Abstract
The preclinical development of anticancer drugs including immunotherapeutics and targeted agents relies on the ability to detect minimal residual tumor burden as a measure of therapeutic efficacy. Real-time quantitative (qPCR) represents an exquisitely sensitive method to perform such an assessment. However, qPCR-based applications are limited by the availability of a genetic defect associated with each tumor model under investigation. Here, we describe an off-the-shelf qPCR-based approach to detect a broad array of commonly used preclinical murine tumor models. In particular, we report that the mRNA coding for the envelope glycoprotein 70 (gp70) encoded by the endogenous murine leukemia virus (MuLV) is universally expressed in 22 murine cancer cell lines of disparate histological origin but is silent in 20 out of 22 normal mouse tissues. Further, we detected the presence of as few as 100 tumor cells in whole lung extracts using qPCR specific for gp70, supporting the notion that this detection approach has a higher sensitivity as compared with traditional tissue histology methods. Although gp70 is expressed in a wide variety of tumor cell lines, it was absent in inflamed tissues, non-transformed cell lines, or pre-cancerous lesions. Having a high-sensitivity biomarker for the detection of a wide range of murine tumor cells that does not require additional genetic manipulations or the knowledge of specific genetic alterations present in a given neoplasm represents a unique experimental tool for investigating metastasis, assessing antitumor therapeutic interventions, and further determining tumor recurrence or minimal residual disease.
Introduction
Understanding the early processes that promote metastasis in animal models of tumor progression relies upon the ability to detect small numbers of disseminated cancer cells in distant organs. Similarly, accurately evaluating therapeutic responses to experimental interventions in vivo is dependent upon the ability to determine residual tumor burden. In pre-clinical mouse models, cancer cells are nowadays detected by visual inspection or palpation at anatomically assessable sites. This method has poor sensitivity, as it requires a large tumor mass to be appreciable by inspection. Furthermore, such evaluations cannot be easily performed for reduced numbers of malignant cells residing in hardly accessible anatomical locations such as the intra-osseous, intra-cranial, or intra-thoracic compartments. Other cell tracking methods such as fluorescence or bioluminescence tagging allow for the real-time and non-invasive tracking of a small number of cancer cells in vivo. Fluorescent proteins also serve as markers for downstream ex vivo analysis including flow cytometry or immunofluorescence (IF) microscopy-based histology. However, these methods require cancer cells to be manipulated biochemically or genetically in order to incorporate fluorescent dyes or express fluorescent proteins. These manipulations can potentially create undesirable confounding effects on the biology of either cancer and host cells. Although the quantification of tumor burden can be accomplished by flow cytometry, this technique often requires fairly laborious procedures that render it inappropriate for the analysis of a large number of samples. Alternatively, the presence of residual cancer cells in tissues can be determined ex vivo using immunohistochemistry (IHC) on tissue sections. However, this method is neither sensitive nor quantitative, and typically requires the expertise of an excellent histopathologist for proper evaluation. Furthermore, the detection of non-fluorescent cancer cells requires the a priori knowledge of tumor-specific antigens, as well as the availability of antibodies against these diagnostic targets.
Among the methods currently available to detect a malignant outgrowth, the molecular analysis and quantification of tumor-specific biomarkers by real-time quantitative PCR (qPCR) offers a high throughput coupled to extraordinary sensitivity. Despite its superior rapidity and sensitivity as compared with other traditional methods, the ability of qPCR to quantify disease burden is limited by the availability of molecular markers specifically expressed by tumor model under investigation. Few exceptions to this technical limitation include (1) tumors that express viral proteins such as human papillomavirus (HPV)-associated cervical cancers, hepatitis C virus (HPC)-associated liver adenocarcinoma, or Epstein-Barr virus (EBV)-associated lymphoma; and (2) neoplasms bearing well-characterized genetic alterations. Thus, the lack of a common tumor-specific biomarker limits the applicability of qPCR-based approaches to the detection of disease burden in most of commonly employed murine cancer models. With this in mind, the discovery of an off-the-shelf and simple qPCR-based biomarker that is shared by a broad array of rodent tumor models would greatly benefit investigators.
Using a combination of high performance liquid chromatography and tandem mass spectrometry, we have previously identified an immunodominant, H-2Ld-restricted bioactive nanomeric peptide, AH1, containing the sequence SPSYVYHQ (). This peptide is derived from envelope glycoprotein 70 (gp70) of endogenous murine leukemia virus (MuLV), and is expressed by BALB/c-derived CT26 colorectal carcinomas.Citation1,Citation2 Here, we report that the mRNA coding for MuLV gp70 is expressed by a wide array of murine tumors of disparate histological origin and elicited by a variety of oncogenic agents. Furthermore, our results demonstrate that the gp70-coding mRNA can be harnessed for quantifying cancer cells in vivo with high sensitivity. Our findings identify a common tumor-specific biomarker that will aid ongoing pre-clinical studies of neoplastic mechanisms and anticancer therapies in syngeneic murine cancer models.
Figure 1. MuLV genomic structure and primer design for the detection of gp70-coding transcripts. The annealing sites of both of the primer pairs used in this study (gp70–1 and gp70–2) are indicated relative to sequence coding for the bioactive, H-2Ld-restricted peptide SPSYVYHQF (AH1, residues 423–431), which localizes near the 3′-end of the murine leukemia virus (MuLV) envelope (env) gp70-coding gene. These primer pairs, gp70–1 and gp70–2, result in a PCR product that is 110 and 103 base pairs in length, respectively.
