Abstract
T-cell receptor (TCR)-based gene immunotherapy has emerged as a promising approach for the treatment of multiple malignancies. We have recently reported an efficient system for the cloning and functional evaluation of TCR-coding cDNAs. This system, which we named hTEC10, allows for the determination of TCR antigen specificity in less than 10 days, and may therefore constitute a fast and powerful platform for the development of new TCR-based anticancer therapies.
T cell receptor (TCR)-based gene immunotherapy is a promising strategy for the treatment of various cancers. Despite its great potential, this approach is still limited to specific tumor-associated antigens (TAAs) and to patients bearing common MHC class I alleles. Generally, characterizing the genes that encode TAA-specific TCRs requires the establishment of TAA-specific T-cell clones, which can take up several months. Furthermore, the screening of large amounts of T-cell clones is laborious. Recently, we have developed a system that allows for the cloning of genes encoding TCR α/β pairs from single TAA-specific T cells and the functional analysis of their antigen-specificity in less than 10 d. We named this system hTEC10, for “human TCR efficient cloning system within 10 days” ().Citation1
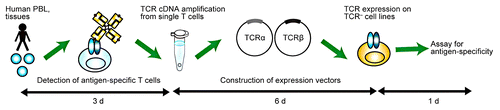
Figure 1. The hTEC10 system. Briefly, the cDNAs coding for human T-cell receptor (TCR) α and β chain are amplified from single T cells and cloned into an expression vector, which is then used to transduce the TCR− T-cell line TG40. The antigen specificity of the TCR can be assessed by staining TCR-expressing TG40 cells with MHC tetramers or by monitoring CD69 expression. Of note, the entire procedure can be performed in less than 10 d. Reproduced with permissions from ref. Citation1.
Using hTEC10, we obtained 73 and 126 α fetoprotein (AFP)-specific TCRα/β-coding cDNA pairs from 2 hepatocellular carcinoma patients who had been successfully treated with an AFP-targeting peptide vaccine. The sequencing of the TCR-coding genes revealed that 199 TCRα/β cDNA pairs were categorized into 3 and 4 cDNA clones, respectively. The functional characterization of 7 AFP-specific TCRs identified one (clone 1–14, obtained from a single T-cell clone out of 199 AFP-specific T-cell clones available) that mediated robust cytotoxic effects against target cells pulsed with AFP-derived peptides. This result suggests that T-cell clones bearing high-affinity TAA-specific TCRs are very rare even among the peripheral blood lymphocytes of patients who had been successfully treated by TAA-targeting vaccines. Using conventional methods, minor T-cell populations such as clone 1–14 would be lost during culture, since large T-cell populations would take over and expand preferentially. Thus, our method is suitable for exploring very small populations of antigen-specific T cells that conventional screening methods may overlook, significantly increasing the possibility to obtain optimal TCR-coding cDNAs for TCR-based gene therapy.
Until recently, numerous studies on the TCR repertoire of antigen-specific T cells have been performed by flow cytometry, based on a panel of monoclonal antibodies specific for TCR β variable fragment (TRBV),Citation2 or by PCR-based methods, using a panel of TRBV-specific primers.Citation3 These methods characterize the TRBV regions of TCRs at the population level, but fail to provide insights into the TCR α variable fragments (TRAVs) as well as into the TRBV regions at a single-cell level. Thus, so far we have not been able to measure the true extent of the clonal diversity within CD8+ cytotoxic T lymphocyte (CTL) populations isolated from cancer patients. In this context, we and others had reported single-cell RT-PCR protocols that permit the simultaneous characterization of the sequences encoding complementarity-determining region 3 (CDR3) α and β in humanCitation4 and mouseCitation5 TCRs. However, these protocols cannot identify TCR α/β pairs, confirm their antigen specificity nor examine their ability to promote cytotoxic effector functions. In contrast, the hTEC10 system may provide us with a new way to analyze the TCR repertoire, as it supplies information of both the TCR α and β chains at the single-cell level and can assess their functional profile. In addition, hTEC10 may provide a useful means to assess the efficacy of anticancer vaccination.
For cancer immunotherapy to be efficient, hence resulting in tumor eradication in vivo, cytotoxic T cells expressing a TCR of sufficiently high avidity are required. In this context, Johnson and colleagues selected CTLs that displayed TAA-specific TCRs with sufficient affinity to induce tumor regression among more than 600 different TAA-specific T cells.Citation6 To obtain T cells with a sufficient avidity to eliminate tumors in vivo, Nauerth and collaborators have recently developed a new assay based on reversible MHC streptamers, allowing for the assessment of the dynamic dissociation (Koff rate) of fluorescently labeled, peptide-loaded MHC class I monomers from TCRs expressed on the surface of living T cells.Citation7 The assay enables a simple, quantitative and reproducible measurement of the Koff rate as a reliable indicator of TCR binding avidity. The combination of the hTEC10 system with this new method may provide us with a valuable approach to selectively retrieve high-affinity T-cell clones for TCR-based gene therapy. Another possible strategy to generate high-affinity TCRs is the genetic alteration of TCR-coding genes. Preliminary clinical trials have already demonstrated that genetically enhanced TCRs can indeed confer improved on-target effector functions to T cells. In this scenario, the hTEC10 system might also contribute by supplying several TCR-coding sequences as starting point for genetic engineering.
