Abstract
Human monoclonal antibodies (mAbs) can routinely be isolated from phage display libraries against virtually any protein available in sufficient purity and quantity, but library design can influence epitope coverage on the target antigen. Here we describe the construction of a novel synthetic human antibody phage display library that incorporates hydrophilic or charged residues at position 52 of the CDR2 loop of the variable heavy chain domain, instead of the serine residue found in the corresponding germline gene. The novel library was used to isolate human mAbs to various antigens, including the alternatively-spliced EDA domain of fibronectin, a marker of tumor angiogenesis. In particular, the mAb 2H7 was proven to bind to a novel epitope on EDA, which does not overlap with the one recognized by the clinical-stage F8 antibody. F8 and 2H7 were used for the construction of chelating recombinant antibodies (CRAbs), whose tumor-targeting properties were assessed in vivo in biodistribution studies in mice bearing F9 teratocarcinoma, revealing a preferential accumulation at the tumor site.
Introduction
Human monoclonal antibodies (mAbs) and their derivatives represent the largest and fastest growing sector of pharmaceutical biotechnology products.Citation1,Citation2 Antibody phage display libraries are useful tools for the isolation of human mAbs for therapeutic and in vivo diagnostic applications.Citation3 Indeed, good-quality binders can be obtained in virtually all cases from selection experiments, whenever the corresponding antigen is available as pure protein in 1–2 milligram quantities.Citation4–Citation11
Adalimumab (Humira®) was the first mAb isolated using phage library technology to be approved for therapeutic applications in Europe, the US and many other countries.Citation12 At present, several phage-derived antibodies are being investigated in clinical trials. Our group has isolated human antibodies against components of the modified extracellular matrix that are currently being investigated in clinical trials either as radiolabeled products or as cytokine fusion proteins.Citation13,Citation14 In particular, the F8 antibody, which is specific to the alternatively-spliced extra domain (EDA) of fibronectin,Citation15 the L19 antibody (specific to the alternatively-spliced EDB of fibronectinCitation16–Citation18) and the F16 antibody (specific to the alternatively-spliced A1 domain of tenascin-CCitation19) have exhibited an impressive potential to selectively localize at neo-vascular structures in solid tumors and in lymphomas.Citation20 The three antibodies were isolated from the ETH2-GOLD synthetic antibody libraryCitation11,Citation21 and affinity-matured using a procedure featuring the combinatorial mutagenesis of residues in CDR1 and CDR2 loops and stringent selections on immobilized antigen. Indeed, the ETH2-GOLD library had been constructed using the most-commonly used VH germline segment (DP47,Citation22) and the frequently used Vκ and Vλ germline segments DPK22 and DPL16,Citation23,Citation24 with combinatorial mutagenesis only for residues of the CDR3 loops of heavy and light chain.Citation11
Over the years, the ETH2-GOLD library has proven to be a reliable source of good-quality human mAbs.Citation11,Citation15,Citation19,Citation25–Citation32 However, as for other libraries, a degree of epitope bias may result from the library design. For example, we were so far unable to isolate further human mAbs from the ETH2-GOLD library that do not overlap with the clinical-stage F8 antibody. We reasoned that the construction of a synthetic antibody library, which closely resembled the design of ETH2-GOLD but which would incorporate charged or hydrophilic residues at central positions of the antigen binding site, could be used for the isolation of mAbs to novel epitopes in target proteins of interest. Indeed, a similar strategy had previously been used to drive antibody selections against certain epitopes on target antigens with defined arrangements of charged residues on their surface.Citation33
Here, we describe the design and construction of a novel antibody phage display library (termed PHILO library) that was used for the isolation of good-quality binding specificities against a variety of different antigens. Interestingly, the PHILO library allowed the isolation of human antibodies specific to an epitope on the EDA domain of fibronectin, which did not overlap with the one recognized by the clinical-stage F8 antibody and which stained tumor neo-vascular structures in frozen cancer sections. One of these antibodies, termed 2H7, was fused to F8 in scFv-scFv formatCitation34,Citation35 by means of two different linkers. The resulting fusion proteins, named chelating recombinant antibodies (CRAbs),Citation34,Citation36 were produced in mammalian cells and characterized for their tumor homing properties using quantitative biodistribution analysis in tumor-bearing mice.
Results
Library design, construction and characterization.
To generate a large, stable and highly diverse library of functional antibody fragments with similar physical properties, we cloned a synthetic antibody library in a phagemid vector. Sequence diversity was restricted to the complementarity-determining region (CDR) 3 loops of the variable heavy (VH) and light chain (VL) domains (). The scFv format was selected because it provides better expression yields and superior phage display properties compared with the Fab format. The scFv antibody scaffold used for the library was based on the germline VH segment DP47 and either the Vκ segment DPK22 or the Vλ segment DPL16 (), which represent 12, 25 and 16%, of the antibody repertoire in humans, respectively.Citation6 This approach offers a number of advantages, e.g., higher thermodynamic stability,Citation37 possible use of Protein A for antibody purification and detection.Citation4 Sequence randomization was limited to the CDR3 loops, which are known to contribute substantially to antigen recognition (). A completely randomized sequence of four, five, six or seven amino acid residues (followed by the conserved Phe-Asp-Tyr sequence) was appended by means of polymerase chain reaction (PCR) to the VH germline segment (), thus forming short CDR3 loops, whereas a partially randomized sequence of six amino acid residues was appended to the VL, forming CDR3 loops that contained at least one proline residue (. The flexible polypeptide Gly4SerGly4SerGly4,Citation38 was used to link the VH chain to the VL chain in the scFv fragments. Additionally, residue 52 in the CDR2 loop of the VH domain was allowed to be either two charged amino acids (Asp, Lys in PHILO1 sub-library) or two hydrogen bond-prone amino acids (Asn, Tyr in PHILO2 sub-library). The sub-libraries could be used together by mixing the corresponding phage preparations, resulting in the PHILO library, which contained 3.1 × 109 different antibody clones.
