Abstract
Recently, we provided the first genetic evidence for the requirement of tomato PLC4 and PLC6 genes in defense activation and disease resistance. The encoded enzymes were catalytically active as they were able to degrade phosphatidylinositol (PI), thereby producing diacylglycerol (DG). Here we report differential PLC gene expression following the initiation of defense signaling by the interaction between Cladosporium fulvum resistance (R) protein Cf-4 and its matching effector Avr4 in tomato hybrid seedlings that express both Cf-4 and Avr4. Furthermore, we observed that PLC3 and PLC6 gene expression is upregulated by elevated temperature in the control seedlings. This upregulation coincides with an increase in the levels of phosphatidic acid (PA) and a decrease in the levels of PI and phosphatidylinositol phosphate (PIP). The decrease in PI and PIP levels matches with the activation of PLC. In addition, the levels of the structural phospholipids phosphatidylcholine (PC), phosphatidylethanolamine (PE) and phosphatidylglycerol (PG) declined transiently during recovery after the exposure to elevated temperature., Further studies will be required to explain the mechanism causing the sustained accumulation of PA during recovery, combined with a reduction in the levels of structural phospholipids.
In an incompatible interaction between a host plant and a pathogenic microbe, in which the plant is resistant and the pathogen is avirulent, the matching products of a resistance (R)-avirulence (Avr) gene combination trigger immune signaling in the plant. The interaction between R and Avr proteins occurs immediately upon pathogen ingress and allows a swift detection of the invader and immediate execution of effective defense responses by the host plant, including the induction of a hypersensitive response (HR) during which programmed cell death takes place.Citation1,Citation2 The activation of the innate immune system of the plant eventually results in the restriction of pathogen outgrowth. Recent studies on plant innate immunity revealed that, similar to mammals, plants have the ability to exploit membrane phospholipid modifications as a means to relay defense signals rapidly after pathogen recognition. Phospholipid-modifying enzymes are therefore considered to be key players in successful defense of plants against intruders.Citation3-Citation5
Several studies showed that cellular defense signaling requires phosphatidylinositol-specific phospholipase C (PI-PLC) enzyme activity. PI-PLC enzymes are considered to be signal transducers, mainly due to the signaling roles attributed to their substrates and reaction products. PLC activity essentially leads to the hydrolysis of phosphatidylinositol-4,5-bisphosphate (PtdIns(4,5)P2) into diacylglycerol (DG) and inositol-1,4,5-triphosphate (Ins(1,4,5)P3).Citation6,Citation7 Subsequent metabolism of these products affects the final outcome of the responses. Ins(1,4,5)P3 is essential for calcium entry from internal and external stores in mammals. In plants, Ins(1,4,5)P3 and its further phosphorylated forms IP4, IP5 and IP6, of which the latter is referred to as phytate, might have similar cellular functions, in addition to a role in auxin signaling.Citation8-Citation10 DG can be phosphorylated by diacylglycerol kinase (DGK) to generate phosphatidic acid (PA). DG is known to activate protein kinase C (PKC) in mammals while PA also plays an important role in cellular signaling in mammals and appears to have similar role in plants.Citation11-Citation16 Phospholipase D (PLD) enzymes also hydrolyse phospholipids, thereby generating PA. For PLDs this occurs in a single step, by hydrolysis of structural phospholipids such as phosphatidylcholine (PC), phosphatidylethanolamine (PE) and phosphatidylglycerol (PG),.Citation7,Citation17 PLDs are involved in plant growth and development, in addition to a multitude of stress responses including biotic and abiotic stress.Citation17-Citation19 Phosphorylation of PA by PA kinaseCitation20 occurs in plants and leads to the generation of diacylglycerol pyrophosphate (DGPP).Citation21 DGPP is a common plant phospholipid which is present at trace amounts under resting conditions and accumulates under general stress conditions, in the presence of elicitors and during pathogen infection processes.Citation3,Citation4,Citation22,Citation23
