mannose and deglycosylated forms in −E. The appearance of emerged Tm3 peaks at pH 6.5 and above could be because the unfolding transitions of Fab and CH3 regions differentiate as pH increases, i.e., from pH 5.5–7.5, there is about 1.1–1.5 °C decrease of Tm2, while Tm3 remains the same. Thus, two transitions are separated and a new peak is shown at around 82.5 °C.
Protease digestion
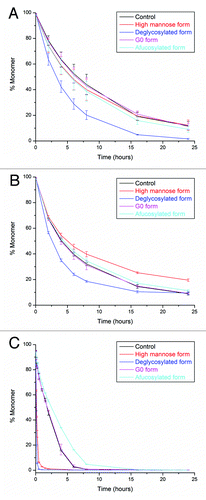
After understanding the glycosylation pattern effect on the thermal unfolding of CH2, CH3, and Fab regions, we used three proteases, human neutrophil elastase (HNE), papain, and pepsin as probes to further characterize the subtle changes in the CH2 domain caused by glycan variation. Both HNE and papain digest antibodies to Fab and Fc fragments while pepsin divides antibodies to F(ab′)2 and Fc fragments. In the sequence of SCDKTHTCPPCPAPELLGGPSV from the heavy chain, HNE predominantly cleaves between Thr and histidine (His); papain primarily cuts between His and Thr; while pepsin mainly digests between the two leucine residues.Citation26 Thus, following the order of HNE, papain, and pepsin, the protease digestion site approaches closer to the glycosylation site. During the digestion process (−C), we used SEC to monitor the decrease of the full-length antibody (monomer) peak. When various glycoforms are digested by the same protease, they all exhibit similar degradation products regardless the glycosylation patterns. On the other hand, the antibody shows slightly different digestion patterns when treated with different proteases. It is most likely due to the variation in digestion sites and the specificity of proteases. During the HNE digestion (), most antibody glycoforms have similar digestion rates except the deglycosylated form, which degrades much faster. During the papain digestion (), digestion rates among antibody glycoforms start to differentiate: the high mannose form shows the slowest digestion rate while the deglycosylated form exhibits the fastest digestion rate. In a recent report, it was shown that the terminal sugar (GlcNAc, β-galactose, or sialic acid) of the glycan plays an important role in the resistance to papain digestion.Citation27 Our findings further support this conclusion, which suggests that the terminal mannose could be essential for mAb stability against papain. In addition, the local steric exclusion effect might also contribute to the increase of the stability of the high mannose glycoform against papain. (pepsin digestion) shows even more obvious differentiation of the digestion rate among antibody glycoforms: the afucosylated form is more stable while both high mannose and deglycosylated forms become less stable compared with G0 and control forms. The increased effect from glycosylation on the resistance to pepsin could be related to the low pH (4.0) of the digestion condition. There may be substantial local conformational changes at pH 4.0 and glycosylation pattern is critical in determining the accessibility to pepsin under this condition. The increased resistance of the afucosylated form needs to be related to the lack of the core fucose and warrants further investigations. The reduced stability of high mannose or deglycosylated forms under pepsin digestion could be related to the effect of glycans on the stability of the CH2 domain. Since the pepsin digestion site is closer to the CH2 domain compared with HNE and papain digestion sites, the stability of the CH2 domain could play a more critical role in pepsin digestion rates. As a result, the less stable CH2 domains for high mannose and deglycosylated forms lead to a faster pepsin digestion rates.
