Abstract
The genus Xanthomonas comprises a large group of plant-pathogenic bacteria. The infection and bacterial multiplication in the plant tissue depends on the type III secretion system and other virulence determinants. Recent studies revealed that bacterial virulence is also controlled at the post-transcriptional level by small non-coding RNAs (sRNAs). In this review, we highlight our current knowledge about sRNAs and RNA-binding proteins in Xanthomonas species.
Introduction
Gram-negative γ-proteobacteria of the genus Xanthomonas are economically important pathogens of many plant species, including crop plants, and are grouped into pathovars (pv.) according to their host range.Citation1 During infection, the bacteria enter the plant tissue via natural openings like stomata and hydathodes or wounds. The bacteria multiply either locally in the intercellular spaces, as in case of Xanthomonas campestris pv. vesicatoria (Xcv), X. oryzae pv. oryzicola (Xoc), and X. axonopodis pv. citri (Xac), or spread systemically via the plant vascular system, e.g., X. campestris pv. campestris (Xcc) and X. oryzae pv. oryzae (Xoo).Citation2 While Xanthomonas spp. only infect plants, another member of the Xanthomonadaceae family, Stenotrophomonas maltophilia (Smal), is an opportunistic human pathogen.Citation3
Pathogenicity of Xanthomonas spp. relies on a type III secretion system (T3SS), which is encoded by the chromosomal hrp (hypersensitive response and pathogenicity)-gene cluster.Citation4 The T3SS is induced during infection and translocates bacterial effector proteins (T3Es) directly into the plant cell.Citation2 In addition, other protein secretion systems contribute to virulence of Xanthomonas spp.Citation5
In our laboratory, we study the sequenced model pathogen Xcv and its interaction with pepper and tomato plants.Citation6 Multiplication of Xcv in susceptible host plants causes bacterial spot symptoms, that later become necrotic. By contrast, resistant host cultivars recognize a certain bacterial T3E, which often induces the hypersensitive response (HR). The HR is a rapid local plant cell death, accompanied by the arrest of bacterial multiplication.Citation7
In the last two decades, bacterial non-coding RNAs (ncRNAs) have emerged as important post-transcriptional regulators, because they affect mRNA stability and/or translation and the activity of proteins.Citation8 ncRNAs encompass trans-encoded small RNAs (sRNAs), protein-binding sRNAs, cis-encoded antisense RNAs, and cis-regulatory mRNA elements, i.e., riboswitches.Citation8 In many bacteria, sRNAs were shown to contribute to environmental adaptation, e.g., carbon utilization and the bacterial response to stress.Citation9 In addition, sRNAs in animal- and human-pathogenic bacteria are known to affect quorum sensing and the expression of virulence genes,Citation10,Citation11 e.g., RNAIII in Staphylococcus aureus and sRNAs of the Qrr family in Vibrio cholerae.Citation12-Citation14 To date, the knowledge of sRNAs in plant-pathogenic bacteria is limited to Agrobacterium tumefaciens, Pseudomonas syringae pv. tomato, and members of the genera Erwinia and Xanthomonas.Citation15-Citation24
This review focuses on sRNAs and the RNA-binding proteins (RBPs) Hfq and CsrA/RsmA in Xanthomonas spp. We will only discuss sRNAs, which have been validated by northern blot analyses and apologize to all colleagues whose data were not discussed in detail for space reasons.
Identification of sRNAs in Xanthomonas spp.
In members of the genus Xanthomonas, sRNAs were so far identified in Xcv, Xcc, and Xoo.Citation19-Citation23 The next paragraphs are subdivided accordingly.