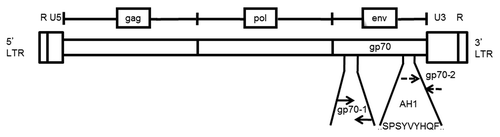
Results
The mRNA coding for the MuLV envelope protein gp70 is expressed in a wide array of murine tumor cell lines
MuLV gp70 is an endogenous viral envelope glycoprotein encoded in the mouse genome that is transcriptionally silent in normal tissues. We have previously shown that the expression of gp70 is reactivated in a few cancer cell lines including CT26 colorectal cancer and B16 melanoma cells.Citation1 Other investigators have demonstrated the presence of MuLV-encoded molecules in murine lymphomas and leukemias as well as in Simian Virus 40 (SV40)-transformed NIH-3T3 cells.Citation3-Citation6 To assess gp70 expression (at the transcriptional level) among other common murine cancer cell lines, we performed quantitative reverse transcription PCR (qPCR) on a total of 44 murine samples, using 2 primer sets: a first one (gp70–1) designed to amplify a 110-bp fragment of the gp70-coding mRNA located near the AH1-coding sequence and the second one (gp70–2) spanning the AH1-coding sequence (). We tested 22 mouse cell lines (including A17, P0, K7, K7M2, MM1, P815, B16 F0, B16 F10, CT26, 4T1, 4T07, 67NR, ALL302, EL4, ICN1, NBL 975A2, RMS 76–9, RMS M3–9-M, and NIH-3T3 cells), and 25 normal mouse tissue samples.Citation7-Citation19 Twenty-two out of the 25 normal tissues were obtained from BALB/c mice and included bone, brain, cerebellum, cervix, colon, cortex, eye, heart, kidney, liver, lung, lymph node, mammary gland, ovary, pancreas, skin, small intestine, spleen, stomach, testis, thymus, and thyroid specimens. Three additional samples were derived from C57BL/6 mice, and including lung, spleen, and inguinal lymph node specimens. Prior to qPCR analysis, samples were subjected to standard, non-quantitative RT-PCR to assess the presence of the gp70-coding mRNA. All tumor cell lines tested in this manner (A17, B16, K7, K7M2, MM1, P0, and P815 cells) showed a 698-bp amplification product corresponding to the gp70-coding mRNA. Conversely, this amplification product was undetectable in all normal tissue samples, with the exception of the cerebellum and testis, for which faint bands were detected (data not shown). We subsequently performed high sensitivity qPCR assays using the gp70–1 primer set on a larger panel of cancer cell lines (A17, P0, K7, K7M2, MM1, P815, B16 F0, B16 F10, CT26, 4T1, 4T07, ALL302, EL4, 67NR, ICN1, NBL 975A2, RMS 76–9, RMS M3–9-M, and NIH-3T3 cells) and normal tissues. With the exception of non-transformed NIH-3T3 fibroblasts, all murine cancer cell lines tested expressed variable but generally abundant levels of the gp70-coding transcripts ( and ). This analysis also confirmed that the murine cerebellum and testis have detectable levels of gp70-coding transcripts while the rest of murine normal tissues express gp70 to nearly undetectable levels ( and ). Similar results were obtained with the gp70–2 primer set, spanning the region of the gp70 mRNA that encodes the bioactive AH1 peptide (Fig. S1, black bars). Nine human cancer cell lines (1 of cervical and 8 of osteal origin) tested negative for the presence of gp70-coding transcripts, confirming that the expression of MuLV is mouse-specific (data not shown).
Table 1. gp70 expression levels in murine tumor cell lines
Figure 2. Relative expression levels of gp70-coding transcripts in mouse cancer cell lines and normal tissues. (A and B) Quantitative reverse-transcriptase PCR (qPCR) assays were performed using the gp70–1 primer pair (see also ) and results were combined to assess the relative amount of gp70-coding transcripts in a panel of common murine cancer cell lines (A) and normal mouse tissues (B). Gene expression was quantified by the 2−ΔΔCt method using cytochrome c1 (Cyc1) as housekeeping gene for normalization. The expression levels of gp70 in ALL302 cells was used as a reference for both neoplastic cells (A) and normal tissues (B). Relative gp70 mRNA levels are plotted on the Y-axis in logarithmic scale in panel A and in linear scale in panel B. Asterisks denote samples with undetectable gp70 expression.
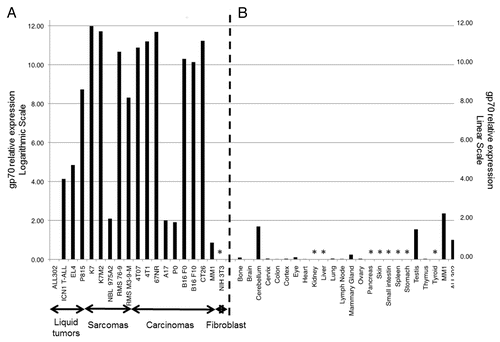
Table 2. gp70 expression levels in normal mouse tissues
Sensitivity of gp70-dependent, qPCR-based detection of cancer cells in whole lungs
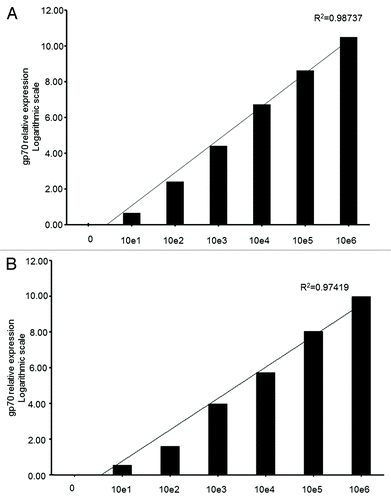
In order to determine the sensitivity of our qPCR-based approach to detect the gp70-coding mRNA as a marker of murine cancer cells, we performed titration experiments by spiking in various amounts of either colon carcinoma CT26 or osteosarcoma K7M2 cells into whole lung cell extracts ex vivo. Replicates of whole lung extracts from naïve BALB/c mice to which known quantities of intact CT26 and K7M2 cells were added (100, 101, 102, 103, 104, 105, and 106 cells/extract) were collected and homogenized. Total RNA was then extracted, digested with DNase I to eliminate contaminating genomic DNA, and reverse-transcribed into cDNA. qPCR analysis of these spiked samples using the gp70–1 primer pair () revealed an exponential increase of the gp70-coding mRNA in a cancer cell dose-dependent manner. Both K7M2 and CT26 cells contained high amounts of the gp70-coding transcript ( and ), such that we were able to detect a specific qPCR signal in a whole lung cell extracts containing as few as 100 cancer cells (ct =30 and 29.46, respectively) (). When we performed the same ex vivo titration experiment using the mastocytoma cell line P815 (which express moderate levels of the gp70-coding mRNA; ) and whole lungs or spleens, we were able to detect a specific qPCR signal from tissue extracts containing a minimum of 104 cells (data not shown). Our results indicate that gp70 can be detected by qPCR with high sensitivity. This method should be amenable for detecting the presence of relatively low amounts of cancer cells in vivo, especially in those tumor models that express abundant levels of gp70-coding transcripts.