Long-term stability
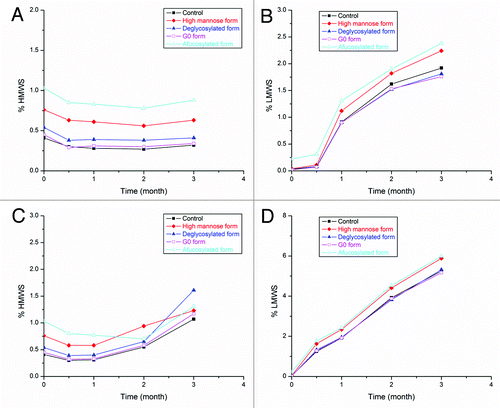
Antibody stability during long-term storage is a critical point to consider during biotherapeutic development. In this study, we evaluated the stability of antibody glycoforms at the storage temperature of 5 °C and at accelerated temperatures of 30 and 40 °C using a low ionic strength formulation buffered at pH 5.5. We used SEC as a stability-indicating assay to monitor monomer loss as well as the formation of high molecular weight species (HMWS; i.e., aggregation) and low molecular weight species (LMWS; i.e., fragmentation) during the storage. We did not observe any substantial monomer loss among all tested antibody glycoforms at 5 °C up to three months (data not shown). At accelerated temperatures (30 and 40 °C), the major degradation pathway of the antibody is fragmentation (−D). These data show that the glycosylation pattern has minimal effect on the stability of the mAb studied here. Our current result is in agreement with a previous report from Gaza-Bulseco, et al.Citation28 that showed that glycosylation does not affect mAb fragmentation in the pH range of 5.0–9.0, which is probably due to the negligible effect on the intra-molecular self-cleavage in the hinge region from glycosylation modifications. However, if the mAb is formulated at low pH, such as pH 4.0, the glycosylation pattern could play an important role in stabilizing the mAb as Gaza-Bulseco et al. had reported,Citation28 such that the fragmentation sites move toward the C-terminal when pH decreases, and glycosylation is essential in protecting against hydrolysis in the Fc region.
Figure 8. Stability profiles of glycoforms measured by SEC. Profiles after three-month storage at 30 °C: (A) HMWS and (B) LMWS. Profiles after three-month storage at 40 °C: (C) HMWS and (D) LMWS. Filled markers in all figures: black squares, control; red diamonds, high mannose form; blue triangles, deglycosylated form. Open markers in all figures: magenta squares, G0 form; cyan triangles, afucosylated form.
Discussion
In this report, we used in vitro and in vivo methods to modify glycosylation patterns of an IgG1 type of mAb and characterized its properties using biophysical and biochemical methods. Both FTIR and fluorescence analysis demonstrated that the glycosylation pattern in the Fc region has a low impact on the conformation of mAbs. The result could be due to the inadequate resolution of these spectroscopic techniques, which are able to provide sufficient information about the structure of the entire molecule but are not very sensitive to the local subtle changes in an individual domain. If there is any conformational change in the CH2 domain induced by glycosylation modifications, isolation of the Fc fragment of each antibody glycoform would help to capture these minor structural changes during the spectroscopic characterization. This work could be continued in a separate study.
On the other hand, thermal stabilities of CH2 domains are affected by modifications of the glycosylation pattern. Previous data showed that oligosaccharides could interact with each other or with proteins, which may contribute to stabilizing CH2 domains.Citation24,Citation29 Our data in this study are in agreement with this theory: the CH2 region of the deglycosylated form has the least thermal stability due to the lack of glycans. The high mannose antibody glycoform is an immature form during the post-translational modification.Citation20 The lower thermal transition temperature of this form (), as compared with the control form, indicates that the additional mannose at the terminus of the oligosaccharide chain may interrupt sugar-sugar or sugar-protein interactions and thus destabilize the thermal stability of the CH2 domain. This explanation is consistent with a previous report that found the distance between the two glycosylation sites, Asn297, on two heavy chains is altered for the high mannose glycoform.Citation20
Furthermore, subtle conformational changes of mAb induced by glycosylation pattern variations could substantially affect the resistance of mAb to external proteolysis. It was reported previously that deglycosylated mAb is less stable to proteolytic cleavage by papain.Citation27,Citation30 Another report using hydrogen-deuterium exchange experiments showed that glycosylation could affect molecular dynamic fluctuation.Citation31 Those authors suggested that the sugar moiety decreases molecular flexibility and may protect the cleavage sites. Our data are consistent with these previous reports and further demonstrate that glycosylation provides a broad protection to the mAb Fc region against proteases, such as HNE, papain, and pepsin. On the