To assess the quality of the PHILO library, a number of tests were performed. First, 96/96 randomly chosen colonies exhibited the presence of an insert of the correct size in PCR screening experiments (). Furthermore, 26 randomly picked clones were shown to contain the correct sequence and randomized residues at the desired positions (Sup. Table 1). A dot blot analysis, based on the detection of the peptidic myc tag at the C-terminus of the recombinant antibodies, confirmed that more than 85% of library clones could be expressed as soluble scFv fragments in the bacterial supernatant ().
The PHILO library was screened against a panel of nine different antigens and in two competition selections, yielding several binders in all cases after two or three rounds of panning (). Except for the first four antigens reported in (GST, BCD, 11A12 and 7B89), which were used as standard selections, most of the different antibodies were further characterized by surface plasmon resonance analysis for KD determination. Some clones specific for fibronectin domain, filtrin, nephrin and BstII were also used as immunohistochemical tools for antigen detection in human tumor sections (data not shown). To assess the epitope variability that can be recognized by our newly designed antibody library, we attempted to isolate antibodies specific for two new epitopes of EDA and EDB of human fibronectin that did not compete with our previously described F8 antibody and L19 antibody, respectively.Citation15–Citation17 In the case of anti-EDB antibodies we selected three different clones from the PHILO library and one from the ETH2-GOLD library (data not shown). For the EDA domain, we isolated one antibody specific for a new epitope from only the PHILO library.
Anti-EDA binders.
A recombinant fibronectin fragment, constituted by the three type-III domains 11, EDA and 12 (hereafter termed “11-A-12”Citation15) was used in panning experiments with both the ETH2-GOLD libraryCitation11 and the PHILO library, using the F8 antibody as competitor during selections to drive the isolation of binders that recognize alternative non-overlapping epitopes. After two rounds of panning, 21/94 ELISA-positive clones were recovered from the PHILO library and 45/94 from the ETH2-GOLD library. Characterization of the mAbs in terms of (1) EDA recognition and (2) absence of competition with F8 indicated that only clone F10 from the PHILO library could bind to EDA without competing with F8 ( and ). and B present the results of competition ELISA and BIAcore experiments with subsequent antibody injections, using the F10 antibody or B7 (a different high-affinity anti-EDA antibody isolated from the phage selectionsCitation15 that maps on the same epitope of the previously described F8) in the presence of F8. Only the F10 antibody could recognize EDA in the presence of F8, while the binding of B7 was completely abrogated. Furthermore, F8 in the small immunoprotein (SIP) format and F10 in scFv format were able to form a triple complex in the presence of 11A12, as assessed by size-exclusion chromatography (SEC) analysis (). The measured KD value for scFv(F10) was 290 nM, prompting the use of an affinity maturation procedure by means of mutagenesis-based technique previously described by our groupCitation11,Citation15 (see also Materials and Methods), yielding clone 2H7 with a Kd = 50 nM dissociation constant to the cognate antigen as measured by BIAcore (). The 2H7 antibody retained the ability of the parental antibody to recognize EDA in the presence of F8 (data not shown).
Production and characterization of chelating recombinant recombinant anti-EDA antibodies (CRAbs).
To generate tandem scFv-scFv molecules that would work as CRAbs specific to the EDA domain of fibronectin, we sequentially fused scFv(F8) and scFv(2H7) using two different flexible polypeptides of 10 and 18 amino acids as linkers joining the two antibodies. The resulting bispecific antibodies were termed CRAb(F8-10aa-2H7) and CRAb(F8-18aa-2H7) (). SEC showed that both anti-EDA CRAbs were able to elute as a main peak at the same retention volume of scFv(F8)-5amino acid linkerCitation15 (). To test the tumor targeting performance of the new bispecific antibodies, purified preparations of CRAb(F8-10aa-2H7) and CRAb(F8-18aa-2H7) were radio-labeled with 125I and injected intravenously (i.v.) in 129SvEv immunocompetent mice bearing subcutaneously-grafted F9 teratocarcinomas. ScFv(F8) and scFv(2H7) in diabody formatCitation15 were used as positive controls of proven targeting ability and comparable molecular weight. Although the absolute tumor uptake of scFv(2H7)-diabody was lower compared with that of scFv(F8)-diabody, this antibody also exhibited a favorable tumor to organ ratio profile (). Both anti-EDA CRAbs displayed a preferential accumulation at the tumor site with comparable levels in the neoplastic masses (). Twenty-four hours after i.v. injection, scFv(F8) and CRAb(F8-18aa-2H7) exhibited comparable tumor to blood ratios of 10.2 and 8.8, respectively. Furthermore, CRAb(F8-18aa-2H7) revealed a long residence time of the antibody fragment at the tumor site, with low background in healthy organs.
Discussion
We have reported the design, construction and characterization of the new synthetic human antibody library (i.e., the PHILO library), containing more than three billion different antibody clones. The previously generated human antibody library ETH2-GOLDCitation11 has been used for isolation of antibodies against more than 100 different antigens. The PHILO synthetic antibody library was generated to expand the structural features and epitope variability that can be recognized by our antibody fragments. The new library design capitalizes on the experience gained with the ETH2-GOLD library, but contains point mutations at position 52 of the VH CDR2 loop that help bias antibody selection towards certain protein epitopes. The PHILO library allowed the isolation of human antibodies specific to a novel epitope in the EDA domain of fibronectin, which we had previously not been able to obtain with existing libraries, and it has now allowed the construction of tumor-targeting CRAbs.