Figure 3. Sensitivity of the qPCR-based detection of gp70-coding transcripts from limiting amounts of cancer cells in total lung extracts. (A and B) Various numbers (100, 101, 10Citation2, 103, 104, 105, and 106) of murine colon carcinoma CT26 (A) or metastatic osteosarcoma K7M2 (B) cells were added to homogenized mouse lung tissue. Following reverse transcription, the gp70–1 primer pair (see also ) was used for quantitative reverse-transcriptase PCR (qPCR) assays. gp70 mRNA levels were then quantified by and the 2−ΔΔCt method using the cytochrome c1 (Cyc1) as housekeeping gene for normalization. Relative gp70 mRNA levels are plotted on the Y-axis in logarithmic scale. The coefficient of determination, R2, was > 0.97 in both cases.
Sensitivity comparison of bioluminescence and gp70 detection in vivo
To directly compare the efficacy of qPCR-, bioluminescence-, and histologically-based approaches for detecting the presence of cancer cells in vivo, we engineered paired osteosarcoma K7 (non-metastatic) and K7M2 (metastatic) cells to express the firefly luciferase (Luc2). BALB/c mice (n = 3) were inoculated with either 106 Luc2-expressing K7 or Luc2-expressing K7M2 cells in the right proximal tibia. Bioluminescence imaging (BLI) was used to monitor the distribution of cancer cells in vivo at the following time points post injection: 30 min, 1 d, 5 d, 10 d, 20 d, and 28 d. BLI tracking showed that pulmonary metastases were detectable between d 10 and 21 in mice injected with either Luc2-expressing K7 or Luc2-expressing K7M2 cells (data not shown). By d 28, mice injected with metastatic Luc2-expressing K7M2 cells displayed a uniform and easily detectable tumor burden. In comparison, mice injected with non-metastatic Luc2-expressing K7 cells exhibited weaker and more variable BLI signals. In some animals of the latter group, the BLI signal was undetectable (). Mice were sacrificed after BLI imaging on d 28 and the right one lung was stored in Trizol for the quantification of gp70-coding transcripts by qPCR, while the left one was embedded in optimal cutting temperature (OCT) compound and frozen for serial sectioning and pathological evaluation by IHC. qPCR analysis of the harvested lungs for gp70 expression showed an excellent linear correlation (as assessed by scatter plot) with the photon emission from the regions of interest (ROIs) of the corresponding BLI images from individual animals ( ; Fig. S2). Linear regression analysis examining the correlation between BLI signal intensity and relative gp70 expression levels as measured by qPCR produced a fitted regression line given by the equation: BLI = 1057.937(qPCR) + 477545.832, with a positive and significant coefficient of determination (R2 = 0.986; P = 0.001). Using the Pearson correlation coefficient, we found a significant correlation between the BLI signal intensity and gp70 expression levels as determined by qPCR (r = 0.993; P = 0.001). Nonparametric correlation analysis using Spearman’s ρ confirmed a significant correlation between BLI signal intensity and gp70 expression levels monitored by qPCR (ρ = 0.900, P = 0.037) (data not shown).
Figure 4. Comparison of gp70-specific qPCR, bioluminescence and histological methods to detect osteosarcoma cells in vivo. (A–C) BALB/c mice (n = 3 per group) were injected with 106 firefly luciferase (Luc2)-expressing K7 or K7M2 cells in the proximal tibia. (A) Relative expression levels of gp70-coding transcripts as detected by quantitative reverse-transcriptase PCR (qPCR) in the whole lung tissues of individual tumor-bearing animals 28 d after the inoculation of cancer cells. (B) Whole mouse bioluminescence imaging (BLI) of the same tumor-bearing mice indicated above, immediately prior to lung harvesting (28 d after cancer cell inoculation). Red circles represent the regions of interest (ROIs) that were employed to quantify BLI signal intensity. (C) Histological examination of tissue sections from the lungs harvested from the same tumor-bearing animals analyzed in panels (A and B). Visible lung tumor nodules were scored in 15 consecutive sections of each individual lung, with “−” means no tumor detected in any of the 15 slides, and the number of “+” represent the maximum number of tumor nodules seen in a single slide.
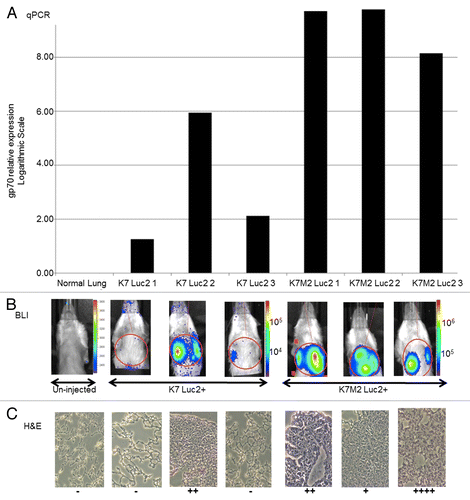
In a parallel analysis, harvested lung tissues were stained with hematoxylin and eosin (H&E) and tumor burden was analyzed from 15 consecutive 10-μm sections of the same tumor-infiltrated lung, an approach that failed to yield good correlation with either the qPCR or the BLI results in individual animals (). As expected, this histological approach had lower global sensitivity in detecting the presence of limiting numbers of tumor cells as compared with the other 2 techniques. Of note, the qPCR-based method readily detected the presence of tumor nodules in the lungs that went undetected by either BLI or histology (e.g., K7 Luc2 #1 and K7 Luc2 #3; ). In a separate experiment, the quantification of gp70 by qPCR revealed the presence of K7M2 cells in the mouse lungs between d 7 and 14 after inoculation, several days prior to the moment in which the pulmonary metastasis could be effectively monitored by whole body BLI (data not shown).