other hand, it indicates that protease digestion is a sensitive method to monitor minor changes in large biological molecules and this feature could lead to a new approach to assess the higher-order structure of the IgG type of macromolecules. For example, information of the higher-order structure of therapeutic mAbs is often needed during regulatory filings. Commonly used analytical assays, such as SEC, ion-exchange chromatography, mass spectroscopy, or other capillary electrophoresis-based methods, are powerful methods to detect changes in a protein’s primary structure, but these data cannot be easily correlated with secondary or tertiary structural alterations. On the other hand, biophysical methods, including circular dichroism, FTIR, or fluorescence spectroscopies, are useful to provide the overall secondary or tertiary structural information of the entire mAb, but are not sensitive enough to detect subtle local conformational alterations, such as glycosylation modification in the CH2 domain, as shown in and 3. The protease digestion method could be a good approach to fill this gap. In addition, the protease digestion method may also be useful for the characterization of antigen-antibody binding or pharmacokinetic studies. Because of the complexity of the protease digestion, additional developmental work will be required before expanding the application of the protease digestion method. For instance, for each specific mAb, it may be necessary to screen multiple proteases to identify the one with the highest sensitivity on minor conformational changes. In addition to the selection of the protease, digestion conditions, including protein concentration, protease concentration, pH, digestion temperature, and digestion duration, need to be optimized to achieve the desired outcome.
Materials and Methods
Production of glycan variants
Protein control
A fully humanized mAb that is a part of the IgG1 subclass was selected as the model protein in this study. It was produced from CHO cells and purified through standard antibody purification procedures, including protein A, cation exchange, and anion exchange chromatography, at Genentech Inc.
Production of the G0 form
To enrich the G0 form, four units of 1–4-galactosidase (ProZyme) were added to 1 g of the antibody, and the solution was incubated in a water bath for 48 h at 37 °C. The antibody was then purified using a Protein A column (GE Healthcare Life Sciences). The pH of the protein A eluted pool was adjusted to 5.5 and then concentrated and buffer exchanged into a buffer at pH 5.5.
Production of the deglycosylated form
PNGase F (New England Biolabs) was used to remove N-linked glycosylation by incubating 1 mg of mAb with 275 units of PNGase F at 37 °C in a water bath for 24 h. The deglycosylated form was purified in the same way as the G0 form.
Generation of the high mannose form
The high mannose form antibody was cultured in growth media in shake flasks using media prepared by Genentech Inc. Kifunensine (Cayman Chemical Company) was added to the cell culture media at a concentration of 1 mg/L as an α-mannosidase inhibitor to prevent the hydrolysis of α-mannosidase linkages during the post-translational modification. Cells were grown for 11 d at 37 °C before harvesting with an amino acid feed performed on day 6. The high mannose form was then purified through standard antibody purification procedures.
Generation of the afucosylated form
The afucosylated antibody glycoform was produced using a knockout cell line in shake flasks with media. Cells were grown for 11 d at 37 °C before harvesting with an amino acid feed performed on day 6. The afucosylated antibody was purified through standard purification procedures.
Liquid chromatography–mass spectrometry
Masses of heavy chains (containing the glycosylation site) were collected to confirm the identity of each antibody glycoform. Samples were diluted to 1 mg/mL and reduced by 1 mg/mL (final concentration) tris (2-carboxyethyl) phosphine hydrochloride (EMD Chemicals) at 60 °C for 10 min. Heavy and light chains were separated after reduction through reverse phase high-performance liquid chromatography (HPLC) and analyzed by electrospray ionization mass spectrometry. Details of the LC-MS method have been published elsewhere.Citation32 In brief, reverse phase HPLC was run on an Agilent 1200 HPLC system with a Varian PLRP-S column (1000 Å, 8 µm, 150 × 4.6 mm). Mass spectroscopy data were collected on a QStar Elite mass spectrometer (Applied Biosystems/MDS Sciex). The detection mode was time-of-flight. The analyst QS 2.0/BioAnalyst 2.0 software (Applied Biosystems/MDS Sciex) was used to obtain and deconvolute the spectrum of the ionized heavy chain.