Several years of extensive research in mammalian systems has expanded our understanding of the important role of PLCs in different aspects of cellular signaling, including immune responses.Citation24-Citation26 Likewise, we recently discovered that also in plants effective innate-immunity requires catalytically active PLC enzymes. Two PLC genes from tomato (Solanum lycopersicum) were found to be essential for the efficient arrest of different types of plant pathogens and for the initiation of immunity-driven cell death.Citation5 Furthermore, differential regulation of gene expression was observed for six PLC genes upon infection of tomato plants by the fungus Cladosporium fulvum, the causal organism of tomato leaf mold disease. This was deduced from infections of susceptible tomato plants, lacking a resistance gene to C. fulvum (Cf-0), resulting in a compatible interaction, and resistant plants carrying the Cf-4 resistance gene, with a C. fulvum strain secreting the Avr4 effector, resulting in an incompatible interaction. Although we observed differential expression patterns for all PLC genes in both susceptible and resistant plants, these patterns were distinct between the incompatible and compatible interactions.Citation5
The interaction between plants and their pathogens is very complex and moreover at first contact, defense signaling will only occur locally at the infection sites, which furthermore appear non-synchronously. As a consequence, only a few cells in the infected tissue will induce a local defense response in a rather non-synchronised way. Therefore, we here studied the expression of the tomato PLC genes in a more synchronised and homogeneous system, where the pathogen is absent and defense signaling is activated solely by the R/Avr interaction, and more importantly, in all cells of the plant. For this purpose we used tomato seedlings expressing both the Cf-4 resistance gene and the matching fungal avirulence gene Avr4.Citation27,Citation28 The immune response in these Cf-4/Avr4-expressing hybrid seedlings is suppressed by growing them at 33°C and 100% relative humidity (RH). Subsequently, defense signaling is synchronously activated by transferring the plants to 20°C and 70% RH. This shift in growth conditions releases the immune response blockade and triggers defense signaling throughout the whole seedling, eventually culminating into the death of the seedling as a result of systemic HR.Citation29 We germinated seeds of a 1:1 mixture of the parental lines (control) and the hybrid Cf-4/Avr4 line in closed transparent containers that were placed in an incubator set to 20°C with a 16 h/day light regime. After one week, one set of seedlings (15–20 seedlings) from the control was left at 20°C to be used as a reference for the effect of the elevated temperature. Other sets, with equal numbers (15–20 seedlings) of the control and Cf-4/Avr4 seedlings, were transferred to a rescue condition, being 33°C/100% RH (16 h/day light regime) and incubated for a period of 2 weeks. Subsequently, the seedlings were shifted from 33°C/100% RH to 20°C/70% RH (16 h/day light regime) which then triggers the Cf4/Avr4-mediated immune responses. Control and Cf-4/Avr4 seedlings were harvested between 0 to 6 h after the temperature and humidity shift by immersing them directly into liquid N2. Total RNA was extracted and cDNA was generated, which was subsequently used for expression analysis of the individual tomato PLC genes as described before.Citation5
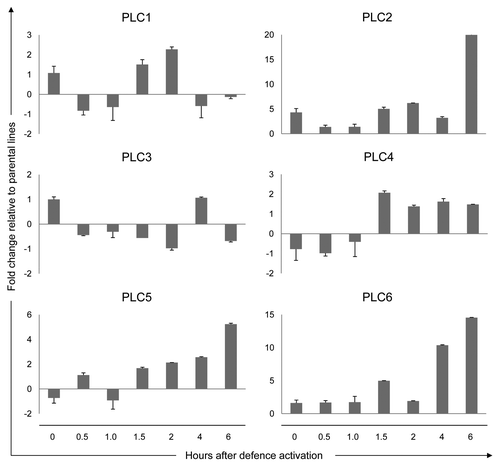
