Abstract
A key challenge facing the cancer immunology field is the discovery of the most suitable targets for therapeutic intervention. We recently reported a novel RNA-interference (RNAi)-based approach for systematic discovery of such targets in the tumor microenvironment in vivo utilizing pooled shRNA libraries as a screening tool. Here, we discuss applying this unbiased method to develop innovative cancer therapeutics.
Why is Drug Discovery In Vivo Important?
Cancer immunotherapy has made substantial progress over the past several years, such that immune-based approaches now provide significant benefit to a subset of cancer patients, even in the face of advanced disease.Citation1-Citation5 Thus, the discovery and prioritization of new targets is among the foremost challenges for the cancer immunotherapy field.
The immune system represents a mobile army whose soldiers—the myriad of functionally and phenotypically specialized subsets of immune cells—continuously patrol the body for threats to the organism. On their journey, immune cells interact with diverse cell populations in distinct microenvironments. Effector T cells play a central role in antitumor immunity and their effectiveness depends on their complex interactions with both cancer cells and many other cell types in various tissue contexts. These include antigen-presenting cells (APCs) in the tumor-draining lymph nodes, tumor-vasculature endothelial cells, and inhibitory immune cells, such as tumor-infiltrating myeloid-derived suppressor cells. These complex interactions are difficult to model using in vitro culture systems. Our hypothesis is that this complexity provides many opportunities for therapeutic intervention, in particular for discovery of genes that selectively affect T-cell behavior in defined tissue microenvironments, such as occurs in tumors.
RNAi-Based Discovery of Negative Regulators of T-cell Function in Tumors
Based on these considerations, we devised an in vivo short hairpin RNA (shRNA) screening approach that enables investigation of candidate molecules regulating T-cell behavior in particular microenvironments, specifically tumors and secondary lymphoid organs.Citation6 The study took advantage of pooled shRNA screening approaches that enable simultaneous investigation of many genes.Citation7,Citation8 Our goal was the discovery of key auto-inhibitory genes expressed by T cells in immunosuppressive tumor microenvironments. When T cells are activated by a microbial antigen in the setting of an acute infection—or in response to tumor antigens—they undergo a fundamental change in cellular state switching from quiescence to rapid cell division and acquisition of effector functions. Such T-cell receptor driven proliferation is inhibited in tumors by a variety of negative signals.Citation9,Citation10 We asked which genes encode key negative regulators of T-cell function in tumors by screening for shRNAs that restored T-cell proliferation following tumor antigen recognition ().
Figure 1. In vivo shRNA screen for the discovery of negative regulators of T-cell function in tumors. (A) Ovalbumin (Ova)-specific T cells were infected with pooled shRNA libraries encoded in a lentiviral vector prior to being injected into mice bearing B16-Ova melanomas. (B) shRNAs that attenuated key negative regulators relieved T cell inhibition, thereby enabling substantial T-cell expansion preferentially within tumors in response to tumor-antigen recognition. (C) Deep sequencing of the shRNA cassette from purified T cells provided a quantitative representation of all shRNAs in the pool across different tissues. shRNAs enabling T-cell proliferation in tumors were substantially enriched due to lentiviral vector integration into the genome of T cells.
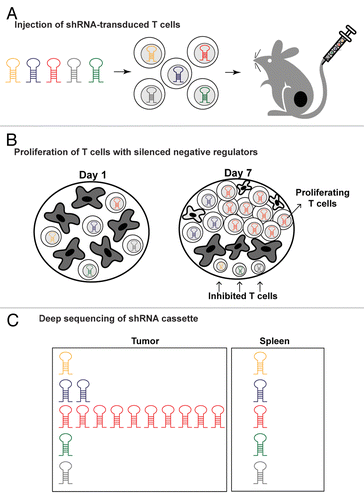
We infected CD8+ T cells with pooled lentiviral shRNA libraries following brief in vitro culture with homeostatic cytokines. Stimulation through the T-cell receptor (TCR)-CD3 complex was intentionally avoided to prevent their differentiation into effector cells. Transduced CD8+ T cells- derived from OT-I ovalbumin-specific TCR transgenic mice- were then transferred to mice bearing B16 tumors that expressed ovalbumin as a surrogate tumor antigen. shRNAs that enabled T-cell accumulation in tumors were discovered by Illumina sequencing of the shRNA cassette from T cells isolated from tumors and other tissues. This approach provided a quantitative representation of all shRNAs in the pool and enabled identification of the shRNAs that were preferentially enriched in tumors but not control organs (irrelevant lymph nodes and spleen). Candidate T-cell inhibitory genes identified from the primary screen were validated by the creation of focused shRNA pools in which each putative T cell regulatory molecule was targeted by approximately 15 shRNAs. We identified 43 genes for which at least three shRNAs induced selective enrichment of T cells in tumors as compared with spleen (≥ 4-fold shRNA enrichment). We further investigated the function of Ppp2r2d (protein phosphatase 2, subunit B delta), a regulatory subunit of the PP2A family of phosphatases. Silencing of this gene substantially reduced T-cell apoptosis in tumors while concomitantly enhancing T-cell proliferation and cytokine production, biological effects culminating in potentiated antitumor activity.Citation6
Important Technical Considerations for Successful shRNA Screens In Vivo
There are several important technical considerations for the successful application of such a screen in vivo. First, it is important to compare the activity of shRNAs across organs, including an organ in which little change is expected. We were particularly interested in shRNAs that were strongly enriched in tumors but not in irrelevant lymph nodes or the spleen. Since shRNAs within a given library may vary in relative abundance, the plotting of the shRNA levels in the organ or tissue of interest, in this case, in tumors, relative to a control organ (e.g., the spleen) addresses this technical issue. Second, each shRNA should be represented many times in the relevant organ for reproducible results. We aimed for > 100-fold representation of each shRNA among tumor-infiltrating T cells. This tissue limits the total number of shRNAs that can be analyzed in a given experiment. We assayed up to ~1000 shRNAs per pool, and 15 recipient mice (per pool) were used to generate sufficient numbers of T cells for DNA isolation and deep sequencing of the integrated shRNA cassette, as well as to reduce potential variability among mice. It may be feasible to utilize larger number of shRNAs per pool, depending on the design of the screen. Third, a large number of control shRNAs within each pool are absolutely essential to assess the frequency of off-target effects.