Size-exclusion chromatography
SEC was performed on an Agilent 1200 HPLC with a TOSOH Bioscience TSK gel G3000 SWXL column (7.8 × 300 mm). The mobile phase contained potassium phosphate and potassium chloride at pH 6.2. Samples were diluted to 2 mg/mL using the SEC mobile phase prior to injection. The injection volume was 25 µL, flow rate was 0.5 mL/min, and detection wavelength was 280 nm.
Fourier transform infrared spectroscopy
IR spectra of antibody glycoforms were collected from a Bruker Tensor 27 IR spectrometer with a mercury cadmium telluride (MCT) detector for the secondary structural analysis. The MCT detector was cooled by liquid nitrogen prior to data acquisition. After setting the resolution of the instrument to 4 cm−1, 50 µL of 20 mg/mL sample were injected into a 6.8-µm path-length AquaSpec transmission cell, and 60 scans were collected for each acquisition. The formulation buffer was injected into the same transmission cell prior to each sample injection and its IR absorbance was subtracted correspondingly. The temperature of the transmission cell was maintained at 25 °C during the entire measurement via a water bath.
Fluorescence spectroscopy
Fluorescence spectra of antibody glycoforms were recorded using a Horiba Jobin Yvon Fluoromax-4 spectrofluorometer for the tertiary structural analysis. Both excitation and emission slit widths were set to 3 nm. The excitation wavelength was set at 295 nm. One mL protein sample at 0.1 mg/mL in formulation buffer at pH 5.5 was injected into a 1-cm path-length quartz cuvette and scanned from 300–450 nm. A blank spectrum of the formulation buffer was subtracted from each sample spectrum. All spectra were collected at 25 °C; temperature was controlled using a water bath. Thermal unfolding of antibody glycoforms was also recorded by collecting emission spectra from 10–85 °C at 2.5 °C increments by the same spectrofluorometer.
Differential scanning calorimetry
The thermal stability of antibody glycoforms was characterized by a MicroCal VP DSC. During each sample measurement, the formulation buffer at pH 5.5 was injected into the reference cell, and 2 mg/mL antibody in the same formulation buffer was injected into the sample cell. Thermograms were collected from 10 to 100 °C at a scan rate of 60 °C⁄h. To avoid the intrinsic difference between reference and sample cells, the formulation buffer was also injected into both reference and sample cells and its thermogram was collected and subtracted from each sample measurement. Origin 7.0 software (OriginLab Corp.) was used for data processing.
To further explore the pH dependency of mAb’s thermal stability, three antibody glycoforms (control, high mannose, and deglycosylated forms) were buffer-exchanged to low ionic strength buffers at pH 4.0, 5.5, 6.5, 7.0, and 7.5 using Zeba spin columns (Thermo Scientific) and diluted to 2 mg/mL. Thermograms of these glycoforms were collected from 10–95 °C at a scan rate of 60 °C⁄h.
Protease digestion
Human neutrophil elastase
Antibody glycoforms were digested by HNE (Athens Research and Technology) in the phosphate buffer at pH 7.5 at 37 °C. The ratio of mAb to HNE was 100:1 (w/w); the mAb concentration during digestion was 2 mg/mL in the phosphate buffer. Digested samples were collected at 2, 4, 6, 8, 16, and 24 h. The digestion was quenched by adding 1 mM (final concentration) 4-(2-aminoethyl) benzenesulfonyl fluoride hydrochloride (Calbiochem). The degradation of antibody glycoforms was monitored by SEC.
Papain
Antibody glycoforms were digested by papain (Sigma) in 20 mM tris HCl, pH 7.0 buffer at 37 °C; the mAb concentration during digestion was 2 mg/mL, and the ratio of mAb to papain was 25:1 (w/w). Digested samples were collected at 2, 4, 6, 8, 16, and 24 h, and reactions were quenched by adding antipain (MP Biomedicals) at a ratio of 40:1 papain:antipain (w/w). SEC was used to measure the digestion of antibody glycoforms.
