Abstract
Mitogen activated protein kinase (MAPK) pathway is one of the most conserved signaling cascade in plants regulating a plethora of cellular processes including normal growth and development, abiotic and biotic stress responses. The perception of external cues triggers the phosphorylation of three tier MAPKKK-MAPKK-MAPK cascade which finally modifies a downstream substrate thereby regulating the cellular processes. Whereas, the transcription regulation by MAPKs, mediated through their substrates is well studied in plants, the transcription and post-transcriptional regulation of the MAPK genes are poorly understood. Previous studies from the animals systems suggested the miRNAs regulate the post-transcriptional regulation of MAPK transcripts. Here we attempt to unravel the post-transcriptional regulation of MAPKs by miRNAs in model crop plant Oryza sativa. Using in silico tools, we predict the miRNAs for 98 out of 99 MAPK transcripts. The predicted miRNAs were validated for the biological relevance of their function. The inverse correlation between relative transcript levels between the MAPKs and their predicted miRNAs validated the in silico prediction. Taken together, this report demonstrates the significance of miRNAs in regulation of the MAPK pathway in plants with a new direction to study the plant signaling molecules.
Introduction
MAP kinases belong to the CMGC (CDK, MAPK, GSK-3, and CKII) group of the protein kinase superfamily and are extensively involved in various stress responses in all the eukaryotes.Citation1,2 Since plants are sessile, they possess a large number of MAP kinase family members to efficiently combat stress conditions. A typical MAP kinase cascade (MAPK cascade) consists of a MAPKKK (MKKK/MEKK/MAP3K) which is phosphorylated and activated by its upstream receptor and in-turn phosphorylates and activates its downstream MAPKK (MKK/MEK/MAP2K). The activated MAPKK then phosphorylates and activates a MAPK (MPK), which ultimately transfers the signal to its specific nuclear or cytoplasmic substrates. Many a times the MAPKs may move into the nucleus to transfer the signals in the form of phosphate groups. The model plant Arabidopsis thaliana consists of 80 MAPKKKs, 10 MAPKKs and 20 MAPKs.Citation3,4 Clearly, in Arabidopsis the signals perceived by 80 MAPKKKs must converge into 10 MAPKKs which is possible by the interaction of each MAPKKs with multiple MAPKKKs. It is also obvious that 20 MAPKs being encoded by the Arabidopsis genome, more than one MPK interacts with a single MAPKK. The same is the case in the model crop plant rice, where the genome encodes 75 MAPKKKs, 8 MAPKKs and 16 MAPKs.Citation4,5
One of the interesting functions of the MAP kinase cascade is the regulation of microRNA biogenesis in humans. MicroRNAs are 21–24 nucleotide smallRNAs molecules that are extensively involved in gene regulation. MKK1-Erk2 signaling phosphorylates TRBP (a dsRNA binding protein), which associates with Dicer to form functional miRNA generating complex.Citation6 In plants, a dsRNA binding protein HYL1 associates with DCL1 to form a functional miRNA generating complex.Citation7 Similar kind of regulation can be anticipated in plants considering the functional conservation of the MAP kinase cascade.
Owing to their large number, size, heterogeneous structure and their limited substrate specificity,Citation8 the knowledge about the functions of MAPKKKs is limited. Out of the 80 MAPKKKs encoded by the Arabidopsis genome only a few of them are functionally characterized. The vast repertoire of MAPKKKs encoded by the plants suggest their diversity and specificity in recognizing the external signals. Therefore, understanding the roles of MAPKKKs and their regulation will complete the knowledge of the whole MAPK cascade itself. Recent studies report the development of artificial microRNA (amiRNA) that specifically targets a MKKK causing reduced sensitivity to ABA during seed germination as compared with wild type.Citation9 Studies from animal systems, especially in C. elegans have clear evidences showing that MAPKKK transcripts are targets of miRNA mediated regulation.Citation10 Studying the similar kind of regulation in plants would reveal exciting details about the conservation of the two major regulatory pathways and the underlying nexus in plant and animal kingdoms. To uncover the functional miRNA mediated-MAP kinase transcript regulation in plants, we chose very important and economic model crop plant rice. In the present study, we predicted the microRNAs targeting rice the three different components MAP kinase cascade transcripts. A single microRNA was identified to target eight different MAPKKK transcripts thereby regulating the complete downstream MAP kinase signaling cascade.
