Abstract
Modification of antibody class and binding properties typically requires cloning of antibody genes, antibody library construction, phage or yeast display and recombinant antibody expression. Here, we describe an alternative “cloning-free” approach to generate antibodies with altered antigen-binding and heavy chain isotype by mimicking the germinal center reaction in antibody-secreting hybridoma cells. This was accomplished by lentiviral transduction and controllable expression of activation-induced cytidine deaminase (AID) to generate somatic hypermutation and class switch recombination in antibody genes coupled with high-throughput fluorescence-activated cell sorting (FACS) of hybridoma cells to detect altered antibody binding properties. Starting from a single established hybridoma clone, we isolated mutated antibodies that bind to a low-temperature structure of polyethylene glycol (PEG), a polymer widely used in nanotechnology, biotechnology and pharmaceuticals. FACS of AID-infected hybridoma cells also facilitated rapid identification of class switched variants of monoclonal IgM to monoclonal IgG. Mimicking the germinal center reaction in hybridoma cells may offer a general method to identify and isolate antibodies with altered binding properties and class-switched heavy chains without the need to carry out DNA library construction, antibody engineering and recombinant protein expression.
Introduction
Monoclonal antibodies (mAbs) play important roles in medicine and biotechnology due to their high specificity and affinity. Elegant mutagenesis and engineering strategies have been developed to alter the antigen-binding properties of antibodies, including complementarity determining region (CDR) mutagenesis,Citation1,Citation2 error-prone PCRCitation3 and DNA shuffling.Citation4 Powerful yeast and mammalian antibody display systems have been developed in parallel to improve antibody properties.Citation5,Citation6 These approaches, however, can be technically challenging, time-consuming and expensive, largely restricting these technologies to laboratories and companies with specialized facilities and expertise.
In the mammalian immune system, progenitor B cells (pro-B cells) and precursor B cells (pre-B cells) in the bone marrow undergo V-D-J gene segment rearrangement at the heavy chain locus and then VL gene rearrangement at the light chain loci to generate the primary B cell receptor repertoire.Citation7 Contact of B cells with cognate antigen in germinal centers along with both membrane-bound and secreted signals provided by T follicular helper cells leads to somatic hypermutation (SHM) by induction of activation-induced cytidine deaminase (AID) dependent deamination of cytidine, followed by error-prone DNA repair that introduces point mutations in the V regions of immunoglobulin genes in centroblast B cells.Citation8,Citation9 Rapidly dividing centroblasts in the dark zone of the germinal center migrate to the light zone to become centrocytes, express surface immunoglobulin and compete for limiting quantities of antigen on follicular dendritic cells (). Centrocytes that display antibodies with higher affinity preferentially interact with antigen and survive to migrate back into the dark zone for further proliferation and SHM. Multiple rounds of SHM and selection coupled with AID-mediated heavy-chain class switch recombination (CSR) results in B cells that secrete high affinity IgG, IgA and IgE antibodies.Citation10
Figure 1. Mimicking the germinal center reaction in vitro. (A) Centroblast B cells in germinal centers rapidly proliferate and express activation-induced cytidine deaminase (AID) to initiate immunoglobulin somatic hypermutation (SHM) and class switch recombination (CSR). B cells that successfully compete for limiting antigen in the light zone can undergo multiple rounds of SHM and selection, leading to generation of plasma cells that secrete high affinity IgG antibodies. (B) The germinal center reaction can be mimicked by lentiviral transduction of the AID gene into hybridoma cells followed by multiple rounds of selection with fluorescence-labeled antigen and cell expansion to allow SHM and CSR of immunoglobulin genes, resulting in a hybridoma clone that secretes antibody with the desired characteristics.
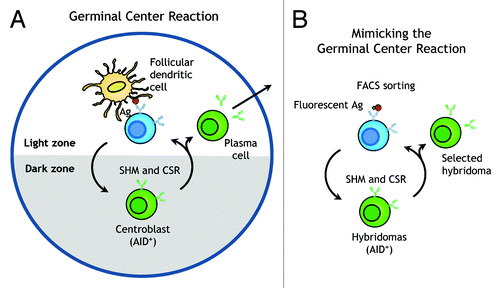
Previous studies have elegantly demonstrated that ectopic expression of AID in hybridoma cells can induce SHM and CSR of antibodies.Citation11,Citation12 Here, we sought to extend these observations to mimic the germinal center reaction directly in antibody-secreting hybridoma cells (). We therefore expressed the mouse AID gene in hybridoma cells to induce SHM and CSR. After periods of cell expansion and SHM, we mimicked centrocyte competition for antigen by incubating the hybridoma population with fluorescence-labeled antigen and selected desirable variants by high-throughput fluorescence-activated cell sorting (FACS) of hybridoma cells ().
We investigated this strategy to isolate antibodies that bind to a conformation of PEG that is preferentially formed at 4°C. PEG is a polymer with repeating subunits of ethylene glycol [HO-(CH2-CH2-O)n-H]. Covalent attachment of PEG (PEGylation) is widely used in the pharmaceutical and biotechnology industries to improve the pharmacokinetic properties of small molecules, nucleotides, peptides, proteins and nanoparticles by increasing drug solubility, reducing renal clearance, enhancing serum half-life, diminishing engulfment by phagocytes and decreasing proteolytic degradation.Citation13 Previous studies suggest that PEG conformation may be temperature-dependent, preferentially existing in a gauche structure at lower temperatures, but possessing a trans conformation at higher temperatures.Citation14,Citation15 However, direct evidence for distinct PEG conformations at different temperatures remains elusive.
Here, we show that mAbs that preferentially bind to PEG at 4°C could be isolated in an anti-PEG hybridoma. We also demonstrate rapid CSR of IgM to IgG antibodies in hybridoma cells. This methodology may represent a general strategy to enhance antibody properties with “on demand” SHM and CSR without the need for antibody engineering and recombinant protein expression.
Results
Surface antibodies can be used to identify antigen-specific hybridoma cells
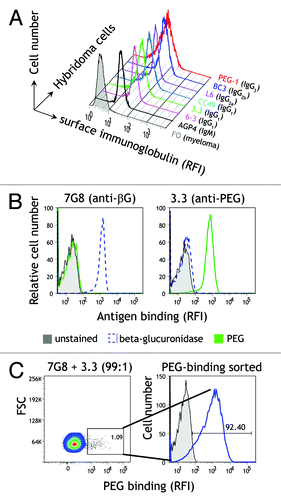
Naive B cells express on their surface multiple copies of antibodies with the identical amino acid sequence and antigen specificity. Upon antigen stimulation and differentiation into plasma cells, membrane antibodies are converted to secreted antibodies by RNA splicing and processing of the primary RNA transcript to remove the hydrophobic transmembrane anchor. However, RNA splicing may be incomplete in hybridoma cells, which would greatly facilitate selection of antibodies with altered binding properties. We therefore used immunofluorescence staining to detect immunoglobulin on the plasma membrane of live hybridoma cells. All tested antibody-secreting hybridoma cells, including those that secrete IgG1, IgG2a, IgG2b, IgG3, and IgM antibodies, displayed moderate to high levels of membrane immunoglobulin on their surface compared with FO myeloma cells, which do not express immunoglobulin (). The functional activity of membrane immunoglobulin on hybridoma cells was examined by labeling 3.3 (anti-PEG) and 7G8 (anti-β-glucuronidase) hybridoma cells with biotinylated polyethylene glycol (PEG) or β-glucuronidase, followed by Alexa Fluor 647-conjugated streptavidin. 3.3 hybridoma cells specifically bound PEG but not β-glucuronidase, whereas 7G8 hybridoma cells bound β-glucuronidase but not PEG (), showing that membrane immunoglobulin displayed the expected antigen-binding specificities. To investigate whether high-throughput FACS of hybridoma cells based on antigen-binding by surface immunoglobulin is feasible, a mixture of 7G8 (anti-β-glucuronidase) and 3.3 (anti-PEG) hybridoma cells (99:1 ratio) was stained with biotinylated PEG and then incubated with Alexa Fluor 647-conjugated streptavidin. Two distinct populations of cells could be clearly differentiated, corresponding to the ratio of the input 7G8 and 3.3 cells (, left panel). The hybridoma cells that bound PEG were collected and cultured for 5 d. Analysis of the expanded hybridoma cells showed that they could bind PEG (, right panel), indicating efficient selection of 3.3 anti-PEG hybridoma cells. We conclude that antibody-secreting hybridoma cells display sufficient functional immunoglobulin on their surface to allow identification and isolation of antigen-specific hybridoma cells.
Figure 2. Analysis of membrane immunoglobulin on hybridoma cells. (A) Membrane-bound immunoglobulin on hybridoma and FO myeloma cells was detected by flow cytometry after staining the cells with goat anti-mouse Ig and rabbit anti-goat FITC. (B) Functional antigen-binding of membrane immunoglobulin on 3.3 anti-PEG and anti-β-glucuronidase 7G8 hybridoma cells was detected on a flow cytometer with biotinylated PEG or β-glucuronidase antigens followed by Alexa Fluor 647-conjugated streptavidin. (C) 7G8 anti-β-glucuronidase and 3.3 anti-PEG hybridoma cells mixed at a ratio of 99:1 were stained with 0.5 nM biotin-PEG and Alexa Fluor 647-conjugated streptavidin. Hybridoma cells that bound biotin-PEG (left panel) were collected with a fluorescence-activated cell sorter, cultured for 5 d and then stained with biotin-PEG and Alexa Fluor 647-streptavidin (right panel).
Controllable induction of SHM
We constructed constitutive and inducible vectors to allow controllable expression of activation-induced cytidine deaminase (AID) in hybridoma cells. These vectors were designed to facilitate termination of AID expression once antibody variants with desirable features are identified. A loxP-flanked constitutive expression cassette (pCMV-AID-loxP) () was used to stably transduce 3.3 anti-PEG hybridoma cells by lentiviral infection, making 3.3/loxP-AID cells. AID was expressed in 3.3/loxP-AID cells as indicated by the fluorescence of the eGFP reporter protein (, left panel). To test whether the expression of AID could be stopped “on demand,” the pLM-mCherry-P2A-Cre plasmid was transferred into 3.3/loxP-AID cells by DNA electroporation. About 23% of the cells expressed Cre recombinase as visualized by mCherry expression, corresponding to the cell population that displayed reduced eGFP expression (, middle panel), indicating deletion of the AID gene cassette. The eGFP-negative cells could be easily isolated by FACS (, right panel). mCherry was transiently expressed and was therefore no longer detectable in these cells. To confirm CRE-dependent deletion of the AID gene, total cell lysates prepared from 3.3 cells, 3.3/loxP-AID cells or sorted Cre-infected 3.3/loxP-AID cells were separated by SDS-PAGE and AID was detected via a C-terminal hemagglutinin (HA) epitope tag sequence on western blots using an anti-HA antibody. AID protein was detected in 3.3/loxP-AID cells but was absent in Cre-treated 3.3/loxP-AID cells (), demonstrating that AID could be stably expressed and conditionally silenced by Cre recombinase mediated gene deletion in hybridoma cells.