gp70-specific PCR is more sensitive than flow cytometry to detect circulating lymphoblasts
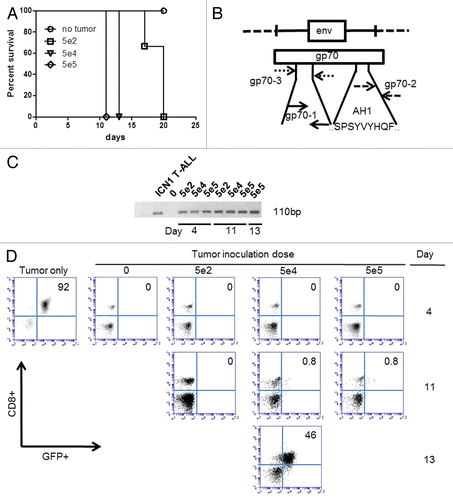
To further demonstrate the utility of a PCR-based gp70 detection method in vivo, we adoptively transferred green fluorescence protein (GFP)-tagged acute lymphoblastic leukemia (ALL) cells intravenously into naïve C57BL/6 mice and analyzed disease burden by gp70-specific PCR of flow cytometry. To this end, we transferred an increasing number of GFP+ murine ICN1 lymphoblasts (5 × 102, 5 × 104, and 5 × 105 cells) from syngeneic donor mice transplanted with GFP-transduced bone marrow progenitor cells 9 weeks earlier to naïve C57BL/6 mice. All C57BL/6 animals receiving ICN1 lymphoblasts succumbed to leukemia within 3 weeks, survival time inversely correlating with the size of the tumor inoculum (). To determine whether the gp70-coding transcript can be detected in the peripheral blood during the first 13 d of disease (sub-clinical phase), we obtained sequential peripheral blood samples and performed nested gp70-specific non-quantitative PCR analysis using the gp70–1 primer set upon an initial amplification with flanking gp70–3 primers (). We were able to detect gp70-coding transcripts in the peripheral blood of mice inoculated with different doses of lymphoblasts. The detection sensitivity on d 4 following tumor cell injection was 100% at the lowest inoculation dose of 500 cancer cells (). In stark contrast, conventional flow cytometry failed to detect GFP+CD8+ lymphoblasts in the peripheral blood of tumor-bearing mice until 24–48 before their death (), and despite the fact that tumor infiltration in the liver, spleen, bone marrow, and lymph nodes was evident at autopsy (data not shown). These results further underscore the sensitivity and specificity of detecting the MuLV gp70-coding mRNA as a biomarker of disease burden in pre-clinical murine tumor models.
Figure 5. The qPCR-based detection of gp70-coding transcripts is more sensitive than flow cytometry to identify T lymphoblasts in the peripheral blood. (A–D) C57BL/6 mice (n = 3 per group) received 5 × 102, 5 × 104, or 5 × 105 green fluorescence protein (GFP)-expressing ICN1 acute lymphoblastic leukemia (ALL) cells and their peripheral blood was collected for subsequent analyses at various time points. (A) Kaplan–Meier survival curves of C57BL/6 mice receiving the indicated number of ALL cells. (B) Map of gp70-coding sequence, demonstrating the annealing sites for the gp70–3 and gp70–1 primer sets employed for the nested PCR-based detection of gp70-coding transcripts in the peripheral blood of tumor-inoculated mice. (C) Detection of gp70-coding transcripts by nested PCR in mice inoculated with the indicated number of ALL cells i.v. 4 d earlier. (D) Serial monitoring of mice receiving the indicated number of ALL cells for the appearance of a GFP+CD8+ cell population (corresponding to ALL cells) in the peripheral blood. Pre-inoculation ALL cells (tumor only) were analyzed for comparative purposes.
Discussion
In the current study, we describe a simple and sensitive method to detect and quantify low quantities of commonly employed murine cancer cells in vivo. This qPCR-based detection method relies on an endogenous murine leukemia retrovirus antigen, MuLV gp70. The gp70-coding gene is transcriptionally silent in the majority of non-transformed mouse tissues, but we found it to be uniformly expressed in virtually all mouse cancer cell lines commonly utilized for experimental aims. Specifically, we analyzed a total of 44 murine samples, surveying 22 mouse cancer cell lines and 22 normal tissues. We detected gp70-coding transcripts in all transformed cell lines but not in the non-cancerous, immortalized NIH-3T3 fibroblasts. Different mouse cancer cell lines expressed gp70-coding transcripts at varying levels. The same analysis performed on RNA derived from normal mouse tissues revealed little to no gp70 expression, with the exception of the cerebellum and testis.
We propose that gp70 may serve as a useful tumor biomarker for the quantification of disease burden in most, if not all, of commonly employed mouse tumor models. Harnessing this biomarker requires neither the a priori knowledge of any additional tumor-specific molecular alteration, nor the introduction of exogenous genetic markers for tracking cancer cells in tissue samples. qPCR-based detection methods are inherently more sensitive and adapt for quantitative purposes than traditional tumor burden monitoring methods such as visual inspection, palpation, BLI, fluorescence in situ hybridization (FISH), cytofluorometry, IHC, and tissue histology. Therefore, measuring gp70 expression at the transcriptional level may allow for the rapid and precise determination of residual tumor burden in vivo when evaluating the pre-clinical efficacy of multiple anticancer drugs, including immunotherapeutics and molecularly targeted agents.
The steps leading to the reactivation of gp70 expression in the course of oncogenic transformation in mice are currently unknown. The reactivation of gp70 was not caused by tissue inflammation alone, as the same qPCR analyses on spontaneous, pre-cancerous adenomatous polyps from in mice lacking SMAD family member 4 (Smad4) and cyclin-dependent kinase inhibitor 1B (Cdkn1b) specifically in the T-cell lineage failed to detect gp70-coding transcripts (data not shown). Similarly, activated T cells (data not shown) and immortalized NIH-3T3 cells did not express gp70 to detectable levels, even though prior studies have shown that these cells reactivate MuLV upon transformation with SV40.Citation6 Clearly, the expression of gp70 in murine cell lines is restricted to transformed cells with a tumorigenic potential. Our study also demonstrates the successful application of gp70-specific qPCR to identify small pulmonary metastases below the detection limits of BLI-based and histological methods (). Moreover, we observed a strong correlation (Pearson correlation coefficient r = 0.993, P = 0.001) between BLI data on pulmonary metastases and gp70 expression levels as measured by qPCR. As expected, both qPCR- and BLI-based methods are more sensitive than the histological assessment of disease burden on a series of tissue sections. Thus, our detection protocol allows for the sensitive and specific detection of low amounts of mouse cancer cells in preparations from normal tissues ex vivo. Due to the apparent expression of gp70 in the non-transformed cerebellum and testis, one limitation of our detection method is that it cannot be used to monitor tumor burden at this anatomical locations. Another consideration is that the sensitivity of our current qPCR-based method is strongly dependent on the relative expression levels of the gp70-coding transcripts in a given cancer cell line. As our titration curve demonstrates, gp70-coding transcripts from as few as 100 tumor cells spiked into the whole lung cell extract can be detected when such cells express high gp70 levels. When the same titration was performed using mouse mastocytoma P815 cells, which express gp70 at a much lower levels than either CT26 or K7M2 cells, the detection threshold increased to 104 cells. In such cases, it may be possible to increase the detection sensitivity of our method by performing a nested PCR.