Pepsin
Pepsin digestion was carried in 20 mM sodium acetate, pH 4.0 buffer at 37 °C; the mAb concentration during digestion was 2 mg/mL. The ratio of mAb to pepsin (Sigma) was 20:1 (w/w). Digested samples were collected at 5, 10, 15, and 30 min, and 1, 1.5, 2, 4, 6, 8, 16, and 24 h. The digestion was quenched by adding 2 M tris base with 1/20 of the total volume. SEC was used to monitor the degradation of antibody glycoforms.
Stability study
All antibody glycoforms were formulated to the formulation buffer at pH 5.5 for the stability study. Samples were filled aseptically into 3-mL type-I glass vials with 1 mL of the filling volume and stored at 5, 30, and 40 °C. Additional samples were stored at –70 °C as control samples. Samples were collected and analyzed by SEC at t = 0, 0.5, 1, 2, and 3-month time points. At each time point and storage condition, an individual vial of each glycoform was opened and analyzed.
| Abbreviations: | ||
| ADCC | = | antibody-dependent cell-mediated cytotoxicity |
| Asn | = | asparagine |
| CDC | = | complement dependent cytotoxicity |
| CHO | = | Chinese hamster ovary |
| DSC | = | differential scanning calorimetry |
| FTIR | = | Fourier transform infrared |
| GlcNAc | = | N-acetylglucosamine |
| His | = | histidine |
| HMWS | = | high molecular weight species |
| HNE | = | human neutrophil elastase |
| IR | = | infrared |
| LC-MS | = | liquid chromatography–mass spectrometry |
| LMWS | = | low molecular weight species |
| mAb | = | monoclonal antibody |
| MCT | = | mercury cadmium telluride |
| Pro | = | proline |
| SEC | = | size-exclusion chromatography |
| Ser | = | serine |
| Thr | = | threonine |
| Trp | = | tryptophan |
| Tyr | = | tyrosine |
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
The authors would like to thank Dr. Darren Brown for the assistance in mass spectroscopy analysis, Dr. Jun Liu, Dr. Thomas M. Scherer, Dr. Vikas K. Sharma, Dr. Yung-Hsiang Kao, and Dr. Jamie M. R. Moore for reviewing the manuscript, and Eileen Y. Ivasauskas for editorial assistance.
References
- Walsh CT, Garneau-Tsodikova S, Gatto GJ Jr.. Protein posttranslational modifications: the chemistry of proteome diversifications. Angew Chem Int Ed Engl 2005; 44:7342 - 72; http://dx.doi.org/10.1002/anie.200501023; PMID: 16267872
- Walsh G, Jefferis R. Post-translational modifications in the context of therapeutic proteins. Nat Biotechnol 2006; 24:1241 - 52; http://dx.doi.org/10.1038/nbt1252; PMID: 17033665
- Van Damme P, Hole K, Pimenta-Marques A, Helsens K, Vandekerckhove J, Martinho RG, Gevaert K, Arnesen T. NatF contributes to an evolutionary shift in protein N-terminal acetylation and is important for normal chromosome segregation. PLoS Genet 2011; 7:e1002169; http://dx.doi.org/10.1371/journal.pgen.1002169; PMID: 21750686
- Yokoyama A, Katsura S, Ito R, Hashiba W, Sekine H, Fujiki R, Kato S. Multiple post-translational modifications in hepatocyte nuclear factor 4α. Biochem Biophys Res Commun 2011; 410:749 - 53; http://dx.doi.org/10.1016/j.bbrc.2011.06.033; PMID: 21708125
- Prigge ST, Kolhekar AS, Eipper BA, Mains RE, Amzel LM. Amidation of bioactive peptides: the structure of peptidylglycine alpha-hydroxylating monooxygenase. Science 1997; 278:1300 - 5; http://dx.doi.org/10.1126/science.278.5341.1300; PMID: 9360928
- Prigge ST, Mains RE, Eipper BA, Amzel LM. New insights into copper monooxygenases and peptide amidation: structure, mechanism and function. Cell Mol Life Sci 2000; 57:1236 - 59; http://dx.doi.org/10.1007/PL00000763; PMID: 11028916
- Brown MA, Stenberg LM. Biopharmaceuticals: post-translational modification carboxylation and hydroxylation. In: Walsh G, ed. Post-translational modification of protein biopharmaceuticals. Weinheim: Wiley-VCH Verlag GmbH & Co., 2009:209-52.