Figure 3. Controllable expression of AID. (A) Schematic representation of the pCMV-AID-loxP vector. A CMV immediate early promoter is followed by the mouse activation-induced cytidine deaminase (AID) gene, a HA epitope-tag, a furin/2A peptide (F2A) bicistronic expression linker and an eGFP reporter gene. The AID cassette is flanked by two loxP motifs for Cre recombinase-mediated gene excision. (B) eGFP (y-axis) and mCherry fluorescence (x-axis) in 3.3 hybridoma cells that stably express pCMV-AID-loxP (3.3/loxP-AID cells, left panel) and after transient transfection with a pLM-mCherry-P2A-Cre plasmid by DNA electroporation (middle panel) and subsequent fluorescence-activated cell sorting of eGFP-negative cells (right panel). (C) Cell lysates prepared from 3.3, 3.3/loxP-AID or Cre-treated 3.3/loxP-AID cells (sorted) were immunoblotted for the HA epitope tag on AID or tubulin as a cell loading control. (D) The autoregulatory pTetOn-AID plasmid is composed of the tetracycline response elements (TRE) promoter, an AID gene, an HA epitope, the furin/2A peptide (F2A) bicistronic expression linker, an eGFP reporter gene, an IRES element and the rtTA-V14 transactivator. (E) Flow cytometry histograms of the eGFP reporter in 3T3/TetOn-AID fibroblasts induced with the indicated doxycycline concentrations for 48 h. (F) DsRed2s contains a premature stop codon at nucleotide position 519 within a hotspot motif of AID. (G) 3T3 (■) or 3T3/TetOn-AID fibroblasts transduced with pDsRed2s were cultured in the presence (▲) or absence (○) of doxycycline and analyzed for expression of DsRed2 by FACS. The fraction of cells expressing red fluorescence (indicating mutation of the stop codon) vs. time is shown.
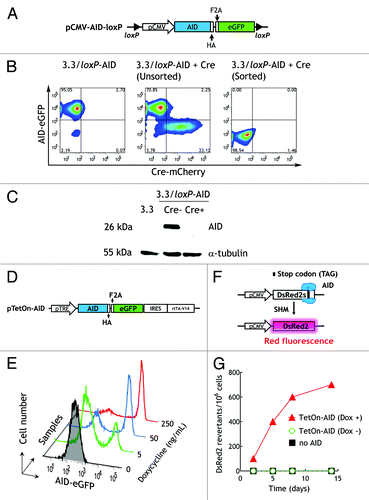
A tetracycline-inducible AID expression cassette (pTetOn-AID) was also constructed as an alternative for controllable induction of AID expression. Doxycycline can bind to rtTA-V14 to form an active transactivator to initiate DNA transcription driven by the pTRE promoter in a feed forward loop ().Citation16,Citation17 Stably infected 3T3/TetOn-AID cells incubated with graded concentrations of doxycycline for two days displayed dose-dependent induction of the eGFP reporter, corresponding to inducible expression of AID (). To examine if AID was functionally competent to induce SHM, 3T3 or 3T3/TetOn-AID cells were stably transfected with a DsRed2 gene, in which tyrosine 173 was mutated to an amber stop codon in the context of a RGYW/WRCY AID consensus hot spot motif ().Citation18 DsRed2 fluorescence was only detected in the 3T3/TetOn-AID transfectants after initiation of SHM by addition of doxycycline (). We conclude that these AID expression cassettes allow controllable expression of AID for induction of SHM.
Mimicking the germinal center reaction in vitro
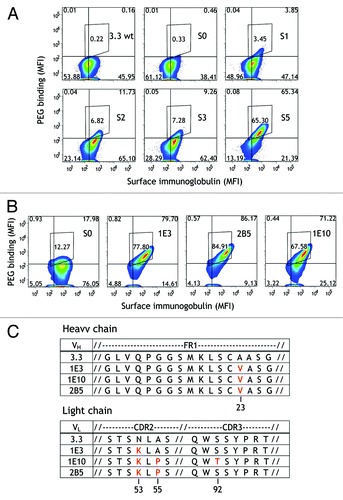
We expressed AID in 3.3 anti-PEG hybridoma cells to isolate anti-PEG antibodies that bind to PEG at low temperatures with high affinity. This was accomplished by expanding 3.3/loxP-AID cells to allow SHM, staining the cells with fluorescence-labeled antigen (PEG) on ice, selecting hybridoma cells that bound the most PEG by FACS and expanding the selected cells for two weeks to allow continued SHM. Initial selection was performed with long-chain multivalent PEG molecules but the PEG length and valency was progressively decreased in subsequent rounds of cell sorting. After five sequential rounds of cell expansion and sorting, hybridoma cell samples collected after each round of sorting were stained with Alexa Fluor 647-PEG5K and analyzed on a flow cytometer (). By the fifth round of sorting, the mean fluorescence intensity (MFI) of Alexa Fluor 647-PEG5K binding to 3.3/loxP-AID hybridoma cells was 233 as compared with a value of 23.2 for the parental 3.3 hybridoma cells, suggesting that these hybridoma cells expressed antibody variants that possessed enhanced PEG affinity. Similar high PEG binding was observed for three hybridoma clones (1E3, 1E10, and 2B5) isolated by single-cell sorting of the 3.3/loxP-AID hybridoma cells after the fifth round of selection (). Sequence analysis of the immunoglobulin variable region genes from the 3.3 subclones revealed that all three antibody variants exhibited a common A23V mutation in framework 1 (FR1) of the VH chain and a common N53K mutation in CDR2 of the VL chain (). In addition, both 1E10 and 2B5 antibodies possessed an A55P mutation in CDR2 of the VL chain while 1E10 exhibited an additional mutation, S92T, in CDR3 of the VL chain.
Figure 4. Isolation of temperature-dependent anti-PEG antibodies by mimicking the germinal center reaction. (A) 3.3/loxP-AID hybridoma cells were enriched for cells binding Alexa Fluor 647-PEG probes (y axis). Surface immunoglobulin was also detected using Alexa Fluor 405-conjugated goat anti-mouse IgG Fc antibody (x axis). The cells were cultured for 2 wk before approximately 3 × 107 cells were sorted each round (S0-S5). Results show binding of fluorescent-labeled PEG vs. surface Ig levels. (B) Binding of PEG-Alexa Fluor 647 to three hybridoma clones (1E3, 1E10 and 2B5) that were isolated from the hybridoma population after round 5 of selection (S5). (C) Amino acid sequences and alignment of immunoglobulin VH (upper panel) and VL (lower panel) gene sequences from 1E3, 1E10 and 2B5 antibodies in comparison to the parental 3.3 antibody. Antibody framework regions and CDRs were assigned according to the Kabat numbering systemCitation61 using the website http://www.bioinf.org.uk/abysis.
Anti-PEG antibody variants selectively bind PEG at low temperatures
We evaluated the temperature-dependent binding of two of the 3.3 antibody variants (1E3 and 2B5) to PEG (10 000 Da) coated in microtiter plates. The 2B5 variant bound PEG with greater apparent affinity than the parental 3.3 antibody whereas 1E3 bound about as well as 3.3 at 4°C, but both variant antibodies displayed progressively reduced binding to PEG at 25°C or 37°C (). Analysis of antibody binding to PEG by surface plasmon resonance showed that 2B5 and 1E3 displayed similar affinity as the parental 3.3 antibody at 4°C, but that the affinity of 1E3 and 2B5 for PEG at 25°C was decreased by 85 and 185-fold, respectively, compared with their affinities at 4°C (). The affinities of 1E3 and 2B5 could not be determined at 37°C due to poor binding to PEG. Similar temperature-selective recognition by 1E3 and 2B5 was observed for other lengths of PEG coated in microtiter plates (Fig. S1). We also observed that 1E3 and 2B5 displayed enhanced binding of long-chain PEG, but reduced binding to short PEG molecules (i.e., 750 Da PEG) compared with the parental 3.3 antibody. 1E3 and 2B5 antibodies were not inherently unstable at elevated temperatures because even after incubation at 37°C for 5 d, they retained full binding activity (). Furthermore, the melting temperatures of 1E3 and 2B5 antibodies (73.6°C and 74.2°C, respectively), as determined by differential scanning calorimetry, were actually higher than the parental 3.3 antibody (70.8°C), indicating that 1E8 and 2B5 antibodies were thermostable (). These results indicate that the AID induced mutations in the immunoglobulin V region genes of 1E3 and 2B5 antibodies conferred the ability to preferentially bind to PEG at 4°C.
Figure 5. Temperature-dependent binding and stability of anti-PEG antibody variants. (A) Graded concentrations of purified 3.3, 1E3, or 2B5 antibodies were added to microplate wells coated with linear amino-PEG (MW 10,000 Da) at the indicated temperatures. After 1 h, the wells were washed and antibody binding was determined by adding HRP-conjugated donkey anti-mouse IgG Fc antibodies, followed by ABTS substrate. The mean absorbance values (405 nm) of triplicate determinations are shown. Bars, SD (B) 3.3 (■), 1E3 (○) and 2B5 (▲) antibodies were incubated at 37°C for the indicated times before their binding to CH3-PEG5k–NH2 in 96-well microtiter plates was determined by ELISA at 4°C. The activity of the antibodies at different times relative to their activities at time zero, determined by measuring 50% maximal responses in ELISA, are shown. (n = 3). Bars, SD (C) Thermal unfolding of 3.3 (black line), 1E3 (short dashed green line) and 2B5 (long dashed red line) as measured by differential scanning calorimetry in PBS at a heating rate of 1°C/min.
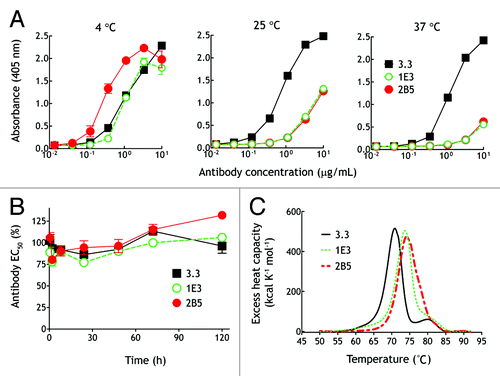
Table 1. Binding kinetics of 3.3, 1E3 and 2B5 antibodies to PEG
Roles of the CDR mutations in temperature-dependent PEG binding
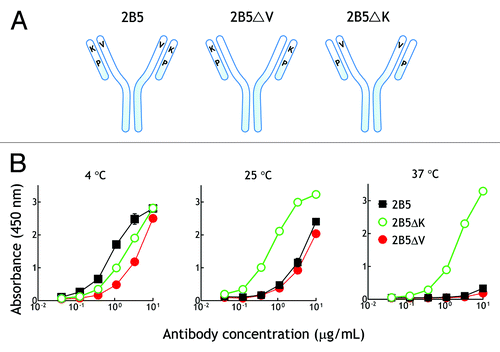
To determine which mutations are responsible for the temperature-dependent binding of the antibody variant 2B5, we individually reverted the conserved amino acid mutations in 2B5 (VH V23A and VL K53N) back to the corresponding amino acids in the parental 3.3 antibody (). Reversion of VH V23 to alanine did not eliminate the temperature dependent binding of 2B5ΔV to PEG (). By contrast, replacement of VL K53 with asparagine abolished the temperature-selective binding of 2B5ΔK to PEG as observed by similar binding of 2B5ΔK to PEG at 4°C, 25°C and 37°C (). We conclude that the VL K53 mutation is responsible for the temperature-dependent binding of 2B5 to PEG.
