Abstract
Eukaryotic RNA polymerase II (RNAP II) has evolved an array of heptad repeats with the consensus sequence Tyr1-Ser2-Pro3-Thr4-Ser5-Pro6-Ser7 at the carboxy-terminal domain (CTD) of its largest subunit (Rpb1). Dynamic phosphorylation of Ser2, Ser5 and Ser7 residues orchestrates the binding of transcription and RNA processing factors to the transcription machinery. Recent studies show that the two remaining potential phosphorylation sites, tyrosine-1 and threonine-4, are phosphorylated as well and contribute to the previously proposed “CTD code“. With the impairment of binding of CTD interacting factors, these novel phosphorylation marks add an accessory layer of regulation to the RNAP II transcription cycle.
Keywords: :
In eukaryotes three different DNA dependent RNA polymerases convert the information encoded in the chromatin template into RNA. RNAP I transcribes rDNA in rRNA, RNAP II transcribes all protein encoding genes in mRNA as well as small, non-coding RNAs like snRNAs, snoRNAs and miRNAs. RNAP III transcribes various short genes, which are coding for structural or catalytic RNAs like tRNAs, 5S rRNA or the U6 snRNA.
All three enzymes are composed of several subunits and have large similarities but show also clear structural differences in their periphery.Citation1 The most eye-catching difference, which separates RNAP II from its relatives, is the carboxy-terminal domain (CTD) of its largest subunit Rpb1.
The CTD consists of multiple heptapeptide repeats with the consensus sequence Tyr1-Ser2-Pro3-Thr4-Ser5-Pro6-Ser7 and is well conserved through evolution, with roughly 10 tandemly repeated heptads in some Protozoa up to 52 heptads in Homo sapiens.Citation2 With rising number of heptads, aberrations from the consensus appear predominantly in the C-terminal part of CTD. Non-consensus repeats deviate mainly at position 7 by replacement of serine to lysine or other amino acids and might fulfill specific tasks. During the transcription cycle, the structural plasticity and dynamic pattern of posttranslational modifications enables CTD to serve as a binding platform for a variety of regulatory factors involved in RNA biogenesis. With Tyr1, Thr4 and Ser2, -5 and -7, five out of seven amino acid residues of the consensus repeat can be target of phosphorylation. Additionally, serine and threonine residues in CTD can be modified by O-linked glycosylationCitation3 and lysine and arginine residues of non-consensus repeats can be target of ubiquitinationCitation4 and methylation,Citation5 respectively. Cis/trans-isomerisation of Pro3 and Pro6 also contributes to changes in CTD structure. The connection of certain phosphorylation patterns to specific stages of the RNAP II transcription cycle support the notion of a CTD code,Citation6 where combinations of modifications are written and read as signatures to regulate the interaction of transcription and RNA processing factors with the transcription machinery.Citation7
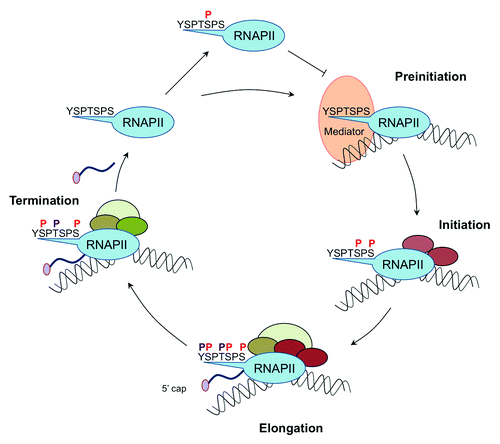
The transcription cycle () starts, when RNAP II with unphosphorylated CTD is recruited by general transcription factors (GTFs) and the Mediator complex to a promoter and forms, together with these factors, a preinitiation complex (PIC). The PIC formation could be inhibited, if the cyclin-dependent kinase (CDK) 8 subunit of the Mediator complex phosphorylates Ser5 beforehand.Citation8 Then a CTD phosphatase has to remove this mark first, before the preinitiation complex can be formed. In the phase of initiation Ser5 and Ser7 are phosphorylated by the TFIIH associated kinase