Results
Identification of the potential miRNAs targeting transcripts of components of MAP kinase cascade
Among several tools available online for miRNA target predictions, only a few can be used for the prediction of targeting miRNAs from the target transcript sequence. One such tool is MicroinspectorCitation11 applicable to the plant systems as well. The online tool Microinspector accepts the user specified DNA/RNA sequence to identify the potential miRNA binding sites from the list of the miRNAs provided. The species can be chosen from the list to specify the optimum hybridization temperature and binding energy for the miRNA-target pair. The miRBase release 17 consisted of 452 mature miRNAs from Oryza sativa. All the mature miRNAs from rice were downloaded and submitted each time along with a single MAP kinase coding sequence to the Microinspector tool. The results obtained were saved for each of the MAP kinase transcript for further analysis.
A total of 99 sequences corresponding to 16 MAPKs, 8 MAPKKs and 75 MAPKKKs were submitted for the identification of potential miRNA binding sites and corresponding miRNAs. Among several predicted miRNAs for a single transcript, only one miRNA displaying the least free energy was selected for a particular transcript. For all the members of 99 MAP kinase cascade component sequences analyzed, the miRNAs for 98 members were predicted except for MKK3 (Supplementary Table 2). Interestingly, it was observed that a single miRNA, miR531 is targeting most of the members of MAPK cascade gene family (25 out of 98). Additionally, by using the target search function of PMRD, miR1429_5p was found to potentially target MPK17–1 transcript. The detailed transcript analysis of miR531 and its predicted targets along with miR1429_5p and MPK17–1 were performed to gain deeper insights into their role ().
Table 1. Selected miRNA: target pairs for validation
Experimental design and validation of predicted targets
The miRNA mediated regulation involves either target transcript degradation or translational repression.Citation7 Therefore by principle, the increase in miRNA transcript expression would decrease the target transcript levels and vice-versa, displaying an inverse correlation between miRNA and corresponding transcript. Such observations of inverse transcript correlation were previously reported in Arabidopsis under diurnal conditions.Citation12 The same was applied to a few of the predicted miRNA: target pairs. The analysis were performed by qRT-PCR using ubiquitin as internal control. The rice plants were subjected to two abiotic stress conditions, namely drought and UV-B rays to study the inverse correlation between miRNA and its targets. Seven time points corresponding to 0h, 0.5h, 1h, 3h, 6h, 12h, and 24h were selected for harvesting stress treated samples. For positive control experiments, validated miRNA:target pairs – miR156a:SPL1 and miR168a:Ago1a reported in rice system were used.Citation13
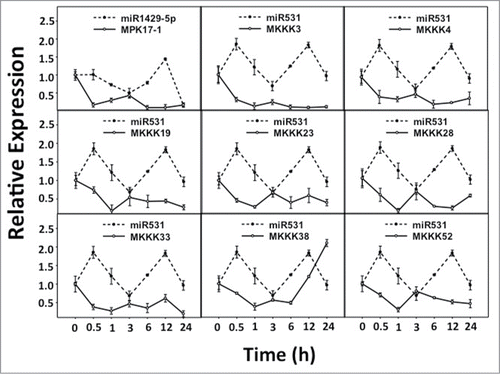
The expression analysis of all the selected miRNAs and the target genes along with control pairs (miR156:SPL1 and miR168:Ago1a) were performed. The RQ values were determined by normalizing against ubiquitin expression. The graphs were plotted by selecting the miRNA with its corresponding target. Clearly, in the control pairs the transcript accumulation were inversely correlated between the miRNA and its targets (Supplementary Figure 1). The correlation coefficients calculated between the expression values of miRNA and its target were negative for the control pairs suggesting the inverse expression pattern. Considering the expression patterns and correlation coefficients of the control pairs in both drought and UV-B stress treatments, it was sought to identify the similar patterns of expression and correlation coefficients in predicted miRNA: target pairs. In nine out of 26 predicted miRNA: target pairs, transcript accumulation pattern between predicted miRNA: target pairs were similar to the control set suggesting their biological significance (). Eight of those pairs included miR531 – predicted by Microinspector, while one was miR1429_5p – predicted by PMRD ( and ). The validated targets of miR531 included only the members of MAPKKK namely, MKKK3, MKKK4, MKKK19, MKKK23, MKKK28, MKKK33, MKKK38 and MKKK52.
Table 2. Correlation coefficients of miRNA: target pairs
Figure 1. Expression patterns of predicted miRNA: target pairs under Drought stress. Relative expression levels of predicted miRNA: target pairs were analyzed under Drought stress for similar correlation patterns as the control set.