Figure 6. VL chain K53 is responsible for the temperature-dependent binding of 2B5 to PEG.(A) Recombinant 2B5 or 2B5 in which VH V23 (2B5ΔV) or VL K53 (2B5ΔK) were replaced by the corresponding amino acid of the parental 3.3 antibody were purified from mammalian cell culture medium. (B) Graded concentrations of recombinant 2B5 (■), 2B5ΔK (○), or 2B5ΔV (●) antibodies were added to microplate wells coated with linear amino-PEG at the indicated temperatures. After 1 h, the wells were washed and antibody binding was determined by adding HRP-conjugated donkey anti-mouse IgG Fc antibodies, followed by ABTS substrate. The mean absorbance values (405 nm) of triplicate determinations are shown. Bars, SD.
2B5 binds to a PEG structure that can be mimicked by crown ether
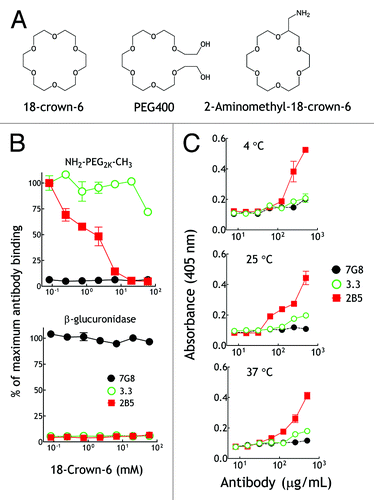
Several studies have reported that PEG can coordinate lysine residues to form crown ether-like structures on the surface of proteins crystals.Citation19-Citation21 We therefore hypothesized that 2B5 may recognize a PEG conformation that can be mimicked by crown ether (). Indeed, 18-crown-6 completely blocked binding of a fixed amount of 2B5 to immobilized methoxy-PEG2k-NH2 at 4°C in a dose-dependent fashion (, upper panel). By contrast, 18-crown-6 was a relatively poor competitor for binding of the parental 3.3 antibody to PEG (, upper panel). 18-crown-6 did not affect the binding of an isotype-matched anti-β-glucuronidase control antibody (7G8) to immobilized β-glucuronidase (, lower panel), demonstrating specificity of the competition reaction. We further reasoned that if 18-crown-6 mimics a PEG structure that forms at 4°C, then 2B5 should bind to the structurally-defined 18-crown-6 at all temperatures. We therefore examined by surface plasmon resonance the binding of 2B5 to 2-aminomethyl-18-crown-6 immobilized on a CM5 chip at three different temperatures (4°C, 25°C and 37°C). 2B5 bound to immobilized 18-crown-6 at all temperatures tested, whereas 3.3 and control 7G8 antibodies showed relative low binding or non-binding to immobilized 18-crown-6, respectively (Fig. S2). Likewise, we detected 2B5 binding to 2-aminomethyl-18-crown-6 coated in 96-well ELISA plates at 4°C, 25°C and 37°C (). We conclude that 2B5 binds to a PEG structure that preferentially forms at 4°C and that can be mimicked by 18-crown-6.
Figure 7. 2B5 can bind to crown ether structure. (A) Illustration of the chemical structures of 18-crown-6, PEG400 and 2-aminomethyl-18-crown-6. (B) Graded concentrations of 18-crown-6 were mixed at 4°C with 10 μg/mL 3.3 (○), 2B5 (■) or 7G8 (●) antibodies prior to addition to 96-well plates coated with amino-PEG2k-NH2 (upper panel) or β-glucuronidase (lower panel). Results show antibody binding as a percentage of maximum binding activity. (n = 3). Bars, SD (C) Graded concentrations of purified 3.3 (○), 2B5 (■) or 7G8 (●) antibodies were incubated at 4°C, 25°C or 37°C in microplate wells coated with 2-aminomethyl-18-crown-6. After 4 h, the wells were washed and antibody binding was determined by adding biotin-conjugated goat anti-mouse IgG Fc antibodies and streptavidin-biotinylated peroxidase complex followed by ABTS substrate. The mean absorbance values (405 nm) of triplicate determinations are shown. Bars, SD.
Thermal affinity purification of PEGylated nanoparticles by 2B5 antibody
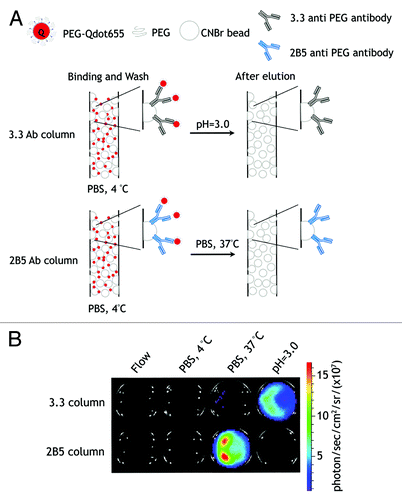
We further explored whether 2B5 anti-PEG antibody could be employed for mild affinity purification of PEGylated compounds. PEG-Qdots in cold PBS (4°C) were loaded onto a column packed with agarose beads with 3.3 or 2B5 antibodies covalently linked to their surface (). After washing the columns with cold PBS, PEG-Qdots were eluted with citrate buffer (pH = 3.0) or with 37°C PBS. Both 3.3 and 2B5 antibodies could capture PEG-Qdots at low temperature. Elution with 37°C PBS released PEG-Qdots from the 2B5 column, whereas elution of the PEG-Qdots from the 3.3 column required acid elution (). These results indicate that 2B5 may be useful for the mild purification of PEGylated compounds by simply thermal cycling between 4°C and 37°C.
Figure 8. Mild affinity purification of PEGylated compounds. (A) Schematic outline of temperature-dependent affinity purification of PEG-Qdot655. (B) Oxidized 3.3 or 2B5 antibodies were immobilized on UltraLink® Hydrazide Resin and packed into columns. PEG-Qdot655 were passed into the columns and then the columns were washed with cold PBS (4°C) and eluted with 37°C PBS or citrate buffer (pH = 3.0). The fluorescence of PEG-Qdot655 was detected on an IVIS 200 optical imaging system (Xenogen).
In situ class switch recombination of antibodies
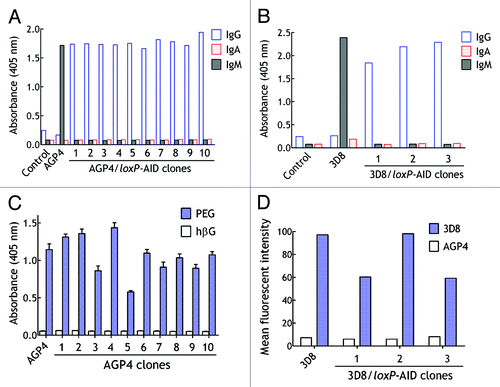
CSR of antibody heavy chain genes from IgM to other antibody classes in the germinal center also depends on AID activity.Citation22 We therefore examined if expression of AID could facilitate isolation of class-switched antibodies. The pCMV-AID-loxP vector was used to stably transduce AGP4 and 3D8 hybridoma cells by lentiviral infection. AGP4 hybridoma cells secrete a monoclonal IgM that binds to PEG whereas 3D8 hybridoma cells secrete a monoclonal IgM that binds to an antigen expressed on the surface of mouse B16F10 melanoma cells.Citation23,Citation24 The hybridoma cells were cultured for four weeks and then live hybridoma cells were stained with fluorescence-labeled PEG (AGP4 hybridoma cells) and PE-conjugated goat anti-mouse IgG (AGP4 and 3D8 hybridoma cells) and individual positive cells were sorted by FACS into individual wells of a 96-well culture plate. The heavy chain class of antibodies in the culture medium of AGP4/loxP-AID and 3D8/loxP-AID clones was examined by ELISA. As expected, only IgM antibodies were detected in the culture medium from the parental AGP4 and 3D8 hybridoma cells (). By contrast, all selected AGP4/loxP-AID and 3D8/loxP-AID hybridoma clones secreted IgG antibodies, confirming class switch from IgM to IgG. Further analysis of the isotype of AGP4 and 3D8 class-switched antibodies revealed that they were all IgG3. The class-switched antibodies retained antigen-binding activity as determined by ELISA (). However, sequencing of the heavy chain variable region genes of the class-switched 3D8 antibodies revealed amino acid changes in AID consensus hot spots (Table S1), indicating that SHM also occurred during CSR. We conclude that mimicking the germinal center reaction can facilitate rapid conversion of IgM to IgG antibodies with concurrent SHM.
Figure 9. Rapid heavy chain class switch in hybridoma cells. (A) AGP4/loxP-AID cells were cultured for 4 wk and then cells that stained positive for surface IgG were cloned by fluorescence-activated cell sorting of individual cells into wells of a 96-well plate. The heavy chain class of antibody in the culture medium from parental AGP4 cells and ten AGP4/loxP-AID clones are shown. (B) 3D8/loxP-AID cells that displayed surface IgG were cloned by fluorescence-activated cell sorting of individual cells into wells of a 96-well plate. The heavy chain class of antibody in the culture medium from parental 3D8 cells and three 3D8/loxP-AID clones are shown. (C) The mean binding of AGP4 IgM or IgG antibodies from the culture medium of ten selected AGP4/loxP-AID clones was determined by ELISA in plates coated with PEG or control β-glucuronidase antigen (n = 2). Bars, SD (D) The binding of antibodies in the culture medium of 3D8, three 3D8/loxP-AID clones or control AGP4 hybridoma cells was determined by FACS using B16F10 cells as antigen source.