CDK7 (Kin28 in yeast) and RNAP II is released from the promoter.Citation9 Phosphorylation of Ser5 is also required for the recruitment of the capping enzyme, as well as other factors involved in mRNA processing and modification of chromatin. Although it is placed during initiation and remains stable until RNAP II reaches the polyadenylation (pA) site, the role of Ser7-P in expression of protein coding genes is still unclear. Ser7-P earned the reputation as the first gene-specific mark of the CTD code for its functional role in the recruitment of Integrator, a complex involved in the expression and processing of snRNAs.Citation10 In contrast to Ser7-P, Ser5-P marks are erased by various CTD phosphatases when RNAP II enters productive elongation and proceeds toward the 3′ end of the gene. The phosphorylation of Ser2 by CDK9 and CDK12 (Bur1 and Ctk1 in yeast) rises downstream of the transcription start site (TSS). During the transition from high levels of Ser5-P to high levels of Ser2-P, both marks are overlapping within the coding region. This double mark is important for interaction with splicing factors and chromatin modifying enzymes. After the removal of Ser5-P, Ser2-P is the prevailing phosphorylation mark during the elongation phase and plays a crucial role also in the recruitment of mRNA 3′ processing and transcription termination factors.
Figure 1. Dynamic CTD phosphorylation patterns during the RNAP II transcription cycle. RNA Polymerase II (RNAP II) with hypophosphorylated CTD forms together with general transcription factors (GTFs) and the Mediator a preinitiation complex (PIC). During initiation, elongation and termination various CTD phosphorylation marks (P) are introduced and erased, successively. These CTD signatures determine the interaction capacity of RNAP II machinery for different transcription- and RNA processing factors (schematically presented by colored ovals).
In the past, the main focus of CTD research laid on the modification of CTD serine residues. The phosphorylation of CTD residues threonine and tyrosine has been reported only very recently. Three studies showed that Thr4 is phosphorylated in vivo in yeast and higher vertebrates.Citation11-Citation13 While the presence of Thr4 is not essential for viability in Saccharomyces cerevisiaeCitation14 and Schizosaccharomyces pombe,Citation15 its replacement either to valine in chickenCitation12 or to alanine in human cellsCitation11 is lethal. Chromatin immunoprecipitation experiments in yeast revealed that Thr4-P is associated to the transcribed region of genes in yeastCitation13 but is low at the pA site, consistent with structural considerations that modification of this CTD residue may impair the binding of the CTD interacting domain (CID) of termination factors like Pcf11 to CTD.Citation16 Chicken DT40 cells expressing a Thr4Val mutant of CTD show insufficient processing of non-polyadenylated histone mRNA whereas transcription of histone genes is not affected.Citation12 In human B-cells, Thr4 phosphorylated RNAP II is enriched in the region downstream of the pA site of genes. The exchange of Thr4 by alanine leads to a global defect in RNA elongation but expression of few genes even gets stimulated, suggesting that Thr4 is also important for the initiation and/or elongation phase in mammalian cells.Citation11 Until know, no Thr4 specific kinase has been identified in yeast, but with PLK3 a kinase has been determined for Thr4 phosphorylation in human cells. Knockdown and induction experiments could confirm PLK3 as a Thr4 specific kinase.Citation11 CDK9 specific inhibitors could also diminish Thr4-P levels in chicken and human cells.Citation11,Citation12 CDK9 is a well characterized kinase for Ser2 in vivo. The inhibitory effect of CDK9 inhibitors on Thr4 phosphorylation could be indirect and caused by blocking phosphorylation of Ser2, if that modification is a requirement for priming Thr4 phosphorylation.Citation11,Citation17
