Abstract
We recently reported that variable expression of immune-response genes distinguishes tumor positive sentinel nodes in melanoma patients with malignant progression from those with non-progressing disease. Our results depict sentinel nodes as sites in which immune functions are associated with metastatic disease and identify CD30 as a host immune-related cancer prognostic marker and potential therapeutic target.
The sentinel lymph node (SN) is the node nearest to a primary tumor on the direct lymphatic drainage pathway and the typical local of the earliest metastases in cancer patients. This site although representing a primary immunological barrier to cancer metastatic spread, shows profound signs of immunosuppression even in the absence of infiltrating tumor cells. Immunological defects affecting various immune cell components have been extensively documented in SN of melanoma and breast carcinoma patients. Decreased frequencies of CD8+ effector T cells and stimulatory CD86+ and CD11c+ dendritic cells, as well as the accumulation of Foxp3+ regulatory T cells (Tregs) and T helper 2 (Th2) cytokines, have been reported in SN relative to immune cell frequency and composition in non-tumor draining nodes.Citation1-Citation3 This altered immune phenotype, possibly permitting, or even promoting, the development of further cancer metastases, supports the premise that the SN is a key component of the tumor microenvironment and a sensor of systemic immune dysfunctions, potentially heralding poor outcome.
We recently reported an exploratory study transcriptionally profiling archived sentinel node biopsies (SNB) from patients with differing disease course.Citation4 Tissue remnants after diagnostic pathology were successfully used by the adaptation of gene expression profiling methods to accurately assay the transcriptome using degraded RNA samples extracted from formalin-fixed paraffin-embedded samples. The study sample set was selected to maximize the differences in terms of disease stage and course.
We found that the gene expression profiles stratify tumor-positive SNB samples obtained from patients with further metastatic nodes and disease progression from patients with good prognosis, irrespective of tumor-positive and tumor-negative status of the SNB. Interestingly, the distinct expression profiles do not appear to reflect differences in tumor burden, as determined by careful histopathological examination to quantify tumor cells, as well as by the expression levels of several melanoma-associated genes. In other words, the changes in the SNB transcriptome profiles do not merely depend upon the presence of tumor cells but rather upon defined immune alterations, possibly reflecting specific antitumor properties.
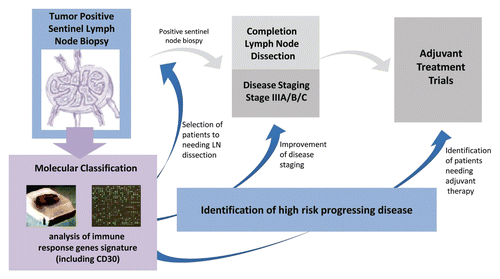
Bioinformatic analysis of the data set with software tools, including Ingenuity Pathway Analysis (IPA) and Gene Set Enrichment Analysis (GSEA), classified differential transcriptional patterns in tumor positive SNB samples mainly according to genes involved in immunity. Among the immune response genes differentially expressed in tumor-positive SNBs isolated from patients with progressive disease, interferon-regulatory factor 1 (IRF1), TNF-receptor associated factor 3 (TRAF3), and lymphotoxin β (LTB) were found to be dowregulated, whereas high mobility group AT-hook protein 1 (HMGA1), tumor necrosis factor receptor superfamily member 8 (TNFRSF8), and basigin (BSG) were upregulated. Taken together, these molecular phenotypes appeared consistent with impaired antitumor immunity in the SNs of patients with progressive disease and poor outcome. Downregulated gene sets included genes involved in (C-X-C) motif receptor 4 (CXCR4) signaling, as well as genes regulating migration and recruitment of immune cells, and downstream targets of the master forkhead family cell-fate regulatory transcription factor FOXP3 in Tregs. Despite the relatively small sample size, our study suggests that the transcriptional profiles of SNB may provide sufficient information to prognosticate metastatic disease behavior and may represent a tool to develop molecular signatures to supplement standard histological approaches for the stratification and treatment of Stage III melanoma patients (see ).
Figure 1. Distinct transcriptional profiles of immune response genes of tumor-positive sentinal node biopsies (SNB) stratify patients with progressing melanoma from those with non-progressing disease. The development of a molecular classification signature to integrate standard pathology assessment of tumor infiltration by sentinel node biopsy may potentially improve disease staging to permit the identification of patients at high risk for recurrence that may benefit from adjuvant therapy. In addition, such a molecular signature could be predictive of the targeted subgroup of patients that may benefit from regional lymph node (LN) dissection, in order to spare overtreatment morbidity and care costs.
Among the different genes involved, we principally focused on TNFRSF8 encoding the activation receptor CD30, a marker reportedly expressed by Tregs during chronic inflammation and autoimmunity,Citation5,Citation6 but not previously investigated in the context of cancer immunity. In SNB from patients with disease progression the increased frequency of CD30+ lymphocytes, particularly localized to the paracortical CD3+ region was confirmed by immunohistology. CD30+ cells typically partially co-express FOXP3, a marker of activated Tregs and programmed cell death-1 (PDCD1, better known as PD-1) a molecule upregulated in anergic lymphocytes. To address the functional consequences of TNFRSF8 differential mRNA expression, CD30 lymphocytes were finely characterized among lymph node cells obtained from tumor invaded regional nodes. Interestingly, higher numbers of CD30+ lymphocytes were detected in tumor-invaded nodes as compared with tumor-free nodes. Phenotypic characterization of the CD30+ lymphocyte population by multiparametric cytofluorimetric analysis revealed that this subset encompasses a variety of cells including CD4-CD8 double negative T cells, CD4+ cells positive for FOXP3 or PDCD1, and CD19+ B lymphocytes. Functionally, CD30+ lymphocytes show impaired proliferative capacity and reduced cytokine production relative to CD30 non-expressing lymphocytes. Of particular note, CD30+ lymphocytes have been detected in the peripheral blood of melanoma patients with advanced regional or systemic disease, primarily among CD3+ and CD4+ subsets of T cells.