Among the various tumor-associated extra-cellular-matrix (ECM) components, oncofetal fibronectin isoforms are often recognized as ideal targets for antibody-based tumor targeting applications due to their high stability, ready accessibility and abundance.Citation39 The EDA and EDB domains of fibronectin are usually absent in both plasma and tissue fibronectin of adults, but they can be inserted into the fibronectin molecule during active tissue remodeling events, such as angiogenesis, wound healing and embryogenesis.Citation40,Citation41 The anti-EDB L19 antibodyCitation16–Citation18 and the anti-EDA F8 antibody have been extensively studied as delivery vehicles in pre-clinical models of cancer and chronic inflammation.Citation41–Citation45 The L19 antibody fused to the pro-inflammatory cytokines IL-2 or TNF or labeled with the β-emitting 131I radionuclide, is currently being evaluated in Phase 2 clinical trials.Citation46–Citation48
In vivo biodistribution experiments have confirmed that both anti-EDA CRAb(F8-10aa-2H7) and CRAb(F8-18aa-2H7) antibodies were able to selectively accumulate at the tumor site. Subtle variations in linker length and amino acid composition have a small but measureable influence on the %ID/g values measured in tumors and in normal organs ().
Bispecific antibodies have been used extensively in biomedical research, particularly with the aim of bridging an effector cell (i.e., cytotoxic T lymphocyte by CD3 co-receptor engagement) with a tumor cell by means of tumor-associated antigens such as CD19,Citation49,Citation50 epithelial cell adhesion molecule (EpCAM),Citation51 carcino-embryonic antigen (CEA),Citation52,Citation53 human epidermal growth factor receptor 2 (HER2)/neuCitation54,Citation55 or epidermal growth factor receptor (EGFR).Citation56,Citation57 Multi-specific antibodies, such as scFv-scFv tandem antibodies, can also be used as modular building blocks for the development of biopharmaceuticals. Multi-specific immunotoxins capable of simultaneous binding to both EpCAM and HER2neu exhibited superior tumor cell killing properties and decreased toxicity in a mouse model of cancer.Citation58 Even though our targeting results obtained with the CRAb(F8-18aa-2H7) are encouraging, further work is needed to evaluate whether CRAbs may display superior targeting performance when compared to homobivalent antibodies. In our biodistribution studies, anti-EDA CRAbs and diabodies exhibited comparable tumor targeting performance ().
In summary, we believe that our new synthetic antibody PHILO library represents a useful source for the isolation of novel mAb fragments specific for virtually any type of antigen. Furthermore, scFv-scFv tandem antibodies may be useful modular building blocks of proven tumor targeting performance for the development of improved anti-cancer biopharmaceuticals.
Materials and Methods
Library construction and cloning.
Two clones from the ETH2-Gold libraryCitation11 were used as template for PCR amplification of DP47, DPK22 and DPL16. Antibody residues are numbered according to TomlinsonCitation22 and Cox.Citation24 First, the point mutation at position 52 of heavy chain was introduced by PCR using primers LMB3long, S52Drev, S52Krev, S52Nrev, S52Yrev, S52fw, i (; all primers used for the construction of the library were purchased from Operon Biotechnologies). The G4SG4SG4 linker between heavy and light chain was not changed compared to the ETH2-GOLD library. Sequence variability in both heavy and light chain was focused in CDR3 loops and introduced by PCR using partially degenerated primers (), essentially as described in Silacci et al.Citation11 Briefly, DP47-based VH domains were randomly mutated from residue 95 to 100; this CDR3 loop was designed to be 4–7 amino acids long. DPK22-based VL domains were randomized between residues 91–96, with a fixed glycine either at position 92 or 93. Furthermore, at least one of the CDR3 loop residues was requested to be a proline.
Randomized CDR3 loops were appended to germline segments (from framework1 to framework3) by PCR using primer pairs a/b1-4 for DP47 heavy chain, c/d1-2 for DPK-22 light chain, and c/e1-5 for DPL-16 light chain ( and ). After gel-purification, heavy and light chain segments were assembled by PCR and further amplified using primers a/f for DPK-22 or a/g for DPL-16. The resulting scFv genes were doubly digested with NcoI/NotI and cloned into the NcoI/NotI-digested expression vector pHEN1.Citation4 The ligation mixture was purified and electroporated into fresh electrocompetent TG-1 cells. Electrocompetent TG-1 cells were prepared by washing the cells twice with 1 mM HEPES/5% glycerol and twice with 10% glycerol in water. Finally, cells were resuspended in 10% glycerol to a density of approximately 2 × 1011 cells. Electroporated cells were spread on 2TY-agar plates (2xTY: 16 g/L bacto-tryptone, 10 g/L bacto-yeast extract, 5 g/L NaCl pH 7.4–15 g/L agar-100 µg/mL ampicillin, 1% glucose) and incubated at 30°C overnight. On the next day, cells were rescued (with 2TY-10% glycerol), used for phage production according to standard protocolCitation21 and stored as glycerol stocks. The sublibraries were electroporated in eight different days, yielding a total library size of 3.1 × 109 antibody clones ().
Library characterization: PCR screening, sequencing of antibody clones, dot blot.
A total of 96 clones were tested by PCR screening with REDTaq Ready Mix PCR reaction mix (Sigma Aldrich GmBH, cat. num. R2523-100RXN) using primers a/i (annealing about 100 bp upstream/downstream of the scFv gene, ). Antibodies were sequenced using Big Dye® Terminator v1.1 Cycle Sequencing kit (Applied Biosystems, cat. num. 4337455) on an ABI PRISM 3130 Genetic analyzer. Termination reactions were performed on PCR screening products using primers a or i (). Dot Blot analysis was performed as described in Villa 2008,Citation15 individual colonies from the plated library were inoculated in 160 µL 2xTY-100 µg/mL ampicillin-0.1% glucose in Nunclon™ Surface 96-well plates (Nunc, cat. num. 163320). The plates were incubated 3 h at 37°C in a shaker incubator. Expression was induced by addition of 1 mM IPTG and cultures were grown overnight at 30°C. ScFv-containing supernatants were blotted onto Protran B85 0.45 µm nitrocellulose membrane (Whatman, cat. num. 10401197) using the ELIFA system (Pierce, cat. num. 77000), which allowed blotting by vacuum in a 96-well format, leading to spots of uniform and reproducible size. ScFv was detected with monoclonal anti-myc tag murine antibody 9E10,Citation59 followed by anti-mouse IgG horseradish peroxidase conjugate (Sigma Aldrich, A2554). Peroxidase activity was detected using the ECL plus western blotting detection system (Amersham Biosciences, GE Healthcare, cat. num. RPN2132) on Amersham Hyperfilm ECL (Amersham GE, cat. num. 28-9068-35).