We obtained similar results by using different primer pairs that hybridize next to sequence encoding the AH1 peptide (residues 423–431), a H-2Ld-restricted, immunodominant tumor-associated antigen derived from the polycistronic gp70-coding mRNA and previously shown to be actively expressed by CT26 colorectal carcinoma cells.Citation1 In this prior study, the adoptive transfer of activated, AH1-specific CD8+ T cells was sufficient to eradicate established CT26 tumors in vivo, confirming the essential role that AH1 – and by extension gp70 – plays in cancer immunosurveillance. One important question originating from our study is whether the abundance of gp70-coding transcripts correlates with the expression and presentation of gp70-derived antigens as targets for tumor-specific CD8+ or CD4+ T cells. One determinant of this phenomenon would be the availability of proteasome machinery within cancer cells, allowing them to process and load AH1, or other gp70-derived peptides, onto the peptide-binding groove of MHC class I and II molecules. One additional factor that may influence how gp70 expression impacts cancer immunosurveillance is the expression levels of MHC molecules and other immunomodulatory molecules by individual cancer cells.
Regarding the translational applications of our current finding, exogenous viral infections contribute to roughly 15–20% of all human cancers.Citation20 For viruses such as the human T-lymphotropic virus 1 (HTLV-1), HPV, human herpesvirus-8 (HHV-8), and hepatitis B and C virus (HBV and HCV), the association between infection and oncogenesis of tumor progression is well established. However, such correlation is not as well established for other viruses including SV40, BK virus (BKv), John Cunningham virus (JCV), human endogenous retroviruses (HERVs), human mammary tumor virus (HMTV), and Torque teno virus (TTV). HERVs (which constitutes about 8% of human DNA) are thought to originate from the integration of exogenous retroviruses into the human germline genome.Citation21 Despite the absence of selective pressures on the host to retain the proviral genome in its intact form, retroviral genes persist and accumulate mutations over time.Citation22 Similar to what we have observed in our current study with murine retrovirus, HERV genes are transcriptionally silent in normal tissues but have been found to be re-expressed in several human neoplasms, including breast cancer, prostate cancer, lung adenocarcinoma, ovarian cancer, and melanoma.Citation23-Citation29 Therefore, HERV gene products could also serve as potential tumor-specific biomarkers for qPCR-based detection.Citation30 In addition, recent findings suggest that retroviral gene products are capable of acting as tumor-associated antigens involved in both T-cell and B-cell responses in breast cancer patients.Citation31 Several drugs aimed at inhibiting HERV-K expression are currently under evaluation in clinical trials as novel therapies for non-Hodgkin lymphoma and multiple sclerosis (source www.clinicaltrials.gov).
In summary, we have developed a sensitive, specific, and technically simple qPCR-based technique to detect and quantify a widely expressed tumor-restricted biomarker in vivo and in vitro. The discovery of a near-to-ubiquitous tumor marker that can be detected with high sensitivity without prior knowledge of tumor-specific genetic alterations provides a unique tool that can be used in many pre-clinical applications. These include the study of metastatic dissemination in vivo, the assessment of the therapeutic efficacy of multiple anticancer agents, the detection of disease recurrence and/or assessment of residual disease. Moreover, considering that gp70 gene products (such as the bioactive AH1 peptide) are exclusively expressed by cancer cells irrespective of the nature of the oncogenic driver, these retroviral molecules may represent potential targets for the development of novel pan tumor-specific immunotherapeutic approaches.
Materials and Methods
Cancer cell lines and tissue sample collection
A panel of 22 mouse cancer cell lines was obtained from various sources and analyzed for gp70 expression by qPCR. Murine cervical cancer TC1P0 cells and their aggressive derivatives TC1A17 (both haplotype H-2b), were a generous gifts from Dr TC Wu at Johns Hopkins. Murine medulloblastoma MM1 cells derived from PTCH1+/−-Trp53−/− mice (H-2b) were a gift from Dr Greg Plautz at the Cleveland Clinic. BALB/c-derived osteosarcoma K7 (parental; H-2d) and K7M2 (metastatic; H-2d) cells were obtained from Dr Chand Khanna at the National Cancer Institute. Spontaneously arising murine breast cancer 4T1 (highly metastatic), 4T07 (invasive but non-metastatic), and 67NR (non-metastatic) cells, from BALB/c mice (H-2d), were obtained from Dr John Letterio at Case Western Reserve University. Syngeneic neuroblastoma NBL 947.4 and NBL 975A2 cells (from C57BL/6 mice; H-2b) were obtained from Dr Remis Orentis at the National Cancer Institute. Mouse rhabdomyosarcoma RMS 76–9 and RMS M3–9-M cells derived from C57BL/6 mice (H-2b) were obtained from Dr Crystal Mackall at the National Cancer Institute. P815 mastocytoma (H-2d) (ATCC #TIB-64), poorly metastatic B16 F0 melanoma cells (H-2b) (ATCC CRL#6322), highly metastatic B16 F10 melanoma cells (ATCC CRL#6475), as well as N-nitroso-N-methylurethane-induced CT26 colorectal carcinoma cells (H-2d) (ATCC CRL#2638), lymphoma EL4 cells (H-2b; ATCC #TIB-39), and NIH-3T3 fibroblasts (ATCC CRL#1658) were obtained from ATCC. ALL302 is a T-cell leukemia cell line derived from Smad3−/− C57BL/6 mice (H-2b) and was obtained from Dr John Letterio at Case Western Reserve University. ICN1 T-cell ALL cells (H-2b) were derived in our lab from a C57BL/6 mouse transplanted with bone marrow progenitors that had been transduced with a lentiviral construct coding for the intracellular portion of Notch1 (ICN1). Additionally, a panel of 25 various normal murine tissues representing 22 different organs (including bone, brain, cerebellum, cervix, colon, cortex, eye, heart, kidney, liver, lung, lymph node, mammary gland, ovary, pancreas, skin, small intestine, spleen, stomach, testis, thymus, and thyroid) were obtained from BALB/c mice. Three additional tissues samples (from lymph nodes, spleen and lung) were obtained from C57BL/6 mice for mRNA isolation and qPCR analysis.