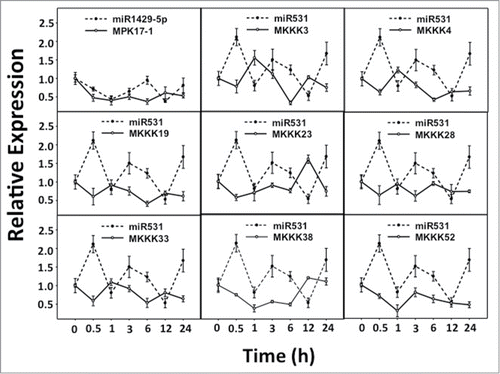
Figure 2. Expression patterns of predicted miRNA: target pairs under UV-B stress. Relative expression levels of predicted miRNA: target pairs were analyzed under UV-B stress for similar correlation patterns as the control set.
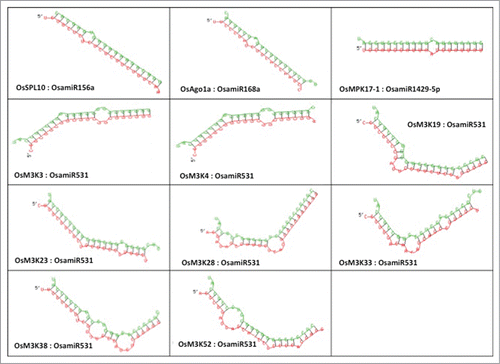
The minimum free energy for hybridization for each of the miRNA-target pair was calculated by submitting microRNA sequence and the RNA sequence of the predicted target to the RNAhybrid server.Citation17 The image generated, depicts the hybridization between miRNA (green) and its corresponding target sequence (red) ().
Discussion
The present work focuses on the identification of miRNAs that target the transcripts of MAP kinase cascade components in Oryza sativa. Two in-silico tools – Microinspector and psRNAtarget (PMRD) were used to predict miRNAs for 99 members of MAPK cascade component transcripts. The miRNAs predicted by both the online tools did not overlap and were different. Nine out of 26 (∼35%) miRNA:target pairs were validated as functional miRNA:target pairs and among them eight targets were MAPKKKs. This fact clearly emphasizes the importance and requirement of miRNA mediated post-transcriptional regulation in MAPK signaling cascade, especially of MAPKKKs. This is in complete agreement with the regulation of RAS (a MAPKKK) by a miRNA let-7 in C. elegans.Citation10 These independent observations from animal and plant systems indicate the functional conservation of the signaling molecules and silencing pathways as well as their intricate networks of regulation in the two major kingdoms of life.
The post-transcriptional regulation of MAPKKK transcripts by miRNAs would only result in the regulation of the complete MAPK cascade downstream of that particular MAPKKK. For instance, OsMKKK3 is the ortholog of CTR1 in Arabidopsis, which negatively regulates MKK9–MPK3/6 cascade.Citation14 Transcriptional inhibition or silencing of OsMKKK3 by miR531 can activate the downstream MKK9–MPK3/6 pathway leading to the stability of EIN3 transcription factor. Similarly, MKKK4 is the ortholog of EDR1 which physically interacts with MKK4/MKK5 and negatively regulates the plant innate immunity.Citation15 Further work is necessary to determine the regulation of the MAPKKK transcripts is due to transcript cleavage or translational inhibition.
Materials and Methods
Sequence retrieval and identification of miRNAs targeting MAP kinase transcripts in rice
All the mature miRNA sequences of rice were downloaded from miRBase Release 17 (http://www.mirbase.org/)Citation16 and all the rice MAP kinase (16 MAPKs, 8 MAPKKs and 75 MAPKKKs) coding sequences were downloaded from Rice genome annotation project (http://rice.plantbiology.msu.edu/)Citation17 database using gene IDs.Citation2-4 Each of the MAP kinase coding sequence along with all the rice miRNA sequences were submitted to the Microinspector online tool (http://bioinfo1.uni-plovdiv.bg/cgi-bin/microinspector/).Citation11 The potential miRNAs targeting the given coding sequence, the position and the resultant target sequence of the CDS, and the free binding energy between the miRNA and the transcript were saved for each of the MAP kinases. Also, the search function of the PMRD (plant microRNA database) (http://bioinformatics.cau.edu.cn/PMRD/) was used to search the targeting miRNAs by querying all the MAP kinase gene IDs.Citation18 This search function of PMRD relies on psRNATarget server.Citation19
Plant growth conditions, maintenance and stress treatments
Rice (Oryza sativa L indica cultivar group var Pusa Basmati 1) was grown in growth chamber (SCILAB instrument, Taiwan) at 28 °C with 16 h light and 8 h dark period or grown in green house at NIPGR. Drought stress was induced by 15% polyethylene glycol-6000 to three week old rice wild type seedlings growing hydroponically with half strength Hoagland solution. UV-B treatment was given by exposing three week old rice wild type seedlings to UV-B tubes (Phillips, Netherland) for ten minutes under normal light conditions. Distance between UV tubes and plants were kept at ∼20 cm. The plant samples were harvested at different time points mentioned in the legend of respective figures by snap freezing in liquid N2 and stored in -80°C for further analysis.