Antibody selection.
All selections were performed as described in Silacci 2005,Citation11 using recombinant antigen with >90% purity as assessed by SDS-PAGE and SEC. Immunotubes (Nunc, cat. num. 470319) were coated with antigen at 10−6 M concentration in phosphate buffered saline (PBS) overnight at room temperature (RT) and blocked for 2 h at RT with 2% w/v skimmed milk in PBS (MPBS). After rinsing with PBS, >1012 phage particles were added to the antigen-coated immunotube in the presence of 2% MPBS, incubated for 30 min at room temperature with shaking (100 rpm), followed by 1.5 h incubation at RT without agitation. In competitive selections, phage particles were added to the antigen-coated immunotube in the presence of 2% MPBS and 10−5 M of competing antibody (SIP(L19) for anti-EDB selection and SIP(F8) for anti-EDA selection). Unbound phage were washed with PBS Tween 0.1% (10 or 20 times) and PBS (10 or 20 times), while bound phage were eluted with 100 mM triethylamine (TEA). Eluted phage were then used for infection of exponentially growing E. coli TG1. After 2 rounds of panning, ELISA screening was performed on 92–94 individual colonies as previously described (Silacci 2005).
ELISA screening and sandwich ELISA.
Bacterial supernatants containing scFv fragments were screened for antigen binding ability by ELISA as described in reference Citation21. Individual colonies were inoculated in 180 µL 2x TY, 100 µg/mL ampicillin (Applichem, cat. num. A0839, 0025), 0.1% glucose (Sigma Aldrich) in NunclonTM Surface 96-well plates (Nunc, cat. num. 163320). The plates were incubated 3 h at 37°C in a shaker incubator. The cells were then induced with isopropyl-thio-galactopyranoside (IPTG; Applichem, cat. num. A1008, 0050) at a final concentration of 1 mM and grown overnight at 30°C. The bacterial supernatants assayed were tested in ELISA experiments as described in reference 60 using the anti-myc tag 9E10 mAb (Sigma, cat. num. M4439) and anti-mouse horseradish peroxidase (HRP) immunoglobulins (Sigma Aldrich, cat. num. A2554) as secondary reagents. For the colorimetric reaction, 100 µL BM-Blue POD substrate (Roche, cat. num. 11484281001) followed by 50 µL 1 M H2SO4 were added to each well. The absorbance was measured using the microtiter plate reader (VersaMax, Molecular Devices) at wavelengths 450 nm and 650 nm. Sandwich ELISA was performed by coating 100 µL (100 µg/mL) anti-EDA antibody SIP(F8)Citation15 directly on Maxisorp (NUNC, cat. num. 439454), followed by addition of 100 µl 11A12 triple domain of fibronectinCitation15 at a concentration equal to 10−5 M. Subsequently, scFv(F10) or scFv(B7),Citation15 were added at a concentration of 50 µg/mL. The anti-myc tag 9E10 antibody and anti-mouse HRP were used for ELISA development.
BIAcore.
Surface plasmon resonance analysis was carried out with BIAcore 3000 system (BIAcore, GE Healthcare) using an 11A12 triple domain of human fibronectin-coated CM5 chip (BIAcore, cat. num. BR-1000-14) at 10 µL/min flow rate. Twenty microliters of SIP(F8) were injected at a 0.1 mg/mL concentration to achieve binding site saturation of the antigen-coated chip. Subsequently, 20 µL of scFv(F10) or scFv(B7) were injected at the same concentration of 0.1 mg/mL. Regeneration of the chip was performed by injecting 20 µL of HCl 10 mM.
Size-exclusion chromatography.
Purified antibody fragments and antigen mixtures were analyzed by SEC on a Superdex 75 10/300 GL column on an ÄKTA FPLC (GE Healthcare, Amersham Biosciences). Before the analysis, column elution volumes were calibrated using different commercially-available protein standards for gel filtration calibration, such as ribonuclease A as 13.7 kDa reference (elution volume (Ve) = 13.346 mL), ovalbumin as 43 kDa reference (Ve = 10.095 mL), BSA as 67 kDa reference (Ve = 9.232 mL) and blue dextran for void volume (V0) calculation (V0 = 7.72 mL) (Amersham Biosciences, cat. num. 28-4038-42). The calibration graph was obtained by plotting Kav (defined as Ve − V0/Vt − V0, given a total column volume (Vt) = 25 mL), versus molecular weights (MW) of the standards. The equation of the trend line approximating the points [MW; Kav] is y = −5E−06x + 0.3702 (R2 = 0.93). Before FPLC analysis, protein mixtures were incubated at room temperature for 20 min to allow binding equilibrium to be reached.
Affinity maturation of scFv(F10).
The affinity maturation of scFv(F10) was performed according to a previously described strategy.Citation15,Citation19,Citation25 An affinity maturation library was cloned by introducing sequence variability in the CDR1 loops of both heavy (at position 31–33) and light chains (at position 31a, 31b, 32). Partially degenerate primers used for library construction are listed in . First, three fragments were obtained by PCR on the parental clone scFv(F10) as template, using primer pairs a/DP74CDR1rev, DP74CDR1fw/DPL16CDR1rev and DPL16CDR1fw/i (). After gel-purification, these three segments were assembled by PCR and further amplified using primers a/i (). The VH-VL combinations were doubly digested with NcoI/NotI and cloned into the NcoI/NotI-digested expression vector pHEN1. ScFv(2H7) was isolated by selection on Immunotube, followed by an ELISA screening procedure as reported above.
CRAbs cloning.
CRAb genes were constructed by PCR amplification of the scFv(F8)Citation15 and scFv(2H7), followed by PCR assembly of the two obtained DNA fragments. ScFv(F8) was amplified using the primers CRAb1/CRAb2 for F8-10aa-2H7 and CRAb1/CRAb3 for CRAb(F8-18aa-2H7).Citation34 The myc-tagged scFv(2H7) gene was amplified using primer CRAb4/CRAb6 (annealing on C-terminal myc-tag) for F8-10aa-2H7 and CRAb5/CRAb6 for CRAb(F8-18aa-2H7). The obtained scFv genes were assembled by PCR, double digested with NheI/HindIII and ligated in double digested pCDNA3.1.