While yeast tolerates the substitution of Thr4, the replacement of Tyr1 with alanine is absolutely lethalCitation15 indicating an essential function of this CTD residue. Phosphorylation of CTD-Tyr1 and the responsible kinase c-Abl were described for human RNAP II almost two decades ago,Citation18 but it was unknown whether this modification is present in other species and if it plays a functional role during the transcription cycle. A recent study shows that Tyr1 is phosphorylated in yeast and that Tyr1-P modification appears at all active genes.Citation13 The levels of Tyr1-P correlate with Ser2-P levels, are low at gene promoters, but rise downstream of the TSS. In contrast to Ser2-P, however, the Tyr1-P mark decreases before RNAP II reaches the pA site suggesting a role of this modification in the recruitment of 3′ processing and termination factors. Indeed, Tyr1-P inhibits the binding of termination factors to transcribing RNAP II in the central region of genes. The gene body is essentially void of termination factors, which peak upstream (Nrd1) or downstream (Pcf11, Rtt103) of Tyr1-P signals. In agreement with this observation, Tyr1-P impairs also CTD binding to the conserved CTD-interacting domain (CID) of termination factors in vitro. Importantly, Tyr1-P does not prevent binding to the CTD-binding domain of elongation factor Spt6, consistent with Spt6 occupancy within the Tyr1-phosphorylated region of genes in vivo. This essential function during transcription can explain why mutation of Tyr1 is lethal and how the recruitment of termination factors is regulated during the RNAP II transcription cycle ().
In summary, with the description and functional analysis Thr4-P and Tyr1-P the initially suggested CTD codeCitation19,Citation20 is now extended to a five letter code. The next urgent questions will be to uncover the language of CTD and how the five letters are used to regulate the association and dissociation of cellular factors to CTD, and whether different sections within CTD can adapt specific modification signatures, and thereby fulfil specific tasks. These questions will require an extensive mass spectroscopy analysis, and the determination of the most common modification signatures occurring in CTD in vivo. The knowledge of these signatures will allow the study of CTD function and its interaction partners in more detail in the future.
Acknowledgments
Work in our laboratory is supported by grants from the Deutsche Forschungsgemeinschaft DFG (SFB/TR5 and SFB684).
We apologize to those colleagues whose work was not directly cited.
References
- Cramer P, Armache KJ, Baumli S, Benkert S, Brueckner F, Buchen C, et al. Structure of eukaryotic RNA polymerases. Annu Rev Biophys 2008; 37:337 - 52; http://dx.doi.org/10.1146/annurev.biophys.37.032807.130008; PMID: 18573085
- Chapman RD, Heidemann M, Hintermair C, Eick D. Molecular evolution of the RNA polymerase II CTD. Trends Genet 2008; 24:289 - 96; http://dx.doi.org/10.1016/j.tig.2008.03.010; PMID: 18472177
- Ranuncolo SM, Ghosh S, Hanover JA, Hart GW, Lewis BA. Evidence of the involvement of O-GlcNAc-modified human RNA polymerase II CTD in transcription in vitro and in vivo. J Biol Chem 2012; 287:23549 - 61; http://dx.doi.org/10.1074/jbc.M111.330910; PMID: 22605332