Discussion
Here, we show that the germinal center reaction can be mimicked in hybridoma cells to facilitate isolation of mAbs with altered binding properties and class-switched heavy chains. This simple procedure entails lentiviral infection of an AID gene into hybridoma cells to induce SHM and CSR followed by rounds of expansion and selection of hybridoma cells that display surface immunoglobulin with the desired binding properties and antibody isotypes. Because each hybridoma cell expresses multiple copies of a single antibody variant on its surface, selection can be rapidly achieved by FACS of a large population of antigen-stained hybridoma cells. We demonstrated the utility of this approach by generating for the first time a mAb that preferentially binds to a conformation of PEG that forms at reduced temperature (4°C) and could be mimicked by crown ether. The variant anti-PEG antibodies were more thermostable than the parental antibody and could be easily produced by conventional hybridoma cell culture. We further demonstrated simple and rapid class switch of monoclonal IgM antibodies to IgG directly in hybridoma cells with the caveat that simultaneous SHM may alter antibody binding properties. Taken together, our study indicates that this approach may help generate improved or class-switched antibodies without the need for antibody library construction, antibody engineering or recombinant antibody production.
AID is both necessary and sufficient to induce SHM in B cells via a process that involves deamination of cytidine followed by error-prone DNA repair to introduce point mutations in the V regions of immunoglobulin genes.Citation9 The power of ectopic AID expression has been recognized in elegant studies demonstrating CSR in hybridoma cells and affinity maturation of antibody libraries in HEK293 cells engineered to express AID.Citation6,Citation12,Citation25,Citation26 Nevertheless, the HEK293 platform requires substantial technical expertise because it entails construction of antibody gene libraries, transfection of antibody libraries into HEK293 cells, selection of the desired antibody variants and mammalian cell production of the final recombinant antibodies. By contrast, we found that antibody variants could be selected by expressing AID in hybridoma cells and directly interrogating immunoglobulins present on the surface of hybridoma cells, presumably due to incomplete splicing of the immunoglobulin heavy chain gene RNA transcript from the transmembrane to the secreted form.Citation27-Citation30 We stained hybridoma cells with fluorescence-labeled antigen to rapidly collect desirable antibody variants by FACS, but other high throughput methods such as antigen-coated magnetic beads may also allow selection of desirable hybridoma cells. A major advantage of this approach is the elimination of tedious and time-consuming procedures, including construction of antibody cDNA libraries, recloning of selected antibody genes and recombinant antibody production steps, which substantially simplifies the technical know-how needed for antibody evolution.
We previously generated several mAbs that bind the repeating ethylene oxide subunits present in PEG.Citation24,Citation31,Citation32 These antibodies have found widespread use in the biotechnology and pharmaceutical industries for the laboratory and clinical analysis of PEGylated therapeutic agents.Citation24,Citation32-Citation34 Previous reports have suggested that PEG tertiary conformation in aqueous solutions may depend on temperature.Citation14,Citation15 We reasoned that antibody variants that preferentially bind the “low temperature” conformation of PEG could be selected by mimicking the germinal center reaction in 3.3 anti-PEG hybridoma cells and selecting variants with fluorescence-labeled PEG at low temperature. Indeed, we isolated antibodies that bound more strongly to PEG at 4°C compared with 37°C. Poor binding at 37°C was not due to antibody instability at elevated temperatures because the new antibodies were at least as stable as the parental 3.3 antibody as determined by differential scanning calorimetry. Rather, the new antibodies were found to bind to a PEG conformation that can be competed by a cyclic ethylene oxide structure (18-crown-6). Mutagenesis studies revealed that a lysine substitution in the light chain variable region of 3.3 conferred the ability to bind to the crown ether-like PEG structure. Our results are consistent with a model in which the parental 3.3 antibody can bind to a form of PEG that is present at all temperatures, whereas 1E3 and 2B5 can selectively bind to a cyclic ethylene oxide structure that is present in PEG molecules at low temperature only. The new lysine residue in 1E3 and 2B5 variant antibodies appears to interfere with the normal binding mode of 3.3 to PEG, resulting in poor binding of the variant antibodies to PEG at 37°C. We speculate that the terminal ammonium group of the new lysine residue in the binding pocket of 1E3 and 2B5 antibodies may coordinate oxygen atoms in PEG chains that form cyclic ethylene oxide structures at lower temperatures (4°C),Citation19-Citation21 thereby compensating for decreased binding via the “standard 3.3” binding mode. The cyclic ethylene oxide structures in PEG molecules appear to form only at low temperatures, otherwise they would be detected by 1E3 and 2B5 at elevated temperatures since these antibodies can bind to a stabilized cyclic ethylene oxide structure (18-crown-6) even at 37°C as demonstrated by ELISA (). The cyclic ethylene oxide structure may represent only a fraction of total PEG structures at 4°C. Thus, 3.3 may bind to a form of PEG found at all temperatures, whereas 1E3 and 2B5 selectively bind to a crown ether-like PEG structure that is preferentially formed at low temperature.
Antibodies able to discriminate PEG conformation in a temperature-dependent fashion may be useful for novel applications such as temperature tunable capture of nanoparticles, creation of temperature controllable gated nanodevices and for gentle bio-separation of PEGylated compounds based on temperature-dependent elution as we demonstrated for a PEGylated nanoparticle.Citation35-Citation37 Our approach may also be useful for developing selective antibodies against new stimulus-responsive peptides and polymers currently under development.Citation38,Citation39
Although the affinity of 2B5 appeared to be increased compared with the parental 3.3 antibody (), the major effect was attainment of the new property of temperature-selective binding to PEG. In the present study, we used progressively less branched and shorter PEG molecules to select hybridoma cells. By contrast, centrocytes in germinal centers are selected on limiting amounts of antigen, which may drive selection of higher affinity antibodies.Citation40 Conditions for rapid affinity maturation are currently under investigation, but preliminary results using progressively decreasing doses of antigen for selection of hybridoma cells during FACS indicates that antibody affinity can indeed be increased by this procedure.
Besides SHM, germinal center B cells also undergo CSR, in which the Cμ heavy chain constant region gene is replaced with downstream Cγ, Cα or Cε genes (i.e., switch from IgM to IgG, IgA or IgE).Citation41 The combination of AID expression and FACS facilitated rapid switching and identification of AGP4 and 3D8 hybridoma cells that switched from secreting IgM antibodies to IgG antibodies. In our study, we sorted IgG-positive hybridoma cells so it not surprising that we did not isolate IgE or IgA antibodies. Interestingly, we only obtained IgG3 antibodies, which are the default subclass for CSR in mouse B cells.Citation42,Citation43 We speculate that it may be possible to obtain other classes and subclasses of antibodies by addition of specific cytokines to the hybridoma cells during CSR, as has been observed for cultured mouse B cells.Citation44-Citation46 It is important to screen class-switched antibodies for retention of antigen-binding activity because SHM also occurs during CSR. We believe that class switching of low affinity IgM to IgG by itself is of limited utility because the resulting IgG is likely to display low functional avidity due to the loss of eight binding sites. Rather, performing simultaneous CSR and selection of high affinity antibodies by our approach may be particularly useful to both affinity mature and class switch low affinity IgM antibodies to high affinity IgG antibodies.
Mimicking the germinal center reaction in hybridoma cells possesses several attractive features. SHM and CSR can be conveniently initiated by lentiviral transduction of hybridoma cells and then stopped after desirable antibodies are identified by removing doxycycline for the Tet-on system or by Cre-mediated excision of the LoxP-flanked AID cassette. Membrane-bound antibodies on the surface of viable hybridoma cells can be used for rapid identification of desirable hybridoma cells whereas secreted antibodies can be collected from the culture medium for further analysis of antibody properties. Controllable expression of AID in myeloma fusion partners may extend this technology to the generation of new hybridoma cells with the built-in capability to perform “on demand” SHM and CSR. We anticipate that this technology may be particularly useful for generating high affinity IgG antibodies against “difficult” antigens such as carbohydrates, haptens, polymers, lipids, and highly homologous proteins and peptides that often produce low affinity IgM responses. Although our study was performed in mouse hybridoma cells, it may be feasible to extend this technology to perform SHM and CSR in human hybridoma cells, for example in those generated from transgenic antibody mice, from immunized or infected human patients or from human immune mice.Citation47-Citation50
Methods
Cell lines and reagents
FO myeloma cells (PTA-11450),Citation51 BALB/3T3 mouse fibroblasts (CCL-163), CC49 (IgG1 mAb against TAG-72, HB-9459), L6 (IgG2a mAb against human L6 antigen, HB-8677), BC3 (IgG2b mAb against human CD3 epsilon chain, HB-10166), and PEG-1–6 (IgG3 mAb against influenza virus, CCL-189) hybridoma cells were purchased from American Type Culture Collection. Hybridoma cell lines AGP4 (IgM mAb against polyethylene glycol), 3.3 and 6–3 (IgG1 mAbs against polyethylene glycol), 7G8 (IgG1 mAb against human β-glucuronidase), and 3D8 (IgM mAb against B16F10 melanoma) were developed in our lab and have been described.Citation24,Citation52,Citation53 Human 293FT cells were kindly provided by Dr. Ming-Zong Lai (Institute of Molecular Biology, Academia Sinica, Taiwan). All cells were cultured in Dulbecco’s modified Eagle’s medium supplemented with 2.98 g/L HEPES, 2 g/L NaHCO3, 10% fetal calf serum (HyClone), 100 U/mL penicillin and 100 μg/mL streptomycin at 37°C in a humidified atmosphere of 5% CO2 in air. Methoxy-PEG750-NH2, methoxy-PEG1K-NH2, methoxy-PEG2K-NH2, methoxy-PEG3K-NH2 hydroxy-PEG5K-NH2, methoxy-PEG10K-NH2, methoxy-PEG20K-NH2 (750, 1000, 2000, 3000, 5000, 10 000, and 20 000 Da, respectively), 4-arm poly(ethylene oxide)10K-NH2, 2-aminomethyl-18-crown-6 and 18-crown-6 were purchased from Sigma-Aldrich.
DNA plasmid construction
The plasmids pAS4w.1.Ppuro (Fig. S3), which contains a tetracycline response element promoter (TRE) for doxycycline-inducible gene expression (Tet-on system), pLKO_AS3w.Ppuro (Fig. S4) and pLKO_AS3w.Pneo (Fig. S5), which contain a CMV early enhancer/chicken β actin (CAG) promoter for cDNA gene expression, and pLKO_AS3w.Ppuro-eGFP (Fig. S6), which contains an eGFP gene, are lentiviral vectors. These plasmids along with pCMVΔR8.91 packaging plasmidCitation54 and pMD.G VSV-G envelope plasmidCitation55 were obtained from the National RNAi Core Facility (Institute of Molecular Biology, Genomic Research Center, Academia Sinica). To generate a stoppable murine AID (AID) expression system, we designed a loxP flanked loxP-CMV-AID-HA-F2A-eGFP-loxP expression cassette (pCMV-AID-loxP). A HA-tagged murine activation-induced deaminase (AID-HA) DNA fragment was cloned from splenocytes isolated from BALB/c mice by RT-PCR. To monitor the expression of AID-HA, a furin-2A (F2A)Citation56 based bicistronic expression strategy was used to link an enhanced green fluorescence protein (eGFP gene) downstream of the AID-HA gene. A HA-F2A-eGFP fragment containing part of the HA tag and eGFP gene was amplified from pLNCX-anti-PEG-eB7.Citation57 The CMV promoter was also cloned by PCR from pLNCX-anti-PEG-eB7. The eGFP fragment was cloned by PCR from pLKO_AS3w.Ppuro-eGFP. The CMV-AID-HA-F2A-eGFP cassette was then created by assembly PCR from CMV, AID-HA and F2A-eGFP fragments and inserted into the pLKO_AS3w.Ppuro plasmid in which the CAG promoter was replaced with the CMV promoter. To introduce loxP sites, annealed oligonucleotides were inserted into a Spe I site upstream of the CMV promoter and in a Pme I site downstream of eGFP, respectively. The resultant plasmid, pCMV-AID-loxP, was co-transfected with pMD.G and pCMVΔR8.91 into 293FT cells to produce recombinant lentivirus, which was used to infect hybridoma cells to introduce the AID gene into the genome.