Tumor cell constructs and in vitro culture conditions
All cell lines were passaged and maintained in RPMI 1640 medium (Thermo Scientific #SH30027.01) supplemented with 10% fetal bovine serum (FBS) (Gemini Bio Products #900–108), 1% penicillin/streptomycin (Thermo Scientific #SV30010), 100 mM sodium pyruvate (Thermo Scientific #SH3023901), and 1% non-essential amino acids (Cellgro #25–025Cl). For bioluminescence imaging, murine osteosarcoma lines K7 and K7M2 cells were transduced with 1 μL of 108 TU/mL Luc2-coding lentiviral particles, according to manufacturer’s recommendation (LentiFire by In Vivo Imaging Solution #48). Luciferase-expressing K7 and K7M2 cells were selected and maintained in media containing 5 μg/mL puromycin.
RNA isolation
Tumor samples were homogenized in Trizol (Life Technologies #15596–026) with the Bullet Blender (Next Advance) and 0.9–2 mm stainless steel beads (Next Advance #SSB14B). Total RNA from cell lines and normal tissues was extracted using Trizol and subsequently purified using RNeasy columns (Qiagen #74106), according to manufacturer’s instructions. Total RNA was extracted from the peripheral blood using the RNeasy kit upon red blood cell (RBC) lysis using the ACK Lysis buffer (Lonza #10–548E). RNA was treated with DNase I (Life Technologies #18068–015) to remove contaminating genomic DNA and retro-transcribed using SuperScriptIII (Life Technologies #18080051) to form cDNA. RNA concentrations were determined by conventional spectrophotometry (NanoDrop Technologies).
Primer design and qPCR analysis
The 3 primer pairs for the detection of gp70 (gp70–1F: AAAGTGACAC ATGCCCACAA, gp70–1R: CCCCAAGAGG CACAATAGAA; gp70–2F: TGACCTTGTC CGAAGTGACC, gp70–2R: TAGGACCCAT CGCTTGTCTT; and gp70–3F: ATCACCCTCT GTGGACTTGG, gp70–3R: GAGACGTATA GGCGCAATCC) and the primer pair for the detection of the housekeeping gene cytochrome c1 (Cyc1F: CTGCCACAGC ATGGATTATG and Cyc1R: GGTCATTGTC AC AGCACCAC) were designed using Primer3 (http://bioinfo.ut.ee/primer3-0.4.0/) with the murine leukemia virus retroviral envelope glycoprotein gene (GenBank: DQ359272.1) as reference sequence. Designed primers were aligned against the corresponding genome sequence using BLAT (http://genome.ucsc.edu/cgi-bin/hgBlat, assembly Dec.2011 (GRCm38/mm10) to confirm specificity. Primers were synthesized by Integrated DNA Technologies (IDT). qPCR reactions were performed on Applied BioSystem 7300 using cycling conditions as follows: 50 °C for 2 min, 95 °C for 10 min, and then 40 cycles of 95 °C for 15 s, 60 °C for 1 min. The qPCR reactions were performed using SYBR Green chemistry (Life Technologies # 4309155) with the primers listed above and analyzed using gp70 Ct values of tumor and tissue samples normalized to Ct values for Cyc1. Quantification results were compared using the ALL302 cell line as a reference. Relative quantification was accomplished by using the 2−ΔΔCt method.Citation32 According to our titration curve in which we spiked known amounts of cancer cells in whole lung, samples with Ct values > 30 were considered non-detectable. Nested RT-PCR was accomplished by two separate and consecutive reactions using gp70–3 as outer primer set and gp70–1 as inner primer set. The RT-PCR was performed using recombinant Taq DNA polymerase (Life Technologies #10342020). Cycling conditions were as follows: 94 °C for 3 min followed by 35 (outer primer set) or 25 cycles (inner primer set) of 94 °C for 45 s, 55 °C for 30 s, 72 °C for 1 min and 30 s. The reaction was incubated for an additional 10 min at 72 °C.
Tumor implantation studies
Eight-week old female BALB/c mice (Jackson Laboratory #000651) were housed and handled according to approved IACUC protocol (Protocol number: 2013–0097) and CWRU Animal Resource Center guidelines. Six animals were injected with either 106 Luc2-expressing K7 cells (3 mice) or 106 Luc2-expressing K7M2 cells (3 mice) in the right proximal tibia. Hair was removed from the chest area by shaving with a hair clipper, and BLI was performed with the Xenogen IVIS-200 apparatus (Caliper Life Sciences) at the following time points: pre-injection and 30 min, 1 d, 5 d, 10 d, 21 d, and 28 d after tumor injection to follow tumor dissemination and growth. All BLI acquisitions were performed after a 200ul intra-peritoneal injection of 15 μg/mL Beetle D-luciferin Potassium Salt (Promega #E1602) 5 min prior to imaging. The animals were anesthetized with 2% isoflurane in O2 at a rate of 2 L/min to achieve adequate sedation during imaging. All images were acquired with an 18.4 cm field of view, medium binning factor, and f/stop of 1. The exposure times ranged from 10 s to 3 min depending on the bioluminescence of primary tumors or metastatic lesions. The emitted light was detected with the IVIS Imaging System, digitized, and electronically displayed as a pseudo-color overlaid onto a gray-scale animal image. ROIs were drawn around tumor sites and quantified as maximum flux (photons per sec (s) per cm2 per steradian (sr) [p/s/cm2/sr]) within a uniform, oval region of interest using the Living Image 2.5 software (Caliper). On d 28, animals were euthanized using CO2 asphyxiation. The left lung was inflated with OCT compound, excised, embedded in OCT compound, and stored at −80 °C for histological evaluation. The right lung was excised and stored immediately in Trizol at −80 °C for qPCR analysis.
For detection of minimal T-ALL cells in vivo, various quantities (5 × 102, 5 × 104, and 5 × 105) of GFP+CD8+ ICN1 T-ALL cells were transferred intravenously into 6–9 week old naïve C57BL/6 mice (Jackson Laboratory #000664) on d 0. Mice were monitored daily for survival to generate Kaplan-Meier curves. Blood samples (approximately 80 μL) were collected via tail-vein bleed twice weekly in order to analyze the presence of GFP+/CD8+ T-lymphoblasts by flow cytometry for gp70 expression by qPCR or nested PCR.
Histological evaluations
A total of 15 serial 10-μm thick, OCT-embedded lung cryosections were stained with H&E (IHCWORLD #IW-3100), according to supplier recommendation. Tumor nodules were enumerated manually and scored as follows: “−,” no tumor nodule in 15 serial sections; “+,” maximum of 1 tumor nodule in any one of 15 serial sections; “++,” maximum of 2 tumor nodules in any one of 15 serial sections; “++++,” ≥ 3 tumor nodules in any one of 15 serial sections.