Differential expression patterns were observed for most tomato PLC genes upon initiating Cf-4/Avr4-mediated defense activation by the shift in growth conditions (). For PLC2, PLC4, PLC5 and PLC6 matching expression patterns, although within a different time-frame, were previously observed for Cf-4 plants inoculated with an avirulent Avr4-expressing strain of C. fulvum.Citation5 In the case of the Cf-4/Avr4 seedlings, transcriptional upregulation of the indicated PLCs occurs at 1.5 h after the temperature and humidity shift. In contrast, such expression patterns were obtained at 5 d after inoculation in Cf-4 tomato inoculated with an avirulent strain of C. fulvum mentioned above. The similarity between these PLC expression profiles is expected to be a consequence of plant defense activation specifically due to the Cf-4/Avr4 interaction and subsequent triggering of downstream signaling. In the Cf-4/Avr4 seedlings, PLC expression might be enhanced due to the activation of defense throughout the plant. Remarkably, changes in PLC expression in the C. fulvum-inoculated plants appeared to be transient, whereas in the Cf-4/Avr4 seedlings, probably due to continuous triggering of defense signaling, these changes become even more pronounced at later time points (). In the Cf-4/Avr4 seedlings, effector recognition occurs simultaneously in every cell as compared with its occurrence in only a limited number of cells present at the infection sites in the C. fulvum-inoculated Cf-4 plants. Furthermore, in C. fulvum-inoculated Cf-4 plants these PLC expression patterns correlate to the fungal growth arrest at 5–7 d upon inoculationCitation5 ().
Figure 1.PLC expression during the activation of defense in the Cf-4/Avr4 tomato seedlings. In Cf-4/Avr4 seedlings, defense signaling is activated by the interaction between the resistance protein Cf-4 and the matching C. fulvum effector Avr4. Defense is initiated by a shift in the growth conditions of the plants (see text for details). The Cf-4/Avr4 seedlings and a 1:1 mixture of the seedlings from the parental Cf-4- and Avr4-carrying lines (control) were harvested at the indicated time points after defense initiation and expression of the different tomato PLCs was analyzed by quantitative Real-Time PCR. The relative PLC expression levels in the Cf-4/Avr4 plants were calculated using the expression of the tomato Actin gene as an internal reference to normalize for the amount of template present in the different samples. PLC expression in the parental lines was used as a control to calculate the fold change in the Cf-4/Avr4 plants for a given time point. Relative expression is presented for the six tomato PLC genes (PLC1 to -6). Error bars represent the standard error of two quantitative PCR samples from the same cDNA archive. The experiment was performed twice, with similar results.
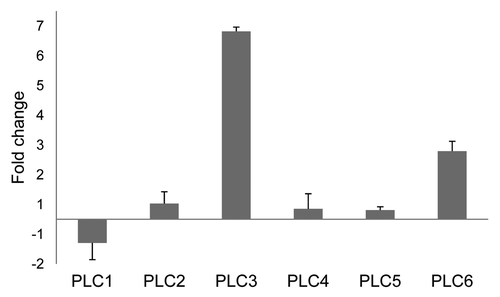
The effect of the shift in growth conditions on the expression of the tomato PLCs in the absence of recognition of Avr4 by Cf-4 was also examined. Now only the 1:1 mixture of the parental seedlings was used, which was either incubated at 33°C/100% RH or at 20°C/70% RH and (16 h/day light regime) after germination at 20°C over a period of one week. After two weeks of subsequent incubation at 33°C, PLC3 expression levels were about 6-fold higher and PLC6 expression levels were about 2-fold higher than those found in seedlings grown at 20°C (n = 2). Other PLC genes were apparently not majorly affected in their expression (). This indicates that the expression of PLC3 and PLC6 is constitutively upregulated due to the shift to the elevated temperature and humidity ().
Figure 2. Effect of elevated temperature on PLC expression in tomato seedlings. Relative expression of the six tomato PLC genes (PLC1 to -6) is presented for a 1:1 mixture of seedlings of the parental lines which were maintained at 33°C. Relative expression was calculated using the expression of the tomato Actin gene as an internal reference and the PLC expression in the seedlings which were maintained at 20°C as control. This allowed determining the changes in PLC expression as a result of elevated temperature. Error bars represent the standard error of two quantitative PCR samples from the same cDNA archive. The experiment was performed twice, with similar results.