We also constructed an inducible AID expression vector. rtTA-M2 was amplified from pRetroX-Tet-On Advanced (Clontech Laboratories, Inc.) by PCR and then mutated using multisite-directed mutagenesisCitation58 to obtain the rtTA-V14 gene, which only requires 10 ng/mL of doxycycline to achieve similar gene induction levels as the wild type rtTA at 1000 ng/mL of doxycycline in the Tet-on system.Citation17 An IRES-rtTA-V14 fragment was generated by assembly PCR. A Nhe I- Pme I digested AID-HA-F2A-eGFP fragment and the IRES-rtTA-V14 fragment were inserted into pAS4w.1.Ppuro to create pAS4w.1.Ppuro-AID-F2A-eGFP-IRES-rtTA-V14, denoted as pTetOn-AID.
A DNA fragment encoding the DsRed2 gene was amplified from pDsRed2 (Clontech Laboratories, Inc.) and inserted into pLKO_AS3w.Pneo to generate pAS3w.Pneo-DsRed2. An amber stop codon was introduced into pAS3w.Pneo-DsRed2 at nucleotide position 519Citation18 by site directed mutagenesis using a QuikChange™ Site-Directed Mutagenesis Kit (Stratagene) to generate pDsRed2s.
Biotinylation of PEG and β-glucuronidase
4arm-PEG10K-NH2, methoxy-PEG5K-NH2 and methoxy-PEG2K-NH2 (Laysan Bio, Arab, AL) dissolved in DMSO at 2 mg/mL were mixed with a 6-fold (for 4arm-PEG10K-NH2) or 2-fold (for methoxy-PEG5K-NH2 and methoxy-PEG2K-NH2) molar excess of EZ-link NHS-LC-Biotin (Pierce) or Alexa Fluor® 647 succinimidyl esters (Invitrogen) (in DMSO) for 2 h at room temperature to produce biotinylated 4arm-PEG10K or Alexa Fluor 647 conjugated methoxy-PEG5K and methoxy-PEG2K, respectively. These compounds were diluted in a 5-fold volume of ddH2O and dialyzed (molecular weight cut-off ~12 000–14 000 daltons) against ddH2O to remove free EZ-link NHS-LC-Biotin or Alexa Fluor 647. Likewise, human β-glucuronidaseCitation59 was dissolved in PBS (pH 8.0) at 2 mg/mL and then mixed with a 20-fold molar excess of EZ-link NHS-LC-Biotin for 2 h at room temperature to produce biotinylated β-glucuronidase. One-tenth volume of 1 M glycine solution was added to stop the reaction. Biotinylated β-glucuronidase was dialyzed against PBS to remove free EZ-link NHS-LC-biotin, sterile filtered and stored at -80°C.
Analysis of membrane-bound immunoglobulin on hybridoma cells
Surface expression of mouse immunoglobulin on live hybridoma cells was measured by staining cells with 2 μg/mL of goat anti-mouse Ig (ICN Pharmaceuticals) or goat anti-E. coli antibody (Abcam) as a negative control in PBS containing 0.05% BSA at 4°C for 1 h. The cells were washed three times with cold PBS and stained with 2 μg/mL FITC-conjugated rabbit anti-(goat IgG F(ab)’2) antibody (ICN Pharmaceuticals). The 3.3 and 7G8 hybridoma cells were also stained with biotinylated 4arm-PEG10K (0.5 nM) or biotinylated β-glucuronidase (5 μg/mL) in Hank's balanced salt solution (HBSS), 2% FBS for 30 min at 4°C followed by Alexa Fluor 647-conjugated streptavidin (2 μg/mL) (Jackson ImmunoResearch Laboratories) for 30 min at 4°C. Unbound probes were removed by washing with cold PBS twice. The surface fluorescence of 104 viable cells was measured on a BD™ LSR II flow cytometer (Becton Dickinson) and analyzed with Flowjo (Tree Star).
Lentiviral transduction of the AID gene into hybridoma cells
Recombinant lentiviral particles were packaged by co-transfection of 7.5 μg pCMV-AID-loxP or pTetOn-AID with 6.75 μg pCMVΔR8.91 packaging plasmidCitation54 and 0.75 μg pMD.G VSV-G envelope plasmidCitation55 using 45 μL TransIT-LT1 transfection reagent (Mirus Bio) in 293FT cells grown in a 10 cm culture dish (90% confluency). After 48 h, lentiviral particles were harvested and concentrated by ultracentrifugation at 50 000xg for 1.5 h at 4°C. Lentiviral particles were suspended in culture medium containing 5 μg/mL polybrene and filtered through a 0.45 μm filter. Hybridoma cells were seeded in 6-well plates (1 × 105 cells/well) one day before viral infection. Lentivirus containing medium was added to the cells, which were then centrifuged for 1.5 h (500xg, 32°C). The cells were selected in complete medium containing puromycin (5 μg/mL) to generate stable 3.3/loxP-AID, AGP4/loxP-AID or 3D8/loxP-AID cells.
Conditional expression of AID in cells
The relative expression level of AID in cells was measured on a BD™ LSR II flow cytometer (Becton Dickinson) by detection of the eGFP reporter in 3.3/loxP-AID and 3T3/TetOn-AID cells in the presence or absence of doxycycline. To examine if AID expression could be stopped, 3.3/loxP-AID hybridoma cells (2.5 × 106 cells) were transfected with 5 μg of pLM-CMV-mCherry-P2A-CreCitation60 DNA (Addgene) in electroporation solution (Mirus Bio) using a BTX electroporator (275 voltage, 15 msec pulse length). The cells were cultured in a 6-well plate for 48 h and then analyzed for eGFP and mCherry fluorescence on a BD™ LSR II flow cytometer. pLM-CMV-mCherry-P2A-Cre transfected 3.3/loxP-AID cells that were negative for eGFP expression were isolated on a FACSAria cell sorter 10 d after DNA transfection. To directly measure AID protein levels in cells, 5 × 106 3.3 or 3.3/loxP-AID hybridoma cells were lysed in 0.5 mL RIPA buffer (1% NP-40, 150 mM NaCl, 0.5% sodium deoxycholate, 0.1% SDS, 50 mM Tris, pH 8.0) for 1 h at 4°C. Fifty μg of total protein from the clarified lysate were analyzed by 12.5% reducing SDS-PAGE, transferred to nitrocellulose paper and sequentially stained with biotinylated goat anti-HA antibody against the HA epitope tag sequence YPYDVPDYA (Vector Laboratories) or rabbit anti-tubulin α antibody (NeoMarkers) followed by streptavidin-HRP and goat anti-rabbit Ig-HRP, respectively (Jackson ImmunoResearch Laboratories). Bands were visualized by ECL detection (Pierce) and analyzed with a LAS-3000 Mini Fujifilm imaging system (FujiFilm).
SHM assay
3T3 or 3T3/TetOn-AID cells were infected with DsRed2s lentivirus to generate 3T3/DsRed2s or 3T3/TetOn-AID x DsRed2s cells. The cells were cultured with or without 500 ng/mL doxycycline. Cells were harvested at defined times and processed for flow cytometry to measure the DsRed2 signal, indicative of mutation of the premature stop codon to allow expression of full-length DsRed2 protein. The percentage of DsRed2-positive cells was calculated and reported as revertants/106 cells.
Isolation of anti-PEG antibody variants by mimicking the germinal center reaction
To isolate anti-PEG antibody variants, 3 × 107 3.3/loxP-AID cells were cultured for two weeks and then stained with biotinylated 4arm-PEG10K (100 pM, 1 × 106 cells/mL) in HBSS, 2% FBS for 30 min at 4°C followed by incubation for 30 min at 4°C with 5 mL of Alexa Fluor 647-conjugated streptavidin (2 μg/mL) and PE-conjugated goat anti-mouse IgG Fc antibody (2 μg/mL) to measure membrane-bound immunoglobulin levels. Unbound probes were removed by washing with cold PBS twice. Cells displaying high Alexa Fluor 647 fluorescence (1% of total cells) were collected on a FACSAria cell sorter and cultured for 2 wk before the cells were sorted again. The PEG chain length and branching of the PEG probe was progressively decreased from 4arm-PEG10K, linear PEG5K and linear PEG2K during subsequent rounds of sorting. After 5 rounds, single 3.3/AID cells were collected into 96-well plates and AID expression was stopped by transient transfection of pLM-CMV-mCherry-P2A-Cre into hybridoma clones.
Antibody production and purification
2.5 × 107 of selected 3.3/loxP-AID variant hybridoma cells (1E3 and 2B5) in 15 mL culture medium (DMEM, 5% FBS) were inoculated into a CELLine CL 1000 two-compartment bioreactor (INTEGRA Biosciences AG). The antibody-containing culture medium was harvested every 7 d and then purified by protein A Sepharose 4 Fast Flow chromatography (GE Healthcare). Collected antibody was dialyzed against PBS and sterile filtered. Antibody concentrations were determined by the bicinchoninic acid (BCA) protein assay (Thermo Scientific).
Site directed mutagenesis of antibody 2B5
The recombinant 2B5 antibody gene was cloned from 2B5 hybridoma cDNA by RT-PCR. The 2B5 light chain and heavy chain DNA were joined by a composite furin-2A bicistronic expression peptide linker in pLNCX-anti-PEG-eB7. An EcoR I- Pme I digested 2B5 IgG fragment was inserted into pAS3w.Ppuro to create pAS3w.Ppuro-2B5. Site-directed mutagenesis of VH V23A and VL K53N was performed in a 50 μL mixture containing 20 ng of pAS3w.Ppuro-2B5 template DNA plasmid, 15 pmole of each primer, 20 nmole of dNTPs, 2 U of Phusion high-fidelity DNA polymerase (Thermo Scientific) in 1 x Phusion buffer. Thermal cycling used an initial denaturation at 95°C for 0.5 min; 18 cycles at 95°C for 0.5 min, 55°C for 1 min and 68°C for 11 min. After cooling to ≤37°C, 2 U of Dpn I restriction enzyme (NEB) was directly added to the amplification reaction at 37°C for 1.5 h. Four microliters of the Dpn I digested sample was used to transform DH5α competent cells by the heat shock method. 3T3 cells that stably secreted 2B5/V23A (2B5ΔV) and 2B5/K53N (2B5ΔK) antibodies were generated by lentiviral transduction and selected in puromycin (10 μg/mL) as described above. The culture medium of cells producing 2B5ΔV and 2B5ΔK recombinant antibodies was harvested from CELLine adhere 1000 bioreactors every 7–10 d and the antibodies were purified by protein A Sepharose 4 Fast Flow chromatography.