- Li H, Zhang Z, Wang B, Zhang J, Zhao Y, Jin Y. Wwp2-mediated ubiquitination of the RNA polymerase II large subunit in mouse embryonic pluripotent stem cells. Mol Cell Biol 2007; 27:5296 - 305; http://dx.doi.org/10.1128/MCB.01667-06; PMID: 17526739
- Sims RJ 3rd, Rojas LA, Beck D, Bonasio R, Schüller R, Drury WJ 3rd, et al. The C-terminal domain of RNA polymerase II is modified by site-specific methylation. Science 2011; 332:99 - 103; http://dx.doi.org/10.1126/science.1202663; PMID: 21454787
- Buratowski S. Progression through the RNA polymerase II CTD cycle. Mol Cell 2009; 36:541 - 6; http://dx.doi.org/10.1016/j.molcel.2009.10.019; PMID: 19941815
- Egloff S, Dienstbier M, Murphy S. Updating the RNA polymerase CTD code: adding gene-specific layers. Trends Genet 2012; a 28:333 - 41; http://dx.doi.org/10.1016/j.tig.2012.03.007; PMID: 22622228
- Hengartner CJ, Myer VE, Liao SM, Wilson CJ, Koh SS, Young RA. Temporal regulation of RNA polymerase II by Srb10 and Kin28 cyclin-dependent kinases. Mol Cell 1998; 2:43 - 53; http://dx.doi.org/10.1016/S1097-2765(00)80112-4; PMID: 9702190
- Akhtar MS, Heidemann M, Tietjen JR, Zhang DW, Chapman RD, Eick D, et al. TFIIH kinase places bivalent marks on the carboxy-terminal domain of RNA polymerase II. Mol Cell 2009; 34:387 - 93; http://dx.doi.org/10.1016/j.molcel.2009.04.016; PMID: 19450536
- Egloff S, Zaborowska J, Laitem C, Kiss T, Murphy S. Ser7 phosphorylation of the CTD recruits the RPAP2 Ser5 phosphatase to snRNA genes. Mol Cell 2012; b 45:111 - 22; http://dx.doi.org/10.1016/j.molcel.2011.11.006; PMID: 22137580
- Hintermair C, Heidemann M, Koch F, Descostes N, Gut M, Gut I, et al. Threonine-4 of mammalian RNA polymerase II CTD is targeted by Polo-like kinase 3 and required for transcriptional elongation. EMBO J 2012; 31:2784 - 97; http://dx.doi.org/10.1038/emboj.2012.123; PMID: 22549466
- Hsin J-P, Sheth A, Manley JL. RNAP II CTD phosphorylated on threonine-4 is required for histone mRNA 3′ end processing. Science 2011; 334:683 - 6; http://dx.doi.org/10.1126/science.1206034; PMID: 22053051
- Mayer A, Heidemann M, Lidschreiber M, Schreieck A, Sun M, Hintermair C, et al. CTD tyrosine phosphorylation impairs termination factor recruitment to RNA polymerase II. Science 2012; 336:1723 - 5; http://dx.doi.org/10.1126/science.1219651; PMID: 22745433
- Stiller JW, McConaughy BL, Hall BD. Evolutionary complementation for polymerase II CTD function. Yeast 2000; 16:57 - 64; http://dx.doi.org/10.1002/(SICI)1097-0061(20000115)16:1<57::AID-YEA509>3.0.CO;2-E; PMID: 10620775
- Schwer B, Shuman S. Deciphering the RNA polymerase II CTD code in fission yeast. Mol Cell 2011; 43:311 - 8; http://dx.doi.org/10.1016/j.molcel.2011.05.024; PMID: 21684186
- Meinhart A, Cramer P. Recognition of RNA polymerase II carboxy-terminal domain by 3′-RNA-processing factors. Nature 2004; 430:223 - 6; http://dx.doi.org/10.1038/nature02679; PMID: 15241417
- Svejstrup JQ. Transcription: another mark in the tail. EMBO J 2012; 31:2753 - 4; http://dx.doi.org/10.1038/emboj.2012.154; PMID: 22617426
- Baskaran R, Dahmus ME, Wang JY. Tyrosine phosphorylation of mammalian RNA polymerase II carboxyl-terminal domain. Proc Natl Acad Sci U S A 1993; 90:11167 - 71; http://dx.doi.org/10.1073/pnas.90.23.11167; PMID: 7504297
- Buratowski S. The CTD code. Nat Struct Biol 2003; 10:679 - 80; http://dx.doi.org/10.1038/nsb0903-679; PMID: 12942140
- Corden JL. Transcription. Seven ups the code. Science 2007; 318:1735 - 6; http://dx.doi.org/10.1126/science.1152624; PMID: 18079391