RNA isolation, cDNA synthesis and qPCR analysis
RNA isolation was performed using TRI reagent® (Sigma) according to the manufacturer's instructions and cDNA was synthesized using RevertAidTM H Minus First strand cDNA synthesis kit (Fermentas). For miRNAs, stem-loop primers were designed as described by Varkonyi-Gasic et al.Citation20 and used in 50 nM final for each of the miRNAs reverse transcribed. Equal concentrations (100ng) of the total RNA was used for the reverse transcription along with oligo(dT)18 and stem-loop primers. qPCR primers for all the MAP kinases were designed from the 3′-UTR regions using Primer Express (Applied Biosystems) software so as to ensure the uniqueness of the primers for each of the studied gene. All the qPCR study was performed on ViiATM 7 (Applied Biosystems) platform using Power SYBR Green PCR master mix (Applied Biosystems). The concentrations of the primers used were first standardized by running the standard curve protocol using successive five dilutions of a cDNA (1:1 till 1:625). Relative expression level of each gene was calculated using ΔΔCT method and by normalizing against Ubiquitin as reference genes.Citation21 The fold change represents the average value of two biological and independent replicates with three technical replicates each. All the primer sequences used are mentioned in the Supplementary Table 1.
RNA hybrid analysis
The minimum free energy for hybridization for each of the miRNA-target pair was calculated by submitting microRNA sequence and the RNA sequence of the predicted target to the RNAhybrid server.Citation22 The image depicting the hybridization between miRNA and its target was also used for analysis.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
972130_Supplemental_Files.zip
Download Zip (105.6 KB)Funding
BR kindly acknowledges Department of Biotechnology, Govt. of India while AS thanks Council of Scientific and Industrial Research, India, for financial support. The work is supported by the core grant of National Institute of Plant Genome Research from Department of Biotechnology, Govt. of India
References
- Stone JM, Walker JC. Plant protein kinase families and signal transduction. Plant Physiol 1995; 108:451-7; PMID:7610156; http://dx.doi.org/10.1104/pp.108.2.451
- Kumar K, Rao KP, Sharma P, Sinha AK. Differential regulation of rice mitogen activated protein kinase kinase (MKK) by abiotic stress. Plant physiology and biochemistry: PPB / Societe francaise de physiologie vegetale 2008; 46:891-7.
- Hamel LP, Nicole MC, Sritubtim S, Morency MJ, Ellis M, Ehlting J, Beaudoin N, Barbazuk B, Klessig D, Lee J, et al. Ancient signals: comparative genomics of plant MAPK and MAPKK gene families. Trends Plant Sci 2006; 11:192-8; PMID:16537113; http://dx.doi.org/10.1016/j.tplants.2006.02.007
- Rao KP, Richa T, Kumar K, Raghuram B, Sinha AK. In silico analysis reveals 75 members of mitogen-activated protein kinase kinase kinase gene family in rice. DNA research: an international journal for rapid publication of reports on genes and genomes 2010; 17:139-53.
- Wankhede DP, Misra M, Singh P, Sinha AK. Rice mitogen activated protein kinase kinase and mitogen activated protein kinase interaction network revealed by in-silico docking and yeast two-hybrid approaches. PLoS One 2013; 8:e65011; PMID:23738013; http://dx.doi.org/10.1371/journal.pone.0065011
- Paroo Z, Ye X, Chen S, Liu Q. Phosphorylation of the human microRNA-generating complex mediates MAPK/Erk signaling. Cell 2009; 139:112-22; PMID: 19804757; http://dx.doi.org/10.1016/j.cell.2009.06.044
- Voinnet O. Origin, biogenesis, and activity of plant microRNAs. Cell 2009; 136:669-87; PMID:19239888; http://dx.doi.org/10.1016/j.cell.2009.01.046
- Wrzaczek M, Hirt H. Plant MAP kinase pathways: how many and what for? Biology of the cell / under the auspices of the European Cell Biology Organization 2001; 93:81-7.