These findings prompted us to study whether we could observe changes in phospholipid profiles under the different growth conditions for the Cf-4/Avr4- and control seedlings. Cold and heat treatments are known to affect phospholipid profiles in Arabidopsis and tobacco cell suspensions.Citation19,Citation30 Initially, we were interested to determine whether PLCs are activated after Cf-4/Avr4-mediated changes in PLC gene expression (). For this, we metabolically labeled the phospholipids in seedlings by feeding them with the radioactive isotope orthophosphate-32 (32P), prior to the temperature shift. Radioactive labeling was conducted by cutting 10-d-old seedlings, again germinated at 20°C, directly above the soil and placing them into a 24 wells micro-titer plate, containing 1 ml of tap water and 100 µCi 32P per well. The plate was incubated overnight at either 20°C or 33°C with a 16 h light/day regime. The phospholipids in the seedlings are 32P-labeled overnight to such an extent that no major changes were anticipated to occur during the period of the sampling.Citation11,Citation31 Seedlings incubated at 33°C were shifted to 20°C and equal numbers of seedlings (3–4 seedlings) were collected after distinct time points. Control samples were collected at the same time points from 32P-fed seedlings which were continuously kept at 20°C. Collected seedlings were flash frozen in a mortar filled with liquid nitrogen and each sample was transferred promptly to a precooled micro tube and placed in liquid nitrogen. Subsequently, phospholipid isolation was performed as described before with minor modifications (see Supplementary Method).Citation3 The phospholipids were dissolved in chloroform and separated by one dimensional thin layer chromatography (TLC) using alkaline conditions. After completion of the run, TLC plates were allowed to dry at room temperature and radioactivity was detected by autoradiography (KODAK T-MAX 100).Citation22
Initial experiments indicated that no differences existed between the phospholipid patterns of the Cf-4/Avr4 and control seedlings when subjected to the temperature shift from 33°C to 20°C, as in both types of seedlings the PA levels increased(data not shown). This increase could result from either the activation of the PLC/DGK pathway or could be caused by increased phospholipase D (PLD) activity. Most likely, both pathways contribute simultaneously to the observed increase in PA levels. From these results we concluded that the effect of the temperature shift dominates the defense-related effectsCitation4 by several orders of magnitude, which makes it difficult to distinguish the latter. To study the phospholipid profile changes upon the temperature shift in more detail, 10-d-old control seedlings germinated at 20°C and grown for three days at 33°C/100% RH and 16 h/day light regime were pre-labeled overnight with 32P at 33°C and then transferred to 20°C/70% RH and 16 h/day light regime. Seedlings were sampled at 0, 1, 2, 4 and 6h after the temperature shift, phospholipids were extracted and analyzed by TLC as described above. Phospholipid profiles were compared with those obtained from seedlings that were kept at 20°C at all steps and that were harvested at the 6h time point. shows that seedlings maintained at 33°C (t = 0) have low levels of phosphatidylinositol (PI) and phosphatidylinositol phosphate (PIP), whereas the levels of the phospholipids PA, DGPP, PC, PE and PG were elevated in comparison to the control seedlings that were maintained at 20°C (; compare the left and right lanes). These results strongly indicate that the phospholipid profile of plants is highly temperature-dependent even at these moderate conditions. After shifting the seedlings from 33°C to 20°C, the levels of PI gradually increased toward the basal levels of plants kept at 20°C. PA levels, already relatively high at 33°C, increased significantly after the temperature downshift. In contrast, PA was only present at very low levels in the seedlings which were maintained at 20°C and harvested at the 6h time point. Most remarkably, we observed a rapid decrease in the levels of the structural phospholipids. Within 2 h, the levels of PC, PE and PG decreased to background levels, whereas under control conditions these typically represent the major radiolabelled species. Their decrease coincided with an increase in PA and DGPP. PLD activation during temperature shifts is a common phenomenon in plants, as demonstrated in various plant systems by PA accumulation and reduced PLD substrate levels.Citation19,Citation30 For example, a recent study showed that PLD activity increased when the temperature was raised. However, in this case this was accompanied by a significant increase in PtdIns(4,5)P2, a phospholipid species that lacks in our observations, probably due to labeling restrictions in whole plants.Citation19 The effects observed also resemble the triggering of PLD activity during water deficiency in whole plants,Citation32 although we anticipated an opposite effect since recovery from an elevated temperature by shifting the seedlings to 20°C is not expected to simulate drought stress. However, it should be noted that the 33°C to 20°C temperature shift is also accompanied by a 100% RH to 70%RH shift. As the seedlings have their stomata fully opened at 33°C/100%RH (results not shown), cutting off the seedlings and lowering the temperature and humidity results in an instantaneous water loss and wilting (results not shown). The seedlings soon recover, however, this short period of drought stress might be the cause of PLD activation. In conclusion, the rapid decrease of PC, PE and PG levels suggests that PLD is involved in the adaptation or recovery process ().