Antibody ELISA
Maxisorp 96-well microplates (Nalge-Nunc International, Roskilde, Denmark) were coated with 0.5 μg/well methoxy-PEG750-NH2, methoxy-PEG1K-NH2 methoxy-PEG2K-NH2, methoxy-PEG3K-NH2 hydroxy-PEG5K-NH2, methoxy-PEG10K-NH2, methoxy-PEG20K-NH2 or 4-arm poly(ethylene oxide)10K-NH2 in 50 μL/well 0.1 M NaHCO3/Na2CO3 (adjusted to pH 8.0 with HCl) buffer for 3 h at 37°C and then blocked with 200 μL/well dilution buffer (5% skim milk in PBS) at 4°C overnight. Antibodies were pre-incubated at 37°C for up to 5 d to check thermal stability. Graded concentrations of antibodies in 50 μL 2% skim milk in PBS were added to the plates at 4°C, RT or 37°C for 1 h. The plates were washed with PBS three times at 4°C, RT or 37°C, respectively. HRP-conjugated donkey anti-mouse IgG Fc (2 μg/mL) in 50 μL dilution buffer were added for 1 h at 4°C, RT or 37°C. The plates were washed as described above. For competition ELISA assays, maxisorp 96-well microplates were coated with 0.5 μg/well amino-PEG2K-NH2 or human β-glucuronidase as described above. 3-fold serial dilutions of 18-crown-6 starting at 120 mM were prepared and mixed 1:1 (v/v) with 20 μg/mL of 3.3, 2B5 or 7G8 antibodies (thus the final concentration of the antibodies was 10 μg/mL). The mixture was added to the plates at 4°C for 1 h. The plates were washed with PBS three times at 4°C and were then incubated with HRP-conjugated donkey anti-mouse IgG Fc (2 μg/mL) at 4°C for 1 h. For crown ether ELISA assays, maxisorp 96-well microplates were coated with 0.5 μmole / well 2-aminomethyl-18-crown-6 as described above. Graded concentrations of antibodies (3.3, 2B5 or 7G8) in 50 μL 2% skim milk in PBS were added to the plates at 4°C, 25°C or 37°C for 4 h. The plates were washed with PBS two times at 4°C, 25°C or 37°C, respectively. Biotin-conjugated goat anti-mouse IgG Fc (2 μg/mL) (Jackson ImmunoResearch Laboratories) in 50 μL dilution buffer was added for 1 h at 4°C, 25°C or 37°C. The plates were washed as described above at different temperatures. Five micrograms of biotinylated peroxidase (Invitrogen) and 10 μg of streptavidin (Thermo Scientific) were pre-incubated in 15 mL PBS for 30 min at room temperature to allow complex formation before 50 μL was added to the wells. Unbound antibodies were washed with PBS four times at 4°C, 25°C or 37°C. In all cases, the bound peroxidase activity was measured by adding 150 μL/well ABTS solution [0.4 mg/mL, 2,2′-azino-bis(3-ethylbenzthiazoline-6-sulfonic acid), 0.003% H2O2, and 100 mM phosphate-citrate, pH 4.0) for 30 min at room temperature. The absorbance (405 nm) of wells was measured in a microplate reader (Molecular Device).
Fab fragmentation of IgG
Papain (Sigma-Aldrich) was dissolved to a final concentration of 0.1 mg/mL in PBS supplemented with 20 mM l-cysteine and 20 mM EDTA (Sigma-Aldrich) and the pH was then adjusted to 7.2. An equal volume of purified 3.3, 1E3 or 2B5 anti-PEG antibody (2 mg/mL) was added to the papain solution and incubated at 37°C for 2.5 h. One-tenth volume of 0.3 M iodoacetamide solution (Sigma-Aldrich) was added to stop the reaction. The 3.3, 1E3 and 2B5 anti-PEG Fab fragments were purified by affinity chromatography on a PEG affinity column, generated by swelling 1g of CNBr-activated Sepharose 4B (GE Healthcare) in 1 mM HCl (pH 3) for 30 min, washing with coupling buffer (0.1 M NaHCO3, pH 8.3) and adding five moles of methoxy-PEG30K-amine (Laysan Bio) per mL gel in coupling buffer for 4 h at 25°C. Remaining active groups on the CNBr-activated Sepharose were blocked by adding 1/10 volume of 1M Tris (pH 8) to the gel at 25°C for 2 h. The PEG coupled Sepharose was washed with 0.1 M acetate buffer (pH 4) containing 0.5 M NaCl followed by 0.1 M Tris (pH 8) containing 0.5 M NaCl. Papain digested antibodies were loaded to the PEG-resin column at 4°C for 45 min and washed with cold PBS to removed papain and Fc fragments. The PEG-resin bound anti-PEG Fab fragments were eluted with 100 mM glycine buffer (pH 3) and dialyzed against PBS.
PEGylation of hen egg white lysozyme
Three mg/mL hen egg white lysozyme (HEL) (Sigma-Aldrich) in PBS (pH 8.0) was mixed with a 4-fold molar excess of methoxy-PEG2k-succinimidylpropionate (Shearwater Polymers) for 2 h at 25°C to produce mono-PEGylated HEL (PEG2k-HEL). One-tenth volume of 1 M glycine solution was added to stop the reaction. Unreacted PEG was removed by gel filtration on a Sephacryl S-300 HR column. The concentration of PEG2k-HEL was determined by the BCA assay (Thermo Scientific) with bovine serum albumin used as the reference protein.
Binding kinetic analysis of anti-PEG antibodies by surface plasmon resonance
The binding kinetics of 3.3, 1E3 and 2B5 anti-PEG Fab fragments was measured on a Biacore T-200 (GE Healthcare) at 4°C, 25°C and 37°C. PEG2k-HEL was immobilized on a CM5 chip (GE Healthcare) by using the standard procedure for amine coupling through the EDC/NHS reaction. A total immobilization of 57 resonance units (RUs) was achieved. Binding was determined at a constant flow rate of 65 μL/min of various concentrations of the antibodies in HEPES buffered saline.
Thermal stability of antibodies
Antibodies were dialyzed in PBS, degassed, and added into the sample chamber of a differential scanning calorimeter (Nano DSC III) (TA Instruments) at concentrations of 0.5 mg/mL. Degassed PBS was injected into the reference chamber. Differential power was monitored as each antibody-buffer pair was heated linearly from 10°C to 110°C at a rate of 1°C per minute under a fixed pressure of 3 atm. Buffer-buffer (degassed PBS) scans were also collected for baseline subtraction using the same procedure as for the antibody samples.
Binding of antibodies to crown ether by surface plasmon resonance
The binding activity of antibodies to 18-crown-6 compounds was measured on a Biacore T-200 (GE Healthcare) at defined temperatures. The 2-aminomethyl-18-crown-6 was immobilized on a CM5 chip by using the standard procedure for amine coupling through the EDC/NHS reaction. Injection of 2-aminomethyl-18-crown-6 (10 mM in 50 mM sodium borate buffer, pH 8.5) to a EDC/NHS activated CM5 chip was performed at a constant flow rate of 10 μL/min for 30 min. The remaining succinimide esters were inactivated by the injection of ethanolamine. A total immobilization of 279.4 resonance units (RUs) was achieved. Antibody binding analysis was performed at a constant flow rate of 50 μL/min of the antibodies at 1 µM in HEPES buffered saline at 4°C, 25°C and 37°C.
Affinity purification of PEGylated nanoparticles by anti-PEG antibodies
PEG-Qdot655 nanoparticles were purified by affinity chromatography on the anti-PEG antibody resin columns. Briefly, 5 mg of 3.3 or 2B5 anti-PEG antibodies in coupling buffer (0.1 M sodium acetate, 0.15 M sodium chloride, pH 5.5) was reacted with 10 mM sodium meta-periodate (Thermo Scientific Pierce) in the dark at room temperature for 30 min. Sodium meta-periodate was removed on a Zeba™ Spin desalting column and the oxidized antibodies were then incubated with 1 mL of UltraLink® Hydrazide Resin containing 0.1 M aniline (Thermo Scientific) at room temperature for 4 h. The anti-PEG resin was packed into a column and washed with PBS 3 times. Five hundred microliters of PEG-Qdot655 solution (32 nM in PBS) was loaded into the 3.3 or 2B5 anti-PEG columns at 4°C. The columns were washed with cold PBS and bound PEG-Qdot655 nanoparticles were eluted with 37°C PBS or 100 mM citrate buffer (pH = 3). The particles were transferred into a Nunc F96 MicroWell black polystyrene plate (200 μL/well) and the fluorescence of PEG-Qdot655 (excitation/emission: 450 nm/660 nm) was detected on an IVIS 200 optical imaging system (Xenogen).
Immunoglobulin ELISA
Maxisorp 96-well microplates were coated with 0.5 μg/well goat anti-mouse IgG+IgA+IgM (Jackson ImmunoResearch Laboratories) in 50 μL/well 0.1 M NaHCO3/Na2CO3 (adjusted to pH 8.0 with HCl) buffer for 3 h at 37°C and then blocked with 200 μL/well dilution buffer (5% skim milk in PBS) at 4°C overnight. Diluted hybridoma supernatants in 50 μL 2% skim milk (1:100) were added to the plates at RT for 1 h. The plates were washed with PBS three times. HRP-conjugated rabbit anti-mouse IgG, IgA, or IgM (MP Biomedicals, 2 μg/mL) in 50 μL dilution buffer were added for 1 h at RT. The plates were washed as described above. The bound peroxidase activity was measured by adding 150 μL/well ABTS solution for 30 min at room temperature. The absorbance (405 nm) of the wells was measured in a microplate reader. The isotypes of class-switched AGP4 and 3D8 antibodies were determined with a Mouse MonoAb-ID kit (Zymed Laboratories) according to the manufacturer’s instructions.