Expression and purification of CRAbs and scFv(F8)-5aa linker in CHO-S cells.
Recombinant antibody fragments were expressed in suspension-adapted CHO-S cells by means of transient gene expression technique.Citation61 Antibody fragments were purified from culture supernatants by affinity chromatography, using protein A Sepharose (GE-Healthcare, cat. num. 17-5280-04), followed by elution either in glycine 100 mM pH 3 or in 100 mM TEA, depending on the pI of the eluted protein. Purified antibody fragments were analyzed by SEC on a Superdex 75 HR 10/30 column (GE-Healthcare, cat. num. 17-5174-01). Protein concentration was estimated from optical density measurements at 280 nm and SDS-PAGE analysis.
Tumor cell line and mouse model.
F9 murine teratocarcinoma cells (CRL-1720, ATCC) were cultured according to ATCC guidelines. F9 tumor-bearing mice were obtained by injecting s.c. 106 F9 teratocarcinoma cells in 10 week-old female 129SvE mice (provided by Taconic, Ry, Denmark), as described in reference Citation15.
Biodistribution studies.
The in vivo targeting performance of anti-EDA antibody fragments was evaluated by quantitative biodistribution analysis as previously described by Tarli et al.Citation18 Briefly, antibodies were first loaded on Superdex 75, and the peaks corresponding to the desired molecular weight (according to the equation reported above) were collected. Purified antibody preparations were then radiolabeled with 125I using the chloramineT methodCitation62 and injected into the tail vein of immunocompetent 129SvEv mice bearing s.c. implanted F9 murine teratocarcinoma (8–10 µg per mouse). Mice were sacrificed 24 and 48 h after injection. Organs were weighed and radioactivity was counted with a PackardCobra gamma counter. Radioactivity content of representative organs was expressed as the percentage of the injected dose per gram of tissue (%ID/g). Data were corrected for tumor growth correction.
Figures and Tables
Figure 1 Design and cloning strategy for the PHILO antibody library. (A) Three-dimensional structure of a scFv antibody fragment. Heavy chain and light chain backbone are represented in dark grey and light grey, respectively. Residues subject to random mutation are DP47 CDR3 position 95–100 (dark grey space-fill representation), light chain position 91 to 96 (light grey space-fill representation) (in more details, for DPK22 CDR3 residue 91–96, and for DPL16 CDR3 position 92–95 and 95b). The scFv structure was displayed using the program PyMol, based on the protein data base (Brookhaven Protein Data Bank) file 1igm. The residue numbers were defined as previously published in reference Citation23 and Citation63. (B) Library cloning strategy. A point mutation was introduced, converting residue S52 of VH to D, K, N or Y. Mutagenesis in the CDR3 regions was generated by PCR using partially degenerate primers. Genes are indicated as rectangles and CDRs as numbered boxes. The VH and VL segment were then assembled by PCR and cloned into the pHEN1 vector.Citation64 Primers used in the amplification and assembly are listed in .
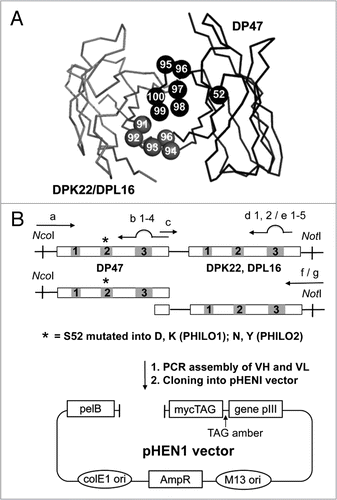
Figure 2 Characterization of the PHILO library. (A) PCR colony screening of 12 clones of each sub-library. As negative control a BirA insert (1,200 bp) of a pHEN1 vector was amplified and a scFv in pHEN1 was amplified as a positive control. All the tested clones showed an insert with the correct size of approximately 1,000 bp. (B) Dot blot analysis of 752 induced supernatants of individual library clones. The soluble scFv fragments were detected with the anti-myc-tag mAb 9E10. More than 90% of the clones express a detectable amount of soluble scFv fragment.
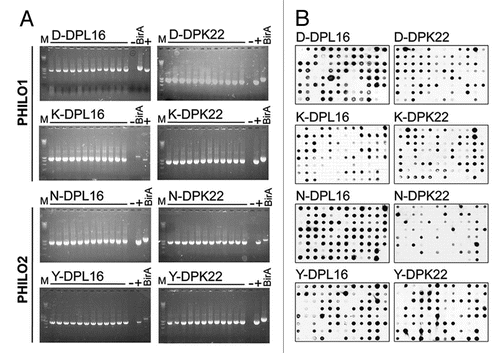
Figure 3 Experimental evidence of non-overlapping epitopes on EDA, recognized by the scFv(F8) and scFv(2H7) antibodies. (A) ELISA and sandwich ELISA results are depicted in dark grey and light grey column, respectively, using microtiter plates coated with the recombinant 11A12 fragment of fibronectin (ELISA) or the F8 antibody (sandwich ELISA). (B) BIAcore experiments were performed on 11A12-coated CM5 chips. The F8 antibody in the small immunoprotein (SIP) formatCitation15 was injected at saturation, followed by injection of either scFv(F10) (black line) or the negative control scFv(B7) antibody (light grey). The EDA epitope recognized by scFv(B7) is identical to the one recognized by the F8 antibody. (C) Triple complex formation of SIP(F8), 11A12 and scFv(F10) in size exclusion chromatography on Superdex 200. Peak 1 corresponds to the triple complex made by SIP(F8) + 11A12 + scFv(F10); peak 2 corresponds to SIP(F8) + 11A12; peak 3 corresponds to scFv(F10) + 11A12; peak 4 corresponds to 11A12 antigen alone.