- Arnold JN, Wormald MR, Sim RB, Rudd PM, Dwek RA. The impact of glycosylation on the biological function and structure of human immunoglobulins. Annu Rev Immunol 2007; 25:21 - 50; http://dx.doi.org/10.1146/annurev.immunol.25.022106.141702; PMID: 17029568
- Cohen P. Signal integration at the level of protein kinases, protein phosphatases and their substrates. Trends Biochem Sci 1992; 17:408 - 13; http://dx.doi.org/10.1016/0968-0004(92)90010-7; PMID: 1333658
- Mann M, Ong SE, Grønborg M, Steen H, Jensen ON, Pandey A. Analysis of protein phosphorylation using mass spectrometry: deciphering the phosphoproteome. Trends Biotechnol 2002; 20:261 - 8; http://dx.doi.org/10.1016/S0167-7799(02)01944-3; PMID: 12007495
- Spiro RG. Protein glycosylation: nature, distribution, enzymatic formation, and disease implications of glycopeptide bonds. Glycobiology 2002; 12:43R - 56R; http://dx.doi.org/10.1093/glycob/12.4.43R; PMID: 12042244
- Varki A. Biological roles of oligosaccharides: all of the theories are correct. Glycobiology 1993; 3:97 - 130; http://dx.doi.org/10.1093/glycob/3.2.97; PMID: 8490246
- Dubé S, Fisher JW, Powell JS. Glycosylation at specific sites of erythropoietin is essential for biosynthesis, secretion, and biological function. J Biol Chem 1988; 263:17516 - 21; PMID: 3182860
- Shields RL, Lai J, Keck R, O’Connell LY, Hong K, Meng YG, Weikert SH, Presta LG. Lack of fucose on human IgG1 N-linked oligosaccharide improves binding to human Fcgamma RIII and antibody-dependent cellular toxicity. J Biol Chem 2002; 277:26733 - 40; http://dx.doi.org/10.1074/jbc.M202069200; PMID: 11986321
- Kukuruzinska MA, Lennon K. Protein N-glycosylation: molecular genetics and functional significance. Crit Rev Oral Biol Med 1998; 9:415 - 48; http://dx.doi.org/10.1177/10454411980090040301; PMID: 9825220
- Krapp S, Mimura Y, Jefferis R, Huber R, Sondermann P. Structural analysis of human IgG-Fc glycoforms reveals a correlation between glycosylation and structural integrity. J Mol Biol 2003; 325:979 - 89; http://dx.doi.org/10.1016/S0022-2836(02)01250-0; PMID: 12527303
- Jefferis R. Glycosylation as a strategy to improve antibody-based therapeutics. Nat Rev Drug Discov 2009; 8:226 - 34; http://dx.doi.org/10.1038/nrd2804; PMID: 19247305
- Kanda Y, Yamada T, Mori K, Okazaki A, Inoue M, Kitajima-Miyama K, Kuni-Kamochi R, Nakano R, Yano K, Kakita S, et al. Comparison of biological activity among nonfucosylated therapeutic IgG1 antibodies with three different N-linked Fc oligosaccharides: the high-mannose, hybrid, and complex types. Glycobiology 2007; 17:104 - 18; http://dx.doi.org/10.1093/glycob/cwl057; PMID: 17012310
- Zhou Q, Shankara S, Roy A, Qiu H, Estes S, McVie-Wylie A, Culm-Merdek K, Park A, Pan C, Edmunds T. Development of a simple and rapid method for producing non-fucosylated oligomannose containing antibodies with increased effector function. Biotechnol Bioeng 2008; 99:652 - 65; http://dx.doi.org/10.1002/bit.21598; PMID: 17680659