Figure 3. Changes in the phospholipid profile are triggered by lowering the temperature. Total phospholipids were labeled in vivo using 32P, isolated and analyzed by alkaline TLC and detected using autoradiography. Alkaline TLC profiles of total phospholipids are shown from a 1:1 mixture of seedlings of tomato parental lines (control) grown at either 20°C (right lane) and 33°C (left lane) and those shifted from 33°C to 20°C, at 1, 2, 4 and 6 h after the shift. The control seedlings were sampled together with the 6 h shifted seedlings. Note the differences in the levels of PA, DGPP, PI and PIP between the seedlings which were grown at 20°C or at 33°C. Also note that after the temperature shift form 33°C to 20°C there is a massive increase in the amount of PA and DGPP and a decrease in PC, PE and PG.
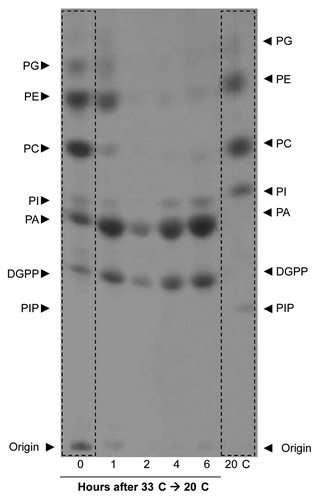
In conclusion, the upregulation of the gene expression of PLC3 and PLC6 at 33°C () shows that PLCs might be involved in the cellular responses of tomato to elevated temperatures. Together with our previous finding that PLC6 is able to degrade PI and produce DG,Citation5 which can be phosphorylated by DGK to generate PA, we conclude that an elevated temperature leads to the activation of the PLC/DGK pathway in tomato and that this is accompanied by transcriptional upregulation of PLC3 and PLC6. Supporting this conclusion is the reduced PI level and depletion concealment of PIP, which is accompanied by increased levels of PA and DGPP in the seedlings growing at 33°C, as compared with those growing at 20°C. We emphasize that PLD activity also might contribute to the PA increase, since the levels of the structural phospholipids PC, PE and PG, which are all PLD substrates, decrease significantly upon the temperature shift, an effect also shown for cold acclimation of Arabidopsis cells.Citation30 This supports a role for PLD in the temperature adaptation process, most likely to replace a pool of structural phospholipids which was formed during high temperature conditions and allowing the synthesis of structural phospholipids with different properties and thus more suitable for membrane stability and fluidity at lower temperatures. Further studies are required to confirm this increased PLD activity, for example by using a transphosphatidylation assay.Citation33 In such an assay, the phosphatidate moiety is preferentially transferred from a structural phospholipid to a primary alcohol leading to the formation of the corresponding phosphatidyl alcohol which is metabolically stable and readily detectable by TLC techniques. We anticipate that molecular differences exist in the structural phospholipid pool generated at various temperatures due to changes in the composition of the acyl chains, as has been reported during cold exposure in Arabidopsis.Citation30 This will modulate membrane properties and the interaction with membrane-localized proteins. It will be intriguing to determine whether such structural membrane changes are required to maintain plant immune responses modulated by temperature. The observed overall increase in PA content as a result of elevated temperature might be a mechanism by which plants recruit different types of PA-interacting protein complexes to be able to cope with acute changes in the ambient temperature. This suggestion is supported by the finding that heat shock protein 90 (Hsp90) was identified as a PA-binding target in plants.Citation34