CD30 is a member of the tumor necrosis factor (TNF) receptor superfamily, expressed primarily on activated T lymphocytes, as well as some B cells and natural killer cells. CD30 ligand (CD30L or CD153) naturally belongs to TNF family and is mainly expressed on the surface of activated CD4+ T cells and B cells. In addition, sCD30, a soluble form of CD30 derived from the extracellular part of membrane-bound CD30 liberated by proteolytical cleavage mediated by ADAM metalloproteases, binds to CD30L, thus acting as a decoy receptor.Citation7 Although the primary biological functions of CD30 signaling remain obscure, they encompass the control of memory cells, providing co-stimulation and trafficking information, and the induction of lymphocyte apoptosis in a microenviroment and cytokine milieu-dependent manner. In fact, similar to other members of the TNFR family, CD30 engagement mainly regulates T-cell survival and attenuates cytolytic capacity. Finally, in B cells, a potential role for CD30 in regulating humoral immunity has also been reported.Citation8
As mentioned above, there has been a paucity of information regarding the role of CD30 in cancer-related immune responses. CD30 is a unique marker for Hodgkin’s lymphoma, anaplastic large cell lymphoma, and germ cells tumors and the target of an agonistic anti-CD30 drug conjugate recently approved by the Federal Drug Adminstration for the treatment of lymphoproliferative malignancies.Citation9 Preliminary experiments have shown that CD30 is upregulated in double negative T cells and in Tregs after co-culture of peripheral blood lymphocytes with melanoma cells, indicating a potential direct role of tumor cells in the induction of CD30 expression. Moreover, in mice bearing B16 or RET melanoma, staining for CD30 indicates high expression levels in tumor-draining lymph nodes and in nodes positive for metastatic melanoma cells. Whether targeting the CD30/CD30L signaling disrupts tumor immunosuppression shall be addressed in our next studies.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Citation: Rodolfo M, Castelli C, Rivoltini L. Immune response markers in sentinel nodes may predict melanoma progression. OncoImmunology 2014; 3:e28498; 10.4161/onci.28498
References
- Cochran AJ, Huang RR, Lee J, Itakura E, Leong SP, Essner R. Tumour-induced immune modulation of sentinel lymph nodes. Nat Rev Immunol 2006; 6:659 - 70; http://dx.doi.org/10.1038/nri1919; PMID: 16932751
- Takeuchi H, Kitajima M, Kitagawa Y. Sentinel lymph node as a target of molecular diagnosis of lymphatic micrometastasis and local immunoresponse to malignant cells. Cancer Sci 2008; 99:441 - 50; http://dx.doi.org/10.1111/j.1349-7006.2007.00672.x; PMID: 18070155
- Zuckerman NS, Yu H, Simons DL, Bhattacharya N, Carcamo-Cavazos V, Yan N, Dirbas FM, Johnson DL, Schwartz EJ, Lee PP. Altered local and systemic immune profiles underlie lymph node metastasis in breast cancer patients. Int J Cancer 2013; 132:2537 - 47; http://dx.doi.org/10.1002/ijc.27933; PMID: 23136075
- Vallacchi V, Vergani E, Camisaschi C, Deho P, Cabras AD, Sensi M, De Cecco L, Bassani N, Ambrogi F, Carbone A, et al. Transcriptional profiling of melanoma sentinel nodes identify patients with poor outcome and reveal an association of CD30+ T lymphocytes with progression. Cancer Res 2014; 74:130 - 40; http://dx.doi.org/10.1158/0008-5472.CAN-13-1672; PMID: 24395820
- Ellis TM, Simms PE, Slivnick DJ, Jäck HM, Fisher RI. CD30 is a signal-transducing molecule that defines a subset of human activated CD45RO+ T cells. J Immunol 1993; 151:2380 - 9; PMID: 8103064
- de Kleer I, Vercoulen Y, Klein M, Meerding J, Albani S, van der Zee R, Sawitzki B, Hamann A, Kuis W, Prakken B. CD30 discriminates heat shock protein 60-induced FOXP3+ CD4+ T cells with a regulatory phenotype. J Immunol 2010; 185:2071 - 9; http://dx.doi.org/10.4049/jimmunol.0901901; PMID: 20631311
- Tarkowski M. Expression and a role of CD30 in regulation of T-cell activity. Curr Opin Hematol 2003; 10:267 - 71; http://dx.doi.org/10.1097/00062752-200307000-00003; PMID: 12799531
- Kennedy MK, Willis CR, Armitage RJ. Deciphering CD30 ligand biology and its role in humoral immunity. Immunology 2006; 118:143 - 52; http://dx.doi.org/10.1111/j.1365-2567.2006.02354.x; PMID: 16771849
- Muta H, Podack ER. CD30: from basic research to cancer therapy. Immunol Res 2013; 57:151 - 8; http://dx.doi.org/10.1007/s12026-013-8464-1; PMID: 24233555