Concerning the adverse effect of high-avidity TCRs, 2 patients receiving affinity-enhanced melanoma antigen family A3 (MAGEA3)-specific T cells have recently died owing to the cross-reaction of adoptively transferred lymphocytes with a protein expressed in the pulsating cardiac tissue.Citation8 Furthermore, T cells expressing TCRs whose affinity is much higher than the physiological one have been shown to mediate increased off-target activity at the expenses of on-target effector functions.Citation9 Thus, methods are needed that allow for refining the affinity/avidity of TCRs to optimal levels, ensuring robust on-target effector functions in the absence of severe side effects.
Finally, in combination with MHC multimer-based staining protocols, the hTEC10 system can detect and retrieve TCR α/β cDNA pairs from CD8+ T cells that secrete specific cytokines or express the activation marker tumor necrosis factor receptor superfamily, member 9 (TNFRSF9, best known as CD137 or 4–1BB) upon stimulation with antigenic peptides. Our findings indicate that the hTEC10 system can be used to isolate T cells from cancer patients without the identification of the corresponding TAAs. Briefly, T cells are stimulated with cancer cells followed by the isolation of interferon γ (IFNγ)-secreting or CD137-expressing CD8+ T cells. Once the TCR-coding sequences of these cells are cloned, their antigen-specificity can be examined by analyzing the response of TCR-transduced T cells against malignant cells. This protocol extends the applicability of the hTEC10 system from the cloning of TCRs with known antigen specificity to the retrieval of TCRs of unknown antigen specificity. In conclusion, the hTEC10 system may provide a fast and powerful approach for the development of novel paradigms of TCR-based gene therapy against cancer.
Disclosure of Potential Conflicts of Interest
Authors have applied a patent of hTEC10 system.
Citation: Kobayashi E, Kishi H, Muraguchi A. A novel system for cloning human TCRs: Cutting short the way to TCR-based anticancer therapy. OncoImmunology 2013; 2:e27258; 10.4161/onci.27258
References
- Kobayashi E, Mizukoshi E, Kishi H, Ozawa T, Hamana H, Nagai T, Nakagawa H, Jin A, Kaneko S, Muraguchi A. A new cloning and expression system yields and validates TCRs from blood lymphocytes of patients with cancer within 10 days. Nat Med 2013; 19:1542 - 6; http://dx.doi.org/10.1038/nm.3358; PMID: 24121927
- Bieganowska K, Höllsberg P, Buckle GJ, Lim DG, Greten TF, Schneck J, Altman JD, Jacobson S, Ledis SL, Hanchard B, et al. Direct analysis of viral-specific CD8+ T cells with soluble HLA-A2/Tax11-19 tetramer complexes in patients with human T cell lymphotropic virus-associated myelopathy. J Immunol 1999; 162:1765 - 71; PMID: 9973440
- Eiraku N, Hingorani R, Ijichi S, Machigashira K, Gregersen PK, Monteiro J, Usuku K, Yashiki S, Sonoda S, Osame M, et al. Clonal expansion within CD4+ and CD8+ T cell subsets in human T lymphotropic virus type I-infected individuals. J Immunol 1998; 161:6674 - 80; PMID: 9862696
- Ozawa T, Tajiri K, Kishi H, Muraguchi A. Comprehensive analysis of the functional TCR repertoire at the single-cell level. Biochem Biophys Res Commun 2008; 367:820 - 5; http://dx.doi.org/10.1016/j.bbrc.2008.01.011; PMID: 18191637
- Dash P, McClaren JL, Oguin TH 3rd, Rothwell W, Todd B, Morris MY, Becksfort J, Reynolds C, Brown SA, Doherty PC, et al. Paired analysis of TCRα and TCRβ chains at the single-cell level in mice. J Clin Invest 2011; 121:288 - 95; http://dx.doi.org/10.1172/JCI44752; PMID: 21135507
- Johnson LA, Morgan RA, Dudley ME, Cassard L, Yang JC, Hughes MS, Kammula US, Royal RE, Sherry RM, Wunderlich JR, et al. Gene therapy with human and mouse T-cell receptors mediates cancer regression and targets normal tissues expressing cognate antigen. Blood 2009; 114:535 - 46; http://dx.doi.org/10.1182/blood-2009-03-211714; PMID: 19451549
- Nauerth M, Weißbrich B, Knall R, Franz T, Dössinger G, Bet J, Paszkiewicz PJ, Pfeifer L, Bunse M, Uckert W, et al. TCR-ligand koff rate correlates with the protective capacity of antigen-specific CD8+ T cells for adoptive transfer. Sci Transl Med 2013; 5:192ra87; http://dx.doi.org/10.1126/scitranslmed.3005958; PMID: 23825303
- Linette GP, Stadtmauer EA, Maus MV, Rapoport AP, Levine BL, Emery L, Litzky L, Bagg A, Carreno BM, Cimino PJ, et al. Cardiovascular toxicity and titin cross-reactivity of affinity-enhanced T cells in myeloma and melanoma. Blood 2013; 122:863 - 71; http://dx.doi.org/10.1182/blood-2013-03-490565; PMID: 23770775
- Schmid DA, Irving MB, Posevitz V, Hebeisen M, Posevitz-Fejfar A, Sarria JC, Gomez-Eerland R, Thome M, Schumacher TN, Romero P, et al. Evidence for a TCR affinity threshold delimiting maximal CD8 T cell function. J Immunol 2010; 184:4936 - 46; http://dx.doi.org/10.4049/jimmunol.1000173; PMID: 20351194