| Abbreviations: | ||
| PI-PLC | = | phosphatidylinositol-specific phospholipase C |
| PLC | = | phospholipase C |
| PLD | = | phospholipase D |
| DGK | = | diacylglycerol kinase |
| PAMPs | = | pathogen-associated molecular patterns |
| R gene | = | resistance gene |
| Avr gene | = | avirulence gene. HR, hypersensitive response |
| PA | = | phosphatidic acid |
| PI | = | phosphatidylinositol |
| PIP | = | phosphatidylinositol phosphate |
| PC | = | phosphatidylcholine |
| PE | = | phosphatidylethanolamine |
| PG | = | phosphatidylglycerol |
| PIP2 | = | phosphatidylinositol-4,5-bisphosphate |
| DG | = | diacylglycerol |
| IP3 | = | inositol-1,4,5-triphosphate |
| PKC | = | protein kinase C |
| DGPP | = | diacylglycerol pyrophosphate |
| TLC | = | thin layer chromatography |
| RH | = | relative humidity |
Acknowledgments
The support by the Dutch Organization for Scienti□c Research (NWO); VIDI grant 10281 to HJGM, VENI grant 863.08.018 to WILT, VIDI grant 864.02.008 to MHAJJ (on which JHV was appointed), and Mosaic grant 017.003.046 to AA, is gratefully acknowledged.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- Dangl JL, Jones JD. Plant pathogens and integrated defence responses to infection. Nature 2001; 411:826 - 33; http://dx.doi.org/10.1038/35081161; PMID: 11459065
- Dodds PN, Rathjen JP. Plant immunity: towards an integrated view of plant-pathogen interactions. Nat Rev Genet 2010; 11:539 - 48; http://dx.doi.org/10.1038/nrg2812; PMID: 20585331
- van der Luit AH, Piatti T, van Doorn A, Musgrave A, Felix G, Boller T, et al. Elicitation of suspension-cultured tomato cells triggers the formation of phosphatidic acid and diacylglycerol pyrophosphate. Plant Physiol 2000; 123:1507 - 16; http://dx.doi.org/10.1104/pp.123.4.1507; PMID: 10938366
- de Jong CF, Laxalt AM, Bargmann BOR, de Wit PJGM, Joosten MHAJ, Munnik T. Phosphatidic acid accumulation is an early response in the Cf-4/Avr4 interaction. Plant J 2004; 39:1 - 12; http://dx.doi.org/10.1111/j.1365-313X.2004.02110.x; PMID: 15200638
- Vossen JH, Abd-El-Haliem A, Fradin EF, van den Berg GC, Ekengren SK, Meijer HJG, et al. Identification of tomato phosphatidylinositol-specific phospholipase-C (PI-PLC) family members and the role of PLC4 and PLC6 in HR and disease resistance. Plant J 2010; 62:224 - 39; http://dx.doi.org/10.1111/j.1365-313X.2010.04136.x; PMID: 20088897
- Meldrum E, Parker PJ, Carozzi A. The PtdIns-PLC superfamily and signal transduction. Biochim Biophys Acta 1991; 1092:49 - 71; http://dx.doi.org/10.1016/0167-4889(91)90177-Y; PMID: 1849017
- Meijer HJ, Munnik T. Phospholipid-based signaling in plants. Annu Rev Plant Biol 2003; 54:265 - 306; http://dx.doi.org/10.1146/annurev.arplant.54.031902.134748; PMID: 14502992