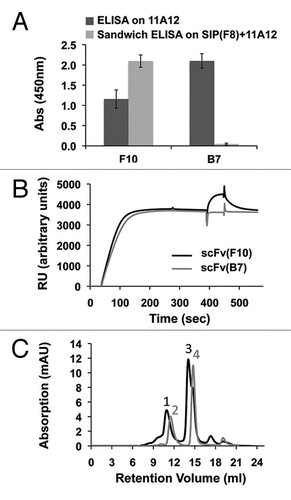
Figure 4 Biodistribution studies in tumor bearing mice of scFv(F8) and scFv(A9) in diabody format (A) and CRAbs (B). Both parts report the schematic representation of antibody structure, the size exclusion analysis of antibody fragments on Superdex75 (Amersham, GE) (all antibodies have a main elution peak around 10 mL, which corresponds to the expected size of 52–54 kDa) and the biodistribution analysis displayed as percent injected dose per gram (%ID/g) in tumor (Tu) and in measured mice organs: liver (Li), lung (Lu), spleen (Sp), heart (He), kidney (Ki), intestine (Int), blood (Blo). Radioactivity in tumor and organs have been counted at 24 (dark grey) and 48 (light grey) hours post intravenous injection of radiolabeled antibodies. In (B), in the SEC profile of F8-10aa linker-2H7, the peak corresponding to the monomeric CRAb is marked. Small grey arrows indicate the retention volumes of the standard proteins (from left to right: blue dextran, bovine serum albumin, ovalbumin, scFv(F8), ribonuclease A).
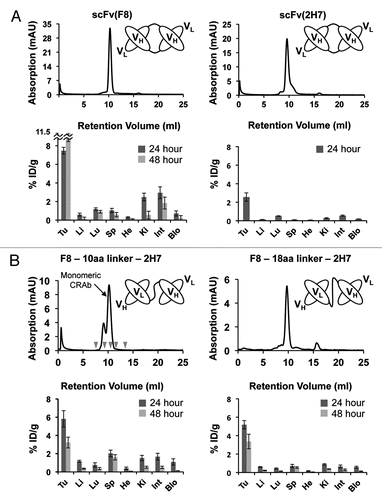
Table 1 Sequence of synthetic primers
Table 2 Summary of test selections with PHILO and ETH2-GOLD libraries
Table 3 Sequences of the scFv CDRs specific to the extra-domain A of fibronectin
Additional material
Download Zip (60.5 KB)Acknowledgments
Financial contributions from the Swiss National Science Foundation, the ETH Zürich, the European Union (ADAMANT and IMMUNO-PDT Projects), the Swiss Cancer League, the Swiss-Bridge Foundation and the Stammbach Foundation are gratefully acknowledged.
References
- Walsh G. Biopharmaceutical benchmarks 2006. Nat Biotechnol 2006; 24:769 - 776
- Walsh G. Biopharmaceutical benchmarks 2010. Nat Biotechnol 2010; 28:917 - 924
- Winter G, Griffiths AD, Hawkins RE, Hoogenboom HR. Making antibodies by phage display technology. Annu Rev Immunol 1994; 12:433 - 455
- Hoogenboom HR, Winter G. By-passing immunisation. Human antibodies from synthetic repertoires of germline VH gene segments rearranged in vitro. J Mol Biol 1992; 227:381 - 388
- Nissim A, Hoogenboom HR, Tomlinson IM, Flynn G, Midgley C, Lane D, et al. Antibody fragments from a ‘single pot’ phage display library as immunochemical reagents. EMBO J 1994; 13:692 - 698
- Griffiths AD, Williams SC, Hartley O, Tomlinson IM, Waterhouse P, Crosby WL, et al. Isolation of high affinity human antibodies directly from large synthetic repertoires. EMBO J 1994; 13:3245 - 3260
- Vaughan TJ, Williams AJ, Pritchard K, Osbourn JK, Pope AR, Earnshaw JC, et al. Human antibodies with sub-nanomolar affinities isolated from a large non-immunized phage display library. Nat Biotechnol 1996; 14:309 - 314
- Knappik A, Ge L, Honegger A, Pack P, Fischer M, Wellnhofer G, et al. Fully synthetic human combinatorial antibody libraries (HuCAL) based on modular consensus frameworks and CDRs randomized with trinucleotides. J Mol Biol 2000; 296:57 - 86
- Soderlind E, Strandberg L, Jirholt P, Kobayashi N, Alexeiva V, Aberg AM, et al. Recombining germline-derived CDR sequences for creating diverse single-framework antibody libraries. Nat Biotechnol 2000; 18:852 - 856
- O'Connell D, Becerril B, Roy-Burman A, Daws M, Marks JD. Phage versus phagemid libraries for generation of human monoclonal antibodies. J Mol Biol 2002; 321:49 - 56
- Silacci M, Brack S, Schirru G, Marlind J, Ettorre A, Merlo A, et al. Design, construction and characterization of a large synthetic human antibody phage display library. Proteomics 2005; 5:2340 - 2350
- den Broeder A, van de Putte L, Rau R, Schattenkirchner M, Van Riel P, Sander O, et al. A single dose, placebo controlled study of the fully human anti-tumor necrosis factor-alpha antibody adalimumab (D2E7) in patients with rheumatoid arthritis. J Rheumatol 2002; 29:2288 - 2298
- Schliemann C, Neri D. Antibody-based vascular tumor targeting. Recent Results Cancer Res 2010; 180:201 - 216
- Schliemann C, Neri D. Antibody-based targeting of the tumor vasculature. Biochim Biophys Acta 2007; 1776:175 - 192
- Villa A, Trachsel E, Kaspar M, Schliemann C, Sommavilla R, Rybak JN, et al. A high-affinity human monoclonal antibody specific to the alternatively spliced EDA domain of fibronectin efficiently targets tumor neo-vasculature in vivo. Int J Cancer 2008; 122:2405 - 2413
- Pini A, Viti F, Santucci A, Carnemolla B, Zardi L, Neri P, et al. Design and use of a phage display library. Human antibodies with subnanomolar affinity against a marker of angiogenesis eluted from a two-dimensional gel. J Biol Chem 1998; 273:21769 - 21776
- Viti F, Tarli L, Giovannoni L, Zardi L, Neri D. Increased binding affinity and valence of recombinant antibody fragments lead to improved targeting of tumoral angiogenesis. Cancer Res 1999; 59:347 - 352