Flow cytometry
Peripheral blood was collected from the tail vein in ACD buffer (136 mM glucose, 75 mM trisodium citrate, 4.2 mM citric acid monohydrate). RBCs were lysed in ACK lysis buffer, and peripheral blood mononuclear cells were then resuspended in FACS buffer (0.1% BSA in PBS, 5 mM EDTA). Cells were incubated with FACS blocking buffer (10% mouse serum in FACS buffer) and stained with anti-CD8 antibodies (eBioscience, clone 53.6–7). Ten thousand events were examined on an Accuri C6 flow cytometer (BD Accuri) and analyzed using Accuri C6 flow cytometry software.
Statistical analyses
Pearson's correlation coefficients were used to delineate the simple linear regression and correlation analysis between gp70 expression levels as determined by qPCR and BLI signal intensity. As a confirmatory non-parametric correlation analysis, Spearman's ρ was employed, when indicated. All statistical analyses were performed using the IBM SPSS Statistics software (IBM Corporation). All statistical tests were two-sided, and P values lower than 0.05 were considered significant.
| Abbreviations: | ||
| ALL | = | acute lymphoblastic leukemia |
| BLI | = | bioluminescent imaging |
| gp70 | = | glycoprotein 70 |
| HCV | = | hepatitis C virus |
| HERV | = | human endogenous retrovirus |
| HPV | = | human papillomavirus |
| ICN1 | = | intracellular portion of Notch1 |
| IHC | = | immunohistochemistry |
| MuLV | = | murine leukemia virus |
| ROI | = | region of interest |
Additional material
Download Zip (205.4 KB)Acknowledgments
The authors would like to acknowledge Deborah Sim Barkauskas for her technical help during the preparation of this manuscript, and the following people for their generous provision of tumor cell lines and tissue samples: Dr John Letterio, MD and Dr Sung-Hee Choi, PhD at Case Western Reserve University, Dr Greg Plautz, MD at The Cleveland Clinic, Dr TC Wu, MD, PhD. at Johns Hopkins University, and Dr Chand Khanna, DVM, PhD, Dr Crystal Mackall, MD and Dr Remis Orentis, PhD at The National Cancer Institute. The authors gratefully acknowledge the following funding support for this work: CWRU Orthopedics T32 (5T32AR7505–25 to FS), the National Cancer Institute (R01CA154656 to AYH; R01CA154656 S1 to FA), the St. Baldrick’s Foundation (AYH), the Cancer Research Institute (AYH), the Alex’s Lemonade Stand Foundation (AYH), the Gabrielle’s Angel Foundation (AYH), the Hyundai Hope-on-Wheels Program (AYH), the Steven G. AYA Foundation (AYH), the Rainbow Board of the Rainbow Babies and Children’s Hospital (IDB).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Citation: Scrimieri F, Askew D, Corn DJ, Eid S, Bobanga ID, Bjelac JA, Tsao ML, Allen F, Othman YS, Wang SG, et al. Murine leukemia virus envelope gp70 is a shared biomarker for the high-sensitivity quantification of murine tumor burden. OncoImmunology 2013; 2:e26889; 10.4161/onci.26889
References
- Huang AY, Gulden PH, Woods AS, Thomas MC, Tong CD, Wang W, Engelhard VH, Pasternack G, Cotter R, Hunt D, et al. The immunodominant major histocompatibility complex class I-restricted antigen of a murine colon tumor derives from an endogenous retroviral gene product. Proc Natl Acad Sci U S A 1996; 93:9730 - 5; http://dx.doi.org/10.1073/pnas.93.18.9730; PMID: 8790399
- Risser R, Horowitz JM, McCubrey J. Endogenous mouse leukemia viruses. Annu Rev Genet 1983; 17:85 - 121; http://dx.doi.org/10.1146/annurev.ge.17.120183.000505; PMID: 6320713
- O’Donnell PV, Stockert E, Obata Y, Old LJ. Leukemogenic properties of AKR dualtropic (MCF) viruses: amplification of murine leukemia virus-related antigens on thymocytes and acceleration of leukemia development in AKR mice. Virology 1981; 112:548 - 63; http://dx.doi.org/10.1016/0042-6822(81)90301-9; PMID: 6266139
- Hartley JW, Wolford NK, Old LJ, Rowe WP. A new class of murine leukemia virus associated with development of spontaneous lymphomas. Proc Natl Acad Sci U S A 1977; 74:789 - 92; http://dx.doi.org/10.1073/pnas.74.2.789; PMID: 191826
- Nowinski RC, Hays EF, Doyle T, Linkhart S, Medeiros E, Pickering R. Oncornaviruses produced by murine leukemia cells in culture. Virology 1977; 81:363 - 70; http://dx.doi.org/10.1016/0042-6822(77)90152-0; PMID: 197696
- Timmons PM, Brickell PM, Latchman DS, Rigby PW. Activation of endogenous retroviral transcription in SV40-transformed mouse cells. Nucleic Acids Res 1991; 19:7215 - 8; http://dx.doi.org/10.1093/nar/19.25.7215; PMID: 1662812
- Lin KY, Guarnieri FG, Staveley-O’Carroll KF, Levitsky HI, August JT, Pardoll DM, Wu TC. Treatment of established tumors with a novel vaccine that enhances major histocompatibility class II presentation of tumor antigen. Cancer Res 1996; 56:21 - 6; PMID: 8548765
- Khanna C, Prehn J, Yeung C, Caylor J, Tsokos M, Helman L. An orthotopic model of murine osteosarcoma with clonally related variants differing in pulmonary metastatic potential. Clin Exp Metastasis 2000; 18:261 - 71; http://dx.doi.org/10.1023/A:1006767007547; PMID: 11315100
- Wetmore C, Eberhart DE, Curran T. Loss of p53 but not ARF accelerates medulloblastoma in mice heterozygous for patched. Cancer Res 2001; 61:513 - 6; PMID: 11212243
- Dunn TB, Potter M. A transplantable mast-cell neoplasm in the mouse. J Natl Cancer Inst 1957; 18:587 - 601; PMID: 13416911