- Streb H, Irvine RF, Berridge MJ, Schulz I. Release of Ca2+ from a nonmitochondrial intracellular store in pancreatic acinar cells by inositol-1,4,5-trisphosphate. Nature 1983; 306:67 - 9; http://dx.doi.org/10.1038/306067a0; PMID: 6605482
- Tan X, Calderon-Villalobos LI, Sharon M, Zheng C, Robinson CV, Estelle M, et al. Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature 2007; 446:640 - 5; http://dx.doi.org/10.1038/nature05731; PMID: 17410169
- Stevenson-Paulik J, Bastidas RJ, Chiou ST, Frye RA, York JD. Generation of phytate-free seeds in Arabidopsis through disruption of inositol polyphosphate kinases. Proc Natl Acad Sci U S A 2005; 102:12612 - 7; http://dx.doi.org/10.1073/pnas.0504172102; PMID: 16107538
- Arisz SA, Testerink C, Munnik T. Plant PA signaling via diacylglycerol kinase. Biochim Biophys Acta 2009; 1791:869 - 75; PMID: 19394438
- Kishimoto A, Takai Y, Mori T, Kikkawa U, Nishizuka Y. Activation of calcium and phospholipid-dependent protein kinase by diacylglycerol, its possible relation to phosphatidylinositol turnover. J Biol Chem 1980; 255:2273 - 6; PMID: 7358670
- Mellor H, Parker PJ. The extended protein kinase C superfamily. Biochem J 1998; 332:281 - 92; PMID: 9601053
- Imagawa W, Bandyopadhyay G, Nandi S. Multifunctional phosphatidic acid signaling in mammary epithelial cells: stimulation of phosphoinositide hydrolysis and conversion to diglyceride. J Cell Physiol 1995; 163:561 - 9; http://dx.doi.org/10.1002/jcp.1041630317; PMID: 7775598
- Testerink C, Munnik T. Phosphatidic acid: a multifunctional stress signaling lipid in plants. Trends Plant Sci 2005; 10:368 - 75; http://dx.doi.org/10.1016/j.tplants.2005.06.002; PMID: 16023886
- Wang X, Devaiah SP, Zhang W, Welti R. Signaling functions of phosphatidic acid. Prog Lipid Res 2006; 45:250 - 78; http://dx.doi.org/10.1016/j.plipres.2006.01.005; PMID: 16574237
- Li M, Hong Y, Wang X. Phospholipase D- and phosphatidic acid-mediated signaling in plants. Biochim Biophys Acta 2009; 1791:927 - 35; PMID: 19289179
- Yamaguchi T, Kuroda M, Yamakawa H, Ashizawa T, Hirayae K, Kurimoto L, et al. Suppression of a phospholipase D gene, OsPLDbeta1, activates defense responses and increases disease resistance in rice. Plant Physiol 2009; 150:308 - 19; http://dx.doi.org/10.1104/pp.108.131979; PMID: 19286937
- Mishkind M, Vermeer JE, Darwish E, Munnik T. Heat stress activates phospholipase D and triggers PIP accumulation at the plasma membrane and nucleus. Plant J 2009; 60:10 - 21; http://dx.doi.org/10.1111/j.1365-313X.2009.03933.x; PMID: 19500308
- Wissing JB, Behrbohm H. Phosphatidate kinase, a novel enzyme in phospholipid metabolism (purification, subcellular localization, and occurrence in the plant kingdom). Plant Physiol 1993; 102:1243 - 9; PMID: 12231900
- Munnik T, de Vrije T, Irvine RF, Musgrave A. Identification of diacylglycerol pyrophosphate as a novel metabolic product of phosphatidic acid during G-protein activation in plants. J Biol Chem 1996; 271:15708 - 15; http://dx.doi.org/10.1074/jbc.271.26.15708; PMID: 8663116
- Meijer HJG, ter Riet B, van Himbergen JA, Musgrave A, Munnik T. KCl activates phospholipase D at two different concentration ranges: distinguishing between hyperosmotic stress and membrane depolarization. Plant J 2002; 31:51 - 9; http://dx.doi.org/10.1046/j.1365-313X.2002.01336.x; PMID: 12100482