- Tarli L, Balza E, Viti F, Borsi L, Castellani P, Berndorff D, et al. A high-affinity human antibody that targets tumoral blood vessels. Blood 1999; 94:192 - 198
- Brack SS, Silacci M, Birchler M, Neri D. Tumor-targeting properties of novel antibodies specific to the large isoform of tenascin-C. Clin Cancer Res 2006; 12:3200 - 3208
- Schliemann C, Wiedmer A, Pedretti M, Szczepanowski M, Klapper W, Neri D. Three clinical-stage tumor targeting antibodies reveal differential expression of oncofetal fibronectin and tenascin-C isoforms in human lymphoma. Leuk Res 2009; 33:1718 - 1722
- Viti F, Nilsson F, Demartis S, Huber A, Neri D. Design and use of phage display libraries for the selection of antibodies and enzymes. Methods Enzymol 2000; 326:480 - 505
- Tomlinson IM, Walter G, Marks JD, Llewelyn MB, Winter G. The repertoire of human germline VH sequences reveals about fifty groups of VH segments with different hypervariable loops. J Mol Biol 1992; 227:776 - 798
- Williams SC, Frippiat JP, Tomlinson IM, Ignatovich O, Lefranc MP, Winter G. Sequence and evolution of the human germline Vlambda repertoire. J Mol Biol 1996; 264:220 - 232
- Cox JP, Tomlinson IM, Winter G. A directory of human germ-line Vkappa segments reveals a strong bias in their usage. Eur J Immunol 1994; 24:827 - 836
- Silacci M, Brack SS, Spath N, Buck A, Hillinger S, Arni S, et al. Human monoclonal antibodies to domain C of tenascin-C selectively target solid tumors in vivo. Protein Eng Des Sel 2006; 19:471 - 478
- Ettorre A, Rosli C, Silacci M, Brack S, McCombie G, Knochenmuss R, et al. Recombinant antibodies for the depletion of abundant proteins from human serum. Proteomics 2006; 6:4496 - 4505
- Ahlskog JK, Schliemann C, Marlind J, Qureshi U, Ammar A, Pedley RB, et al. Human monoclonal antibodies targeting carbonic anhydrase IX for the molecular imaging of hypoxic regions in solid tumours. Br J Cancer 2009; 101:645 - 657
- Pfaffen S, Hemmerle T, Weber M, Neri D. Isolation and characterization of human monoclonal antibodies specific to MMP-1A, MMP-2 and MMP-3. Exp Cell Res 2010; 316:836 - 847
- Sgier D, Zuberbuehler K, Pfaffen S, Neri D. Isolation and characterization of an inhibitory human monoclonal antibody specific to the urokinase-type plasminogen activator, uPA. Protein Eng Des Sel 2010; 23:261 - 269
- Piazza T, Cha E, Bongarzone I, Canevari S, Bolognesi A, Polito L, et al. Internalization and recycling of ALCAM/CD166 detected by a fully human single-chain recombinant antibody. J Cell Sci 2005; 118:1515 - 1525
- Avignolo C, Bagnasco L, Biasotti B, Melchiori A, Tomati V, Bauer I, et al. Internalization via Antennapedia protein transduction domain of an scFv antibody toward c-Myc protein. FASEB J 2008; 22:1237 - 1245
- Lovato V, Roesli C, Ahlskog J, Scheuermann J, Neri D. A monoclonal antibody prevents aggregation of the NBD1 domain of the cystic fibrosis transmembrane conductance regulator. Protein Eng Des Sel 2007; 20:607 - 614
- Kirkham PM, Neri D, Winter G. Towards the design of an antibody that recognises a given protein epitope. J Mol Biol 1999; 285:909 - 915
- Neri D, Momo M, Prospero T, Winter G. High-affinity antigen binding by chelating recombinant antibodies (CRAbs). J Mol Biol 1995; 246:367 - 373
- Wolf E, Hofmeister R, Kufer P, Schlereth B, Baeuerle PA. BiTEs: bispecific antibody constructs with unique anti-tumor activity. Drug Discov Today 2005; 10:1237 - 1244
- Wright MJ, Deonarain MP. Phage display of chelating recombinant antibody libraries. Mol Immunol 2007; 44:2860 - 2869
- Ewert S, Huber T, Honegger A, Pluckthun A. Biophysical properties of human antibody variable domains. J Mol Biol 2003; 325:531 - 553
- Huston JS, Levinson D, Mudgett-Hunter M, Tai MS, Novotny J, Margolies MN, et al. Protein engineering of antibody binding sites: recovery of specific activity in an anti-digoxin single-chain Fv analogue produced in Escherichia coli. Proc Natl Acad Sci USA 1988; 85:5879 - 5883
- Kaspar M, Zardi L, Neri D. Fibronectin as target for tumor therapy. Int J Cancer 2006; 118:1331 - 1339
- Ffrench-Constant C, Van de Water L, Dvorak HF, Hynes RO. Reappearance of an embryonic pattern of fibronectin splicing during wound healing in the adult rat. J Cell Biol 1989; 109:903 - 914
- Schwager K, Kaspar M, Bootz F, Marcolongo R, Paresce E, Neri D, et al. Preclinical characterization of DEKAVIL (F8-IL10), a novel clinical-stage immunocytokine which inhibits the progression of collagen-induced arthritis. Arthritis Res Ther 2009; 11:142 - 1
- Frey K, Schliemann C, Schwager K, Giavazzi R, Johannsen M, Neri D. The immunocytokine F8-IL2 improves the therapeutic performance of sunitinib in a mouse model of renal cell carcinoma. J Urol 2010; 184:2540 - 2548
- Kaspar M, Trachsel E, Neri D. The antibody-mediated targeted delivery of interleukin-15 and GM-CSF to the tumor neovasculature inhibits tumor growth and metastasis. Cancer Res 2007; 67:4940 - 4948