FACS analysis of class switched 3D8 antibody variants
B16F10 cells were stained for 30 min at 4°C with culture supernatant from 3D8 or 3D8/loxP-AID cells. The cells were washed three times with cold PBS and stained with 2 μg/mL PE-conjugated goat anti-mouse IgG Fc antibody (Jackson ImmunoResearch Laboratories). Unbound antibodies were removed by washing with cold PBS twice. The surface fluorescence of 104 viable cells was measured on a BD™ LSR II flow cytometer and analyzed with Flowjo.
| Abbreviations: | ||
| AID | = | activation-induced cytidine deaminase |
| CDR | = | complementarity-determining region |
| CSR | = | class switch recombination |
| eGFP | = | enhanced green fluorescent protein |
| FACS | = | fluorescence-activated cell sorting |
| FR1 | = | framework one of the variable region gene segment |
| HA | = | hemagglutinin |
| HEL | = | hen egg white lysozyme |
| mAbs | = | monoclonal antibodies |
| MFI | = | mean fluorescence intensity |
| PEG | = | polyethylene glycol |
| pre-B cells | = | precursor B cells |
| pro-B cells | = | progenitor B cells |
| pCMV-AID-loxP | = | lentiviral vector for stoppable expression of AID |
| pTetOn-AID | = | lentiviral vector for tetracycline-inducible expression of AID |
| SHM | = | somatic hypermutation |
| V-D-J | = | heavy chain variable, diverse and joining gene segments |
| VL | = | light chain variable region |
Additional material
Download Zip (2.5 MB)Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
This work was supported by Academia Sinica and a research grant from the National Science Council, Taipei, Taiwan (NSC 102–2320-B-001–013-MY3). The authors thank Dr. Shu-Chuan Jao of the Biophysics Core Facility, Scientific Instrument Center at Academia Sinica for assistance in performing Biacore T200 and Nano DSC III experiments. We also thank Ms. Chia-Chen Tai and Ms. Tzu-Wen Tai of the Flow Cytometry Core, Scientific Instrument Center at the Institute of Biomedical Sciences, Academia Sinica, Taipei, Taiwan for help with the LSR II flow cytometer and FACSAria cell sorter. We appreciate helpful discussions with Dr. Mi-Hua Tao of the Institute of Biomedical Sciences and Dr. Andrew H.-J. Wang and Dr. Meng-Chiao Ho of the Institute of Biological Chemistry at Academia Sinica. We appreciate help with lentivirus production by the National RNAi Core Facility, Institute of Molecular Biology / Genomic Research Center, at Academia Sinica, Taipei, Taiwan.
Author Contributions
Y.-C.S. and S.R.R. conceived the project and designed the experiments and analyses. Y.-C.S., T.S.A.Q., H.-Y.T., and B.-M.C. performed experiments. K.-H.C. cloned recombinant antibody genes. Y.-C.S., T.-L.C., and S.R.R. analyzed data and wrote the manuscript.
References
- Söderlind E, Strandberg L, Jirholt P, Kobayashi N, Alexeiva V, Aberg AM, Nilsson A, Jansson B, Ohlin M, Wingren C, et al. Recombining germline-derived CDR sequences for creating diverse single-framework antibody libraries. Nat Biotechnol 2000; 18:852 - 6; http://dx.doi.org/10.1038/78458; PMID: 10932154
- Hoet RM, Cohen EH, Kent RB, Rookey K, Schoonbroodt S, Hogan S, Rem L, Frans N, Daukandt M, Pieters H, et al. Generation of high-affinity human antibodies by combining donor-derived and synthetic complementarity-determining-region diversity. Nat Biotechnol 2005; 23:344 - 8; http://dx.doi.org/10.1038/nbt1067; PMID: 15723048
- Daugherty PS, Chen G, Iverson BL, Georgiou G. Quantitative analysis of the effect of the mutation frequency on the affinity maturation of single chain Fv antibodies. Proc Natl Acad Sci U S A 2000; 97:2029 - 34; http://dx.doi.org/10.1073/pnas.030527597; PMID: 10688877
- Crameri A, Cwirla S, Stemmer WP. Construction and evolution of antibody-phage libraries by DNA shuffling. Nat Med 1996; 2:100 - 2; http://dx.doi.org/10.1038/nm0196-100; PMID: 8564822
- Boder ET, Wittrup KD. Yeast surface display for screening combinatorial polypeptide libraries. Nat Biotechnol 1997; 15:553 - 7; http://dx.doi.org/10.1038/nbt0697-553; PMID: 9181578
- Bowers PM, Horlick RA, Neben TY, Toobian RM, Tomlinson GL, Dalton JL, Jones HA, Chen A, Altobell L 3rd, Zhang X, et al. Coupling mammalian cell surface display with somatic hypermutation for the discovery and maturation of human antibodies. Proc Natl Acad Sci U S A 2011; 108:20455 - 60; http://dx.doi.org/10.1073/pnas.1114010108; PMID: 22158898
- Ramsden DA, van Gent DC, Gellert M. Specificity in V(D)J recombination: new lessons from biochemistry and genetics. Curr Opin Immunol 1997; 9:114 - 20; http://dx.doi.org/10.1016/S0952-7915(97)80167-7; PMID: 9039786
- Crouch EE, Li Z, Takizawa M, Fichtner-Feigl S, Gourzi P, Montaño C, Feigenbaum L, Wilson P, Janz S, Papavasiliou FN, et al. Regulation of AID expression in the immune response. J Exp Med 2007; 204:1145 - 56; http://dx.doi.org/10.1084/jem.20061952; PMID: 17452520
- Odegard VH, Schatz DG. Targeting of somatic hypermutation. Nat Rev Immunol 2006; 6:573 - 83; http://dx.doi.org/10.1038/nri1896; PMID: 16868548
- Jacob J, Przylepa J, Miller C, Kelsoe G. In situ studies of the primary immune response to (4-hydroxy-3-nitrophenyl)acetyl. III. The kinetics of V region mutation and selection in germinal center B cells. J Exp Med 1993; 178:1293 - 307; http://dx.doi.org/10.1084/jem.178.4.1293; PMID: 8376935
- Martin A, Bardwell PD, Woo CJ, Fan M, Shulman MJ, Scharff MD. Activation-induced cytidine deaminase turns on somatic hypermutation in hybridomas. Nature 2002; 415:802 - 6; http://dx.doi.org/10.1038/nature714; PMID: 11823785
- Iglesias-Ussel MD, Fan M, Li Z, Martin A, Scharff MD. Forced expression of AID facilitates the isolation of class switch variants from hybridoma cells. J Immunol Methods 2006; 316:59 - 66; http://dx.doi.org/10.1016/j.jim.2006.08.002; PMID: 16997317
- Harris JM, Chess RB. Effect of pegylation on pharmaceuticals. Nat Rev Drug Discov 2003; 2:214 - 21; http://dx.doi.org/10.1038/nrd1033; PMID: 12612647
- Bjorling M, Karlstrom G, Linse P. Conformational Adaption of Poly(Ethylene Oxide) - a C-13 Nmr-Study. J Phys Chem 1991; 95:6706 - 9; http://dx.doi.org/10.1021/j100170a060
- Shephard JJ, Dickie SA, McQuillan AJ. Structure and conformation of methyl-terminated poly(ethylene oxide)-bis[methylenephosphonate] ligands adsorbed to boehmite (AlOOH) from aqueous solutions. Attenuated total reflection infrared (ATR-IR) spectra and dynamic contact angles. Langmuir 2010; 26:4048 - 56; http://dx.doi.org/10.1021/la903506q; PMID: 19928841
- Markusic D, Oude-Elferink R, Das AT, Berkhout B, Seppen J. Comparison of single regulated lentiviral vectors with rtTA expression driven by an autoregulatory loop or a constitutive promoter. Nucleic Acids Res 2005; 33:e63; http://dx.doi.org/10.1093/nar/gni062; PMID: 15809225
- Centlivre M, Zhou X, Pouw SM, Weijer K, Kleibeuker W, Das AT, Blom B, Seppen J, Berkhout B, Legrand N. Autoregulatory lentiviral vectors allow multiple cycles of doxycycline-inducible gene expression in human hematopoietic cells in vivo. Gene Ther 2010; 17:14 - 25; http://dx.doi.org/10.1038/gt.2009.109; PMID: 19727135
- Klasen M, Spillmann FJ, Lorens JB, Wabl M. Retroviral vectors to monitor somatic hypermutation. J Immunol Methods 2005; 300:47 - 62; http://dx.doi.org/10.1016/j.jim.2005.02.015; PMID: 15936027
- Gouet P, Fabry B, Guillet V, Birck C, Mourey L, Kahn D, Samama JP. Structural transitions in the FixJ receiver domain. Structure 1999; 7:1517 - 26; http://dx.doi.org/10.1016/S0969-2126(00)88342-2; PMID: 10647182
- Hillig RC, Coulie PG, Stroobant V, Saenger W, Ziegler A, Hülsmeyer M. High-resolution structure of HLA-A*0201 in complex with a tumour-specific antigenic peptide encoded by the MAGE-A4 gene. J Mol Biol 2001; 310:1167 - 76; http://dx.doi.org/10.1006/jmbi.2001.4816; PMID: 11502003
- Michalska K, Fernandes H, Sikorski M, Jaskolski M. Crystal structure of Hyp-1, a St. John’s wort protein implicated in the biosynthesis of hypericin. J Struct Biol 2010; 169:161 - 71; http://dx.doi.org/10.1016/j.jsb.2009.10.008; PMID: 19853038
- Muramatsu M, Kinoshita K, Fagarasan S, Yamada S, Shinkai Y, Honjo T. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell 2000; 102:553 - 63; http://dx.doi.org/10.1016/S0092-8674(00)00078-7; PMID: 11007474
- Tsai NM, Chen B-M, Wei S-L, Wu C-W, Roffler SR. Anti-tumor immunoglobulin M increases lung metastasis in an experimental model of malignant melanoma. Clin Exp Metastasis 2003; 20:103 - 9; http://dx.doi.org/10.1023/A:1022616223359; PMID: 12705631
- Su YC, Chen BM, Chuang KH, Cheng TL, Roffler SR. Sensitive quantification of PEGylated compounds by second-generation anti-poly(ethylene glycol) monoclonal antibodies. Bioconjug Chem 2010; 21:1264 - 70; http://dx.doi.org/10.1021/bc100067t; PMID: 20536171
- McConnell AD, Spasojevich V, Macomber JL, Krapf IP, Chen A, Sheffer JC, Berkebile A, Horlick RA, Neben S, King DJ, et al. An integrated approach to extreme thermostabilization and affinity maturation of an antibody. Protein Eng Des Sel 2013; 26:151 - 64; http://dx.doi.org/10.1093/protein/gzs090; PMID: 23173178