- Hauser F, Chen W, Deinlein U, Chang K, Ossowski S, Fitz J, Hannon GJ, Schroeder JI. A genomic-scale artificial microRNA library as a tool to investigate the functionally redundant gene space in Arabidopsis. Plant Cell 2013; 25:2848-63; PMID:23956262; http://dx.doi.org/10.1105/tpc.113.112805
- Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A, Labourier E, Reinert KL, Brown D, Slack FJ. RAS is regulated by the let-7 microRNA family. Cell 2005; 120:635-47; PMID:15766527; http://dx.doi.org/10.1016/j.cell.2005.01.014
- Rusinov V, Baev V, Minkov IN, Tabler M. MicroInspector: a web tool for detection of miRNA binding sites in an RNA sequence. Nucleic Acids Res 2005; 33:W696-700; PMID:15980566; http://dx.doi.org/10.1093/nar/gki364
- Siré C, Moreno AB, Garcia-Chapa M, López-Moya JJ, San Segundo B. Diurnal oscillation in the accumulation of Arabidopsis microRNAs, miR167, miR168, miR171 and miR398. FEBS Lett 2009; 583:1039-44; PMID:19236868; http://dx.doi.org/10.1016/j.febslet.2009.02.024
- Zhou L, Liu Y, Liu Z, Kong D, Duan M, Luo L. Genome-wide identification and analysis of drought-responsive microRNAs in Oryza sativa. J Exp Bot 2010; 61:4157-68; PMID:20729483; http://dx.doi.org/10.1093/jxb/erq237
- Yoo SD, Cho YH, Tena G, Xiong Y, Sheen J. Dual control of nuclear EIN3 by bifurcate MAPK cascades in C2H4 signalling. Nature 2008; 451:789-95; PMID: 18273012; http://dx.doi.org/10.1038/nature06543
- Zhao C, Nie H, Shen Q, Zhang S, Lukowitz W, Tang D. EDR1 physically interacts with MKK4/MKK5 and negatively regulates a MAP kinase cascade to modulate plant innate immunity. PLoS Genet 2014; 10:e1004389; PMID: 24830651; http://dx.doi.org/10.1371/journal.pgen.1004389
- Kozomara A, Griffiths-Jones S. miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res 2011; 39:D152-7; PMID: 21037258; http://dx.doi.org/10.1093/nar/gkq1027
- Ouyang S, Zhu W, Hamilton J, Lin H, Campbell M, Childs K, Thibaud-Nissen F, Malek RL, Lee Y, Zheng L, et al. The TIGR Rice Genome Annotation Resource: improvements and new features. Nucleic Acids Res 2007; 35:D883-7; PMID:17145706; http://dx.doi.org/10.1093/nar/gkl976
- Zhang Z, Yu J, Li D, Zhang Z, Liu F, Zhou X, Wang T, Ling Y, Su Z. PMRD: plant microRNA database. Nucleic Acids Res 2010; 38:D806-13; PMID: 19808935; http://dx.doi.org/10.1093/nar/gkp818
- Dai X, Zhao PX. psRNATarget: a plant small RNA target analysis server. Nucleic Acids Res 2011; 39:W155-9; PMID:21622958; http://dx.doi.org/10.1093/nar/gkr319
- Varkonyi-Gasic E, Wu R, Wood M, Walton EF, Hellens RP. Protocol: a highly sensitive RT-PCR method for detection and quantification of microRNAs. Plant Methods 2007; 3:12; PMID:17931426; http://dx.doi.org/10.1186/1746-4811-3-12
- Patel RK, Jain M. PlantRGS: a web server for the identification of most suitable candidate reference genes for quantitative gene expression studies in plants. DNA research: an international journal for rapid publication of reports on genes and genomes 2011; 18:463-70.
- Krüger J, Rehmsmeier M. RNAhybrid: microRNA target prediction easy, fast and flexible. Nucleic Acids Res 2006; 34:W451-4; PMID:16845047; http://dx.doi.org/10.1093/nar/gkl243