- Fidler IJ. Biological behavior of malignant melanoma cells correlated to their survival in vivo. Cancer Res 1975; 35:218 - 24; PMID: 1109790
- Wang M, Bronte V, Chen PW, Gritz L, Panicali D, Rosenberg SA, Restifo NP. Active immunotherapy of cancer with a nonreplicating recombinant fowlpox virus encoding a model tumor-associated antigen. J Immunol 1995; 154:4685 - 92; PMID: 7722321
- Aslakson CJ, Miller FR. Selective events in the metastatic process defined by analysis of the sequential dissemination of subpopulations of a mouse mammary tumor. Cancer Res 1992; 52:1399 - 405; PMID: 1540948
- Gorer PA. Studies in antibody response of mice to tumour inoculation. Br J Cancer 1950; 4:372 - 9; http://dx.doi.org/10.1038/bjc.1950.36; PMID: 14801344
- Li X, Gounari F, Protopopov A, Khazaie K, von Boehmer H. Oncogenesis of T-ALL and nonmalignant consequences of overexpressing intracellular NOTCH1. J Exp Med 2008; 205:2851 - 61; http://dx.doi.org/10.1084/jem.20081561; PMID: 18981238
- Stauffer JK, Orentas RJ, Lincoln E, Khan T, Salcedo R, Hixon JA, Back TC, Wei JS, Patidar R, Song Y, et al. High-throughput molecular and histopathologic profiling of tumor tissue in a novel transplantable model of murine neuroblastoma: new tools for pediatric drug discovery. Cancer Invest 2012; 30:343 - 63; http://dx.doi.org/10.3109/07357907.2012.664670; PMID: 22571338
- Weigel BJ, Rodeberg DA, Krieg AM, Blazar BR. CpG oligodeoxynucleotides potentiate the antitumor effects of chemotherapy or tumor resection in an orthotopic murine model of rhabdomyosarcoma. Clin Cancer Res 2003; 9:3105 - 14; PMID: 12912962
- Meadors JL, Cui Y, Chen QR, Song YK, Khan J, Merlino G, Tsokos M, Orentas RJ, Mackall CL. Murine rhabdomyosarcoma is immunogenic and responsive to T-cell-based immunotherapy. Pediatr Blood Cancer 2011; 57:921 - 9; http://dx.doi.org/10.1002/pbc.23048; PMID: 21462302
- Todaro GJ, Green H. Quantitative studies of the growth of mouse embryo cells in culture and their development into established lines. J Cell Biol 1963; 17:299 - 313; http://dx.doi.org/10.1083/jcb.17.2.299; PMID: 13985244
- Parkin DM. The global health burden of infection-associated cancers in the year 2002. Int J Cancer 2006; 118:3030 - 44; http://dx.doi.org/10.1002/ijc.21731; PMID: 16404738
- Löwer R, Boller K, Hasenmaier B, Korbmacher C, Müller-Lantzsch N, Löwer J, Kurth R. Identification of human endogenous retroviruses with complex mRNA expression and particle formation. Proc Natl Acad Sci U S A 1993; 90:4480 - 4; http://dx.doi.org/10.1073/pnas.90.10.4480; PMID: 8506289
- Gifford R, Tristem M. The evolution, distribution and diversity of endogenous retroviruses. Virus Genes 2003; 26:291 - 315; http://dx.doi.org/10.1023/A:1024455415443; PMID: 12876457
- Wang-Johanning F, Frost AR, Jian B, Epp L, Lu DW, Johanning GL. Quantitation of HERV-K env gene expression and splicing in human breast cancer. Oncogene 2003; 22:1528 - 35; http://dx.doi.org/10.1038/sj.onc.1206241; PMID: 12629516
- Wang-Johanning F, Frost AR, Jian B, Azerou R, Lu DW, Chen DT, Johanning GL. Detecting the expression of human endogenous retrovirus E envelope transcripts in human prostate adenocarcinoma. Cancer 2003; 98:187 - 97; http://dx.doi.org/10.1002/cncr.11451; PMID: 12833471
- Kahyo T, Tao H, Shinmura K, Yamada H, Mori H, Funai K, Kurabe N, Suzuki M, Tanahashi M, Niwa H, et al. Identification and association study with lung cancer for novel insertion polymorphisms of human endogenous retrovirus. Carcinogenesis 2013; 34:2531 - 8; http://dx.doi.org/10.1093/carcin/bgt253; PMID: 23872666
- Wang-Johanning F, Liu J, Rycaj K, Huang M, Tsai K, Rosen DG, Chen DT, Lu DW, Barnhart KF, Johanning GL. Expression of multiple human endogenous retrovirus surface envelope proteins in ovarian cancer. Int J Cancer 2007; 120:81 - 90; http://dx.doi.org/10.1002/ijc.22256; PMID: 17013901
- Büscher K, Trefzer U, Hofmann M, Sterry W, Kurth R, Denner J. Expression of human endogenous retrovirus K in melanomas and melanoma cell lines. Cancer Res 2005; 65:4172 - 80; http://dx.doi.org/10.1158/0008-5472.CAN-04-2983; PMID: 15899808
- Muster T, Waltenberger A, Grassauer A, Hirschl S, Caucig P, Romirer I, Födinger D, Seppele H, Schanab O, Magin-Lachmann C, et al. An endogenous retrovirus derived from human melanoma cells. Cancer Res 2003; 63:8735 - 41; PMID: 14695188
- Serafino A, Balestrieri E, Pierimarchi P, Matteucci C, Moroni G, Oricchio E, Rasi G, Mastino A, Spadafora C, Garaci E, et al. The activation of human endogenous retrovirus K (HERV-K) is implicated in melanoma cell malignant transformation. Exp Cell Res 2009; 315:849 - 62; http://dx.doi.org/10.1016/j.yexcr.2008.12.023; PMID: 19167380
- Pichon JP, Bonnaud B, Mallet F. Quantitative multiplex degenerate PCR for human endogenous retrovirus expression profiling. Nat Protoc 2006; 1:2831 - 8; http://dx.doi.org/10.1038/nprot.2006.475; PMID: 17406542
- Wang-Johanning F, Radvanyi L, Rycaj K, Plummer JB, Yan P, Sastry KJ, Piyathilake CJ, Hunt KK, Johanning GL. Human endogenous retrovirus K triggers an antigen-specific immune response in breast cancer patients. Cancer Res 2008; 68:5869 - 77; http://dx.doi.org/10.1158/0008-5472.CAN-07-6838; PMID: 18632641
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001; 25:402 - 8; http://dx.doi.org/10.1006/meth.2001.1262; PMID: 11846609