- Bowers PM, Neben TY, Tomlinson GL, Dalton JL, Altobell L, Zhang X, Macomber JL, Wu BF, Toobian RM, McConnell AD, et al. Humanization of antibodies using heavy chain complementarity-determining region 3 grafting coupled with in vitro somatic hypermutation. J Biol Chem 2013; 288:7688 - 96; http://dx.doi.org/10.1074/jbc.M112.445502; PMID: 23355464
- Alt FW, Bothwell AL, Knapp M, Siden E, Mather E, Koshland M, Baltimore D. Synthesis of secreted and membrane-bound immunoglobulin mu heavy chains is directed by mRNAs that differ at their 3′ ends. Cell 1980; 20:293 - 301; http://dx.doi.org/10.1016/0092-8674(80)90615-7; PMID: 6771018
- Kehry M, Ewald S, Douglas R, Sibley C, Raschke W, Fambrough D, Hood L. The immunoglobulin mu chains of membrane-bound and secreted IgM molecules differ in their C-terminal segments. Cell 1980; 21:393 - 406; http://dx.doi.org/10.1016/0092-8674(80)90476-6; PMID: 6773668
- Rogers J, Early P, Carter C, Calame K, Bond M, Hood L, Wall R. Two mRNAs with different 3′ ends encode membrane-bound and secreted forms of immunoglobulin mu chain. Cell 1980; 20:303 - 12; http://dx.doi.org/10.1016/0092-8674(80)90616-9; PMID: 6771019
- Price PW, McKinney EC, Wang Y, Sasser LE, Kandasamy MK, Matsuuchi L, Milcarek C, Deal RB, Culver DG, Meagher RB. Engineered cell surface expression of membrane immunoglobulin as a means to identify monoclonal antibody-secreting hybridomas. J Immunol Methods 2009; 343:28 - 41; http://dx.doi.org/10.1016/j.jim.2009.01.005; PMID: 19187782
- Cheng TL, Wu PY, Wu MF, Chern JW, Roffler SR. Accelerated clearance of polyethylene glycol-modified proteins by anti-polyethylene glycol IgM. Bioconjug Chem 1999; 10:520 - 8; http://dx.doi.org/10.1021/bc980143z; PMID: 10346886
- Cheng TL, Cheng CM, Chen BM, Tsao DA, Chuang KH, Hsiao SW, Lin YH, Roffler SR. Monoclonal antibody-based quantitation of poly(ethylene glycol)-derivatized proteins, liposomes, and nanoparticles. Bioconjug Chem 2005; 16:1225 - 31; http://dx.doi.org/10.1021/bc050133f; PMID: 16173802
- Bailon P, Won C-Y. PEG-modified biopharmaceuticals. Expert Opin Drug Deliv 2009; 6:1 - 16; http://dx.doi.org/10.1517/17425240802650568; PMID: 19236204
- Cheng T-L, Chuang K-H, Chen B-M, Roffler SR. Analytical measurement of PEGylated molecules. Bioconjug Chem 2012; 23:881 - 99; http://dx.doi.org/10.1021/bc200478w; PMID: 22242549
- Lee SB, Mitchell DT, Trofin L, Nevanen TK, Söderlund H, Martin CR. Antibody-based bio-nanotube membranes for enantiomeric drug separations. Science 2002; 296:2198 - 200; http://dx.doi.org/10.1126/science.1071396; PMID: 12077410
- Climent E, Bernardos A, Martínez-Máñez R, Maquieira A, Marcos MD, Pastor-Navarro N, Puchades R, Sancenón F, Soto J, Amorós P. Controlled delivery systems using antibody-capped mesoporous nanocontainers. J Am Chem Soc 2009; 131:14075 - 80; http://dx.doi.org/10.1021/ja904456d; PMID: 19739626
- Stuart MA, Huck WT, Genzer J, Müller M, Ober C, Stamm M, Sukhorukov GB, Szleifer I, Tsukruk VV, Urban M, et al. Emerging applications of stimuli-responsive polymer materials. Nat Mater 2010; 9:101 - 13; http://dx.doi.org/10.1038/nmat2614; PMID: 20094081
- Löwik DW, Leunissen EH, van den Heuvel M, Hansen MB, van Hest JC. Stimulus responsive peptide based materials. Chem Soc Rev 2010; 39:3394 - 412; http://dx.doi.org/10.1039/b914342b; PMID: 20523948
- Stuart MAC, Huck WTS, Genzer J, Müller M, Ober C, Stamm M, Sukhorukov GB, Szleifer I, Tsukruk VV, Urban M, et al. Emerging applications of stimuli-responsive polymer materials. Nat Mater 2010; 9:101 - 13; http://dx.doi.org/10.1038/nmat2614; PMID: 20094081
- Boder ET, Wittrup KD. Optimal screening of surface-displayed polypeptide libraries. Biotechnol Prog 1998; 14:55 - 62; http://dx.doi.org/10.1021/bp970144q; PMID: 10858036
- Honjo T, Kinoshita K, Muramatsu M. Molecular mechanism of class switch recombination: linkage with somatic hypermutation. Annu Rev Immunol 2002; 20:165 - 96; http://dx.doi.org/10.1146/annurev.immunol.20.090501.112049; PMID: 11861601
- Deenick EK, Hasbold J, Hodgkin PD. Switching to IgG3, IgG2b, and IgA is division linked and independent, revealing a stochastic framework for describing differentiation. J Immunol 1999; 163:4707 - 14; PMID: 10528168
- Deenick EK, Hasbold J, Hodgkin PD. Decision criteria for resolving isotype switching conflicts by B cells. Eur J Immunol 2005; 35:2949 - 55; http://dx.doi.org/10.1002/eji.200425719; PMID: 16180247
- Morawetz RA, Gabriele L, Rizzo LV, Noben-Trauth N, Kühn R, Rajewsky K, Müller W, Doherty TM, Finkelman F, Coffman RL, et al. Interleukin (IL)-4-independent immunoglobulin class switch to immunoglobulin (Ig)E in the mouse. J Exp Med 1996; 184:1651 - 61; http://dx.doi.org/10.1084/jem.184.5.1651; PMID: 8920855
- McIntyre TM, Klinman DR, Rothman P, Lugo M, Dasch JR, Mond JJ, Snapper CM. Transforming growth factor beta 1 selectivity stimulates immunoglobulin G2b secretion by lipopolysaccharide-activated murine B cells. J Exp Med 1993; 177:1031 - 7; http://dx.doi.org/10.1084/jem.177.4.1031; PMID: 8459202
- Sonoda E, Matsumoto R, Hitoshi Y, Ishii T, Sugimoto M, Araki S, Tominaga A, Yamaguchi N, Takatsu K. Transforming growth factor beta induces IgA production and acts additively with interleukin 5 for IgA production. J Exp Med 1989; 170:1415 - 20; http://dx.doi.org/10.1084/jem.170.4.1415; PMID: 2677210
- Mompó SM, González-Fernández Á. Antigen-Specific Human Monoclonal Antibodies from Transgenic Mice. Human Monoclonal Antibodies: Springer, 2014:245-76.
- Yu X, Tsibane T, McGraw PA, House FS, Keefer CJ, Hicar MD, Tumpey TM, Pappas C, Perrone LA, Martinez O, et al. Neutralizing antibodies derived from the B cells of 1918 influenza pandemic survivors. Nature 2008; 455:532 - 6; http://dx.doi.org/10.1038/nature07231; PMID: 18716625
- Shultz LD, Brehm MA, Garcia-Martinez JV, Greiner DL. Humanized mice for immune system investigation: progress, promise and challenges. Nat Rev Immunol 2012; 12:786 - 98; http://dx.doi.org/10.1038/nri3311; PMID: 23059428
- Becker PD, Legrand N, van Geelen CM, Noerder M, Huntington ND, Lim A, Yasuda E, Diehl SA, Scheeren FA, Ott M, et al. Generation of human antigen-specific monoclonal IgM antibodies using vaccinated “human immune system” mice. PLoS One 2010; 5:e13137; http://dx.doi.org/10.1371/journal.pone.0013137; PMID: 20957227
- de StGroth SF, Scheidegger D. Production of monoclonal antibodies: strategy and tactics. J Immunol Methods 1980; 35:1 - 21; http://dx.doi.org/10.1016/0022-1759(80)90146-5; PMID: 7009747
- Chen KC, Cheng TL, Leu YL, Prijovich ZM, Chuang CH, Chen BM, Roffler SR. Membrane-localized activation of glucuronide prodrugs by beta-glucuronidase enzymes. Cancer Gene Ther 2007; 14:187 - 200; http://dx.doi.org/10.1038/sj.cgt.7700999; PMID: 16977328
- Tsai NM, Chen BM, Wei SL, Wu CW, Roffler SR. Anti-tumor immunoglobulin M increases lung metastasis in an experimental model of malignant melanoma. Clin Exp Metastasis 2003; 20:103 - 9; http://dx.doi.org/10.1023/A:1022616223359; PMID: 12705631
- Zufferey R, Nagy D, Mandel RJ, Naldini L, Trono D. Multiply attenuated lentiviral vector achieves efficient gene delivery in vivo. Nat Biotechnol 1997; 15:871 - 5; http://dx.doi.org/10.1038/nbt0997-871; PMID: 9306402
- Kahl CA, Marsh J, Fyffe J, Sanders DA, Cornetta K. Human immunodeficiency virus type 1-derived lentivirus vectors pseudotyped with envelope glycoproteins derived from Ross River virus and Semliki Forest virus. J Virol 2004; 78:1421 - 30; http://dx.doi.org/10.1128/JVI.78.3.1421-1430.2004; PMID: 14722297
- Fang J, Qian J-J, Yi S, Harding TC, Tu GH, VanRoey M, Jooss K. Stable antibody expression at therapeutic levels using the 2A peptide. Nat Biotechnol 2005; 23:584 - 90; http://dx.doi.org/10.1038/nbt1087; PMID: 15834403
- Chuang KH, Wang HE, Cheng TC, Tzou SC, Tseng WL, Hung WC, Tai MH, Chang TK, Roffler SR, Cheng TL. Development of a universal anti-polyethylene glycol reporter gene for noninvasive imaging of PEGylated probes. J Nucl Med 2010; 51:933 - 41; http://dx.doi.org/10.2967/jnumed.109.071977; PMID: 20484433
- Sawano A, Miyawaki A. Directed evolution of green fluorescent protein by a new versatile PCR strategy for site-directed and semi-random mutagenesis. Nucleic Acids Res 2000; 28:E78; http://dx.doi.org/10.1093/nar/28.16.e78; PMID: 10931937
- Wu CH, Balasubramanian WR, Ko YP, Hsu G, Chang SE, Prijovich ZM, Chen KC, Roffler SR. A simple method for the production of recombinant proteins from mammalian cells. Biotechnol Appl Biochem 2004; 40:167 - 72; http://dx.doi.org/10.1042/BA20030184; PMID: 14725509
- Papapetrou EP, Lee G, Malani N, Setty M, Riviere I, Tirunagari LM, Kadota K, Roth SL, Giardina P, Viale A, et al. Genomic safe harbors permit high β-globin transgene expression in thalassemia induced pluripotent stem cells. Nat Biotechnol 2011; 29:73 - 8; http://dx.doi.org/10.1038/nbt.1717; PMID: 21151124
- Kabat EA, Te Wu T, Gottesman KS, Foeller C. Sequences of proteins of immunological interest. Collingdale, PA Diane Publishing, 1992.