- Sommavilla R, Pasche N, Trachsel E, Giovannoni L, Roesli C, Villa A, et al. Expression, engineering and characterization of the tumor-targeting heterodimeric immunocytokine F8-IL12. Protein Eng Des Sel 2010; 23:653 - 661
- Trachsel E, Bootz F, Silacci M, Kaspar M, Kosmehl H, Neri D. Antibody-mediated delivery of IL-10 inhibits the progression of established collagen-induced arthritis. Arthritis Res Ther 2007; 9:9
- Neri D, Bicknell R. Tumour vascular targeting. Nat Rev Cancer 2005; 5:436 - 446
- Sauer S, Erba PA, Petrini M, Menrad A, Giovannoni L, Grana C, et al. Expression of the oncofetal ED-B-containing fibronectin isoform in hematologic tumors enables ED-B-targeted 131I-L19SIP radioimmunotherapy in Hodgkin lymphoma patients. Blood 2009; 113:2265 - 2274
- Johannsen M, Spitaleri G, Curigliano G, Roigas J, Weikert S, Kempkensteffen C, et al. The tumour-targeting human L19-IL2 immunocytokine: preclinical safety studies, phase I clinical trial in patients with solid tumours and expansion into patients with advanced renal cell carcinoma. Eur J Cancer 2010; 46:2926 - 2935
- Dreier T, Baeuerle PA, Fichtner I, Grun M, Schlereth B, Lorenczewski G, et al. T cell costimulus-independent and very efficacious inhibition of tumor growth in mice bearing subcutaneous or leukemic human B cell lymphoma xenografts by a CD19-/CD3-bispecific single-chain antibody construct. J Immunol 2003; 170:4397 - 4402
- Gruen M, Bommert K, Bargou RC. T-cell-mediated lysis of B cells induced by a CD19 × CD3 bispecific single-chain antibody is perforin dependent and death receptor independent. Cancer Immunol Immunother 2004; 53:625 - 632
- Ren-Heidenreich L, Davol PA, Kouttab NM, Elfenbein GJ, Lum LG. Redirected T-cell cytotoxicity to epithelial cell adhesion molecule-overexpressing adenocarcinomas by a novel recombinant antibody, E3Bi, in vitro and in an animal model. Cancer 2004; 100:1095 - 1103
- FitzGerald K, Holliger P, Winter G. Improved tumour targeting by disulphide stabilized diabodies expressed in Pichia pastoris. Protein Eng 1997; 10:1221 - 1225
- Holliger P, Manzke O, Span M, Hawkins R, Fleischmann B, Qinghua L, et al. Carcinoembryonic antigen (CEA)-specific T-cell activation in colon carcinoma induced by anti-CD3 × anti-CEA bispecific diabodies and B7 × anti-CEA bispecific fusion proteins. Cancer Res 1999; 59:2909 - 2916
- McCall AM, Adams GP, Amoroso AR, Nielsen UB, Zhang L, Horak E, et al. Isolation and characterization of an anti-CD16 single-chain Fv fragment and construction of an anti-HER2/neu/anti-CD16 bispecific scFv that triggers CD16-dependent tumor cytolysis. Mol Immunol 1999; 36:433 - 445
- McCall AM, Shahied L, Amoroso AR, Horak EM, Simmons HH, Nielson U, et al. Increasing the affinity for tumor antigen enhances bispecific antibody cytotoxicity. J Immunol 2001; 166:6112 - 6117
- Haisma HJ, Grill J, Curiel DT, Hoogeland S, van Beusechem VW, Pinedo HM, et al. Targeting of adenoviral vectors through a bispecific single-chain antibody. Cancer Gene Ther 2000; 7:901 - 904
- Kontermann RE. Recombinant bispecific antibodies for cancer therapy. Acta Pharmacol Sin 2005; 26:1 - 9
- Stish BJ, Chen H, Shu Y, Panoskaltsis-Mortari A, Vallera DA. Increasing anticarcinoma activity of an anti-erbB2 recombinant immunotoxin by the addition of an anti-EpCAM sFv. Clin Cancer Res 2007; 13:3058 - 3067
- Chan HS, Gallie BL, DeBoer G, Haddad G, Ikegaki N, Dimitroulakos J, et al. MYCN protein expression as a predictor of neuroblastoma prognosis. Clin Cancer Res 1997; 3:1699 - 1706
- Marks JD, Hoogenboom HR, Bonnert TP, McCafferty J, Griffiths AD, Winter G. By-passing immunization. Human antibodies from V-gene libraries displayed on phage. J Mol Biol 1991; 222:581 - 597
- Wulhfard S, Tissot S, Bouchet S, Cevey J, De Jesus M, Hacker DL, et al. Mild hypothermia improves transient gene expression yields several fold in Chinese hamster ovary cells. Biotechnol Prog 2008; 24:458 - 465
- Trussel S, Dumelin C, Frey K, Villa A, Buller F, Neri D. New strategy for the extension of the serum half-life of antibody fragments. Bioconjug Chem 2009; 20:2286 - 2292
- Tomlinson IM, Cox JP, Gherardi E, Lesk AM, Chothia C. The structural repertoire of the human Vkappa domain. EMBO J 1995; 14:4628 - 4638
- Hoogenboom HR, Griffiths AD, Johnson KS, Chiswell DJ, Hudson P, Winter G. Multi-subunit proteins on the surface of filamentous phage: methodologies for displaying antibody (Fab) heavy and light chains. Nucleic Acids Res 1991; 19:4133 - 4137
- Borsi L, Balza E, Bestagno M, Castellani P, Carnemolla B, Biro A, et al. Selective targeting of tumoral vasculature: comparison of different formats of an antibody (L19) to the ED-B domain of fibronectin. Int J Cancer 2002; 102:75 - 85