Future Applications in Immunotherapeutic Discovery
We believe that there are many applications for this non-biased approach, both in the investigation of immune cell function in specific tissue microenvironments in vivo, as well as in the discovery of new therapeutic targets. This general approach should be amenable to the study of other important immune cell populations in a physiological context and to discover genes that control key aspects of behavior in vivo, including survival, tissue homing, activation, and (or) acquisition of key effector functions. Considering the integral involvement of these cell properties in immunity, there are also exciting applications for our screening method in the development of novel cancer immunotherapies. A major application is the discovery of shRNAs that enhance the therapeutic activity of adoptively transferred human T cells. To this end, we have initiated such efforts and experiments are underway using human T cells in a xenotransplantation model. Our approach may also impact the field by guiding the pursuit of selected targets for the development of antibody or small molecule based inhibitors, depending on their cellular localization and function. We believe that the discovery of therapeutic targets on the basis of studies performed in vivo will provide new and exciting avenues for the development of cancer immunotherapeutics.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Citation: Zhou P, Wucherpfennig KW. Discovering cancer immunotherapy targets in vivo. OncoImmunology 2014; 3:e28500; 10.4161/onci.28500
References
- Hodi FS, O’Day SJ, McDermott DF, Weber RW, Sosman JA, Haanen JB, Gonzalez R, Robert C, Schadendorf D, Hassel JC, et al. Improved survival with ipilimumab in patients with metastatic melanoma. N Engl J Med 2010; 363:711 - 23; http://dx.doi.org/10.1056/NEJMoa1003466; PMID: 20525992
- Topalian SL, Hodi FS, Brahmer JR, Gettinger SN, Smith DC, McDermott DF, Powderly JD, Carvajal RD, Sosman JA, Atkins MB, et al. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N Engl J Med 2012; 366:2443 - 54; http://dx.doi.org/10.1056/NEJMoa1200690; PMID: 22658127
- Brahmer JR, Tykodi SS, Chow LQ, Hwu WJ, Topalian SL, Hwu P, Drake CG, Camacho LH, Kauh J, Odunsi K, et al. Safety and activity of anti-PD-L1 antibody in patients with advanced cancer. N Engl J Med 2012; 366:2455 - 65; http://dx.doi.org/10.1056/NEJMoa1200694; PMID: 22658128
- Kalos M, June CH. Adoptive T cell transfer for cancer immunotherapy in the era of synthetic biology. Immunity 2013; 39:49 - 60; http://dx.doi.org/10.1016/j.immuni.2013.07.002; PMID: 23890063
- Korman AJ, Peggs KS, Allison JP. Checkpoint blockade in cancer immunotherapy. Adv Immunol 2006; 90:297 - 339; http://dx.doi.org/10.1016/S0065-2776(06)90008-X; PMID: 16730267
- Zhou P, Shaffer DR, Alvarez Arias DA, Nakazaki Y, Pos W, Torres AJ, Cremasco V, Dougan SK, Cowley GS, Elpek K, et al. In vivo discovery of immunotherapy targets in the tumour microenvironment. Nature 2014; 506:52 - 7; http://dx.doi.org/10.1038/nature12988; PMID: 24476824
- Luo B, Cheung HW, Subramanian A, Sharifnia T, Okamoto M, Yang X, Hinkle G, Boehm JS, Beroukhim R, Weir BA, et al. Highly parallel identification of essential genes in cancer cells. Proc Natl Acad Sci U S A 2008; 105:20380 - 5; http://dx.doi.org/10.1073/pnas.0810485105; PMID: 19091943
- Zender L, Xue W, Zuber J, Semighini CP, Krasnitz A, Ma B, Zender P, Kubicka S, Luk JM, Schirmacher P, et al. An oncogenomics-based in vivo RNAi screen identifies tumor suppressors in liver cancer. Cell 2008; 135:852 - 64; http://dx.doi.org/10.1016/j.cell.2008.09.061; PMID: 19012953
- Shiao SL, Ganesan AP, Rugo HS, Coussens LM. Immune microenvironments in solid tumors: new targets for therapy. Genes Dev 2011; 25:2559 - 72; http://dx.doi.org/10.1101/gad.169029.111; PMID: 22190457
- Nishikawa H, Sakaguchi S. Regulatory T cells in tumor immunity. Int J Cancer 2010; 127:759 - 67; PMID: 20518016