- Crispin M, Bowden TA, Coles CH, Harlos K, Aricescu AR, Harvey DJ, Stuart DI, Jones EY. Carbohydrate and domain architecture of an immature antibody glycoform exhibiting enhanced effector functions. J Mol Biol 2009; 387:1061 - 6; http://dx.doi.org/10.1016/j.jmb.2009.02.033; PMID: 19236877
- Zheng K, Bantog C, Bayer R. The impact of glycosylation on monoclonal antibody conformation and stability. MAbs 2011; 3:568 - 76; http://dx.doi.org/10.4161/mabs.3.6.17922; PMID: 22123061
- Fabian H, Naumann D. Methods to study protein folding by stopped-flow FT-IR. Methods 2004; 34:28 - 40; http://dx.doi.org/10.1016/j.ymeth.2004.03.004; PMID: 15283913
- Surewicz WK, Mantsch HH. New insight into protein secondary structure from resolution-enhanced infrared spectra. Biochim Biophys Acta 1988; 952:115 - 30; http://dx.doi.org/10.1016/0167-4838(88)90107-0; PMID: 3276352
- Mimura Y, Church S, Ghirlando R, Ashton PR, Dong S, Goodall M, Lund J, Jefferis R. The influence of glycosylation on the thermal stability and effector function expression of human IgG1-Fc: properties of a series of truncated glycoforms. Mol Immunol 2000; 37:697 - 706; http://dx.doi.org/10.1016/S0161-5890(00)00105-X; PMID: 11275255
- Ionescu RM, Vlasak J, Price C, Kirchmeier M. Contribution of variable domains to the stability of humanized IgG1 monoclonal antibodies. J Pharm Sci 2008; 97:1414 - 26; http://dx.doi.org/10.1002/jps.21104; PMID: 17721938
- Ryan MH, Petrone D, Nemeth JF, Barnathan E, Björck L, Jordan RE. Proteolysis of purified IgGs by human and bacterial enzymes in vitro and the detection of specific proteolytic fragments of endogenous IgG in rheumatoid synovial fluid. Mol Immunol 2008; 45:1837 - 46; http://dx.doi.org/10.1016/j.molimm.2007.10.043; PMID: 18157932
- Raju TS, Scallon B. Fc glycans terminated with N-acetylglucosamine residues increase antibody resistance to papain. Biotechnol Prog 2007; 23:964 - 71; http://dx.doi.org/10.1002/bp070118k; PMID: 17571902
- Gaza-Bulseco G, Liu H. Fragmentation of a recombinant monoclonal antibody at various pH. Pharm Res 2008; 25:1881 - 90; http://dx.doi.org/10.1007/s11095-008-9606-3; PMID: 18473123
- Feige MJ, Walter S, Buchner J. Folding mechanism of the CH2 antibody domain. J Mol Biol 2004; 344:107 - 18; http://dx.doi.org/10.1016/j.jmb.2004.09.033; PMID: 15504405
- Raju TS, Scallon BJ. Glycosylation in the Fc domain of IgG increases resistance to proteolytic cleavage by papain. Biochem Biophys Res Commun 2006; 341:797 - 803; http://dx.doi.org/10.1016/j.bbrc.2006.01.030; PMID: 16442075
- Rudd PM, Joao HC, Coghill E, Fiten P, Saunders MR, Opdenakker G, Dwek RA. Glycoforms modify the dynamic stability and functional activity of an enzyme. Biochemistry 1994; 33:17 - 22; http://dx.doi.org/10.1021/bi00167a003; PMID: 8286336
- Ren D, Zhang J, Pritchett R, Liu H, Kyauk J, Luo J, Amanullah A. Detection and identification of a serine to arginine sequence variant in a therapeutic monoclonal antibody. J Chromatogr B Analyt Technol Biomed Life Sci 2011; 879:2877 - 84; http://dx.doi.org/10.1016/j.jchromb.2011.08.015; PMID: 21900054