- van Schooten B, Testerink C, Munnik T. Signalling diacylglycerol pyrophosphate, a new phosphatidic acid metabolite. Biochim Biophys Acta 2006; 1761:151 - 9; PMID: 16469533
- Chiang CY, Veckman V, Limmer K, David M. Phospholipase Cγ-2 and intracellular calcium are required for lipopolysaccharide-induced Toll-like receptor 4 (TLR4) endocytosis and interferon regulatory factor 3 (IRF3) activation. J Biol Chem 2012; 287:3704 - 9; http://dx.doi.org/10.1074/jbc.C111.328559; PMID: 22158869
- Wang D, Feng J, Wen R, Marine JC, Sangster MY, Parganas E, et al. Phospholipase Cgamma2 is essential in the functions of B cell and several Fc receptors. Immunity 2000; 13:25 - 35; http://dx.doi.org/10.1016/S1074-7613(00)00005-4; PMID: 10933392
- Takata M, Homma Y, Kurosaki T. Requirement of phospholipase C-gamma 2 activation in surface immunoglobulin M-induced B cell apoptosis. J Exp Med 1995; 182:907 - 14; http://dx.doi.org/10.1084/jem.182.4.907; PMID: 7561693
- Cai X, Takken FL, Joosten MH, De Wit PJ. Specific recognition of AVR4 and AVR9 results in distinct patterns of hypersensitive cell death in tomato, but similar patterns of defence-related gene expression. Mol Plant Pathol 2001; 2:77 - 86; http://dx.doi.org/10.1046/j.1364-3703.2001.00053.x; PMID: 20572994
- Stulemeijer IJ, Joosten MH, Jensen ON. Quantitative phosphoproteomics of tomato mounting a hypersensitive response reveals a swift suppression of photosynthetic activity and a differential role for hsp90 isoforms. J Proteome Res 2009; 8:1168 - 82; http://dx.doi.org/10.1021/pr800619h; PMID: 19178300
- de Jong CF, Takken FLW, Cai X, de Wit PJGM, Joosten MHAJ. Attenuation of Cf-mediated defense responses at elevated temperatures correlates with a decrease in elicitor-binding sites. Mol Plant Microbe Interact 2002; 15:1040 - 9; http://dx.doi.org/10.1094/MPMI.2002.15.10.1040; PMID: 12437302
- Ruelland E, Cantrel C, Gawer M, Kader JC, Zachowski A. Activation of phospholipases C and D is an early response to a cold exposure in Arabidopsis suspension cells. Plant Physiol 2002; 130:999 - 1007; http://dx.doi.org/10.1104/pp.006080; PMID: 12376663
- Arisz SA, van Himbergen JA, Musgrave A, van den Ende H, Munnik T. Polar glycerolipids of Chlamydomonas moewusii. Phytochemistry 2000; 53:265 - 70; http://dx.doi.org/10.1016/S0031-9422(99)00505-1; PMID: 10680181
- Frank W, Munnik T, Kerkmann K, Salamini F, Bartels D. Water deficit triggers phospholipase D activity in the resurrection plant Craterostigma plantagineum. Plant Cell 2000; 12:111 - 24; PMID: 10634911
- Arisz SA, Valianpour F, van Gennip AH, Munnik T. Substrate preference of stress-activated phospholipase D in Chlamydomonas and its contribution to PA formation. Plant J 2003; 34:595 - 604; http://dx.doi.org/10.1046/j.1365-313X.2003.01750.x; PMID: 12787242
- Testerink C, Dekker HL, Lim ZY, Johns MK, Holmes AB, Koster CG, et al. Isolation and identification of phosphatidic acid targets from plants. Plant J 2004; 39:527 - 36; http://dx.doi.org/10.1111/j.1365-313X.2004.02152.x; PMID: 15272872