Xcv—Differential RNA-Seq Analysis Revealed 24 sRNAs
In 2012 we provided a first insight into the transcriptome of Xcv strain 85-10 using a differential RNA-seq (dRNA-seq) approach, which permits the discrimination of primary and processed transcripts, and thus, enables the identification of transcription start sites (TSSs).Citation22 In contrast to previous studies of other bacterial species, which were based on the tedious manual inspection of dRNA–seq data, we applied an automated statistical approach and mapped 1421 TSSs in Xcv.Citation22 Eight hundred and thirty-one of these presumably represent the TSSs of 17.35% of all annotated coding sequences (CDSs). Interestingly, 14% of the identified mRNAs lack a 5′-untranslated region (UTR), whereas 13% carry unusually long 5′-UTRs ranging from 150–300 bp, including six mRNAs that encode T3Es (xopAA, xopB, avrBs1, xopC, xopD, and xopN).Citation22 The long UTRs might harbor cis-regulatory elements, such as riboswitches, or serve as binding sites for post-transcriptional regulators, i.e., sRNAs or proteins. Notably, the CDSs of some effector genes might be larger than annotated in the genome, as recently demonstrated for xopD.Citation6,Citation25 The dRNA–seq analysis also revealed a plethora of antisense RNAs in Xcv, some of which are associated with virulence genes that encode components (HrcC) or substrates of the T3SS (AvrBs1, XopAA, XopB, XopD, XopE2, and XopO).Citation22 It is tempting to speculate that these antisense RNAs could prevent the intracellular accumulation of potentially cytotoxic T3Es under conditions that induce the hrp–regulon, but do not allow for T3S, as in case of Xcv grown in XVM2 medium.Citation26 However, this hypothesis remains to be tested. Overall, the dRNA–seq study revealed that 22% of all nucleotides, which belong to annotated CDSs, are covered by antisense reads.Citation22 It has been hypothesized that antisense transcription over a great portion of the genome might play a role in fine-tuning of gene expression.Citation27
The major aim of the dRNA–seq analysis of Xcv was the identification of sRNAs. In total, 23 potential regulatory RNAs and 6S RNA were confirmed by northern blot analyses, 15 of which represent sRNAs (sX1 to 15) and eight represent cis-encoded antisense RNAs (asX1 to 7 and PtaRNA1; ).Citation19,Citation22 Except for sX6, which was shown to encode a small protein, the identified RNAs are non-coding. Bioinformatic analyses suggest that one of the abundant antisense RNAs, PtaRNA1 (plasmid-transferred antisense RNA1), represents a chromosomally encoded RNA-regulator of a novel toxin–antitoxin locus.Citation19 Such type I toxin–antitoxin loci employ an antisense RNA to post-transcriptionally repress the synthesis of toxic proteins.Citation28 However, the regulatory function of PtaRNA1 and the presumed toxic character of the potential target mRNA (XCV2162) awaits further analyses. The Xcv transcriptome analysis also provided evidence for a role of sRNAs in virulence, because the abundance of five sRNAs, including sX12, and three antisense RNAs was affected by the key regulators of the T3SS, HrpG, and HrpX.Citation22 Further analysis revealed that sX12 promotes Xcv virulence, but does not affect in planta growth and T3S in vitro.Citation22 So far, potential targets of sX12 are unknown and the functional mechanism needs to be explored. To date, the Xcv sRNA sX13 is the most extensively studied sRNA in Xanthomonas spp. with respect to potential targets and physiological functions and is discussed below in detail. Altogether, the dRNA–seq approach in Xcv provided a first comprehensive insight into the transcriptome of a Xanthomonas strain and significantly contributed to the improvement of the genome annotation.Citation6,Citation22
Table 1. Rfam classification of verified sRNAs from Xanthomonadaceae.
Xcc—Shotgun Cloning and RNA–Seq Revealed 12 sRNAs
In 2010 Jiang et al. reported the identification of four sRNAs in Xcc strain 8004 using shotgun cloning and sequencing of cDNA derived from size-selected RNA (50–500 nt).Citation20 The respective sRNA candidates were validated by northern blot and termed sRNA-Xcc1 () to 4. Although potential mRNA-targets of the Xcc sRNAs were predicted,Citation20 the presumed base-pairing function of these transcripts is questionable, since Rfam searches (Rfam 11.0; Aug. 2012)Citation29 suggest that sRNA-Xcc2, -Xcc3, and -Xcc4 represent orthologs of the conserved 6S RNA, SRP RNA, and 5S rRNA.
In another study, an RNA–seq approach of Xcc strain 8004 was used to identify genes, whose expression is affected by mutation of rpf (regulation of pathogenicity factors)-genes.Citation23 The Rpf-regulatory system in Xcc controls the production and perception of the quorum-sensing autoinducer DSF (diffusible signal factor) and contributes to biofilm formation and the production of secreted virulence factors, i.e., extracellular enzymes and the extracellular polysaccharide xanthan.Citation30,Citation31 The RNA–seq study suggests the existence of previously not-annotated genes, including about 150 potential virulence factors and 24 sRNA candidates.Citation23 Considering that the approach did not allow strand-specific assignment of sequencing reads, it is possible that some of the sRNA candidates are derived from processed mRNAs or represent cis-encoded antisense RNAs. Eight sRNAs were validated by northern blot, which showed that the expression of three sRNAs (sRNA-Xcc15, -Xcc16, and -Xcc28), was Rpf-dependent. Intriguingly, the virulence of an Xcc triple mutant deleted in the three sRNA genes was reduced compared with the wild-type, whereas virulence of single mutants was not altered.Citation23
Xoo—Shotgun Cloning Identified Eight sRNAs
In 2011 Liang et al. described the identification of eight sRNAs in Xoo strain PXO99 based on computational prediction, shotgun cloning, and sequencing of cDNA derived from size-selected RNA (50–500 nt).Citation21 The expression of these sRNAs, termed sRNA-Xoo1 to 8, was verified by northern blot. Xoo single mutants deleted in each of the seven sRNA genes (sRNA-Xoo1 to 7) did not exhibit phenotypic changes compared with the wild-type with respect to in vitro growth, secretion of extracellular enzymes, and virulence.Citation21 Proteome analyses suggest that sRNA–Xoo1 might be involved in the regulation of amino acid metabolism and transport.Citation21 Such a role was also demonstrated for the Salmonella sRNA GcvB.Citation32,Citation33 In addition, the proteome data imply a role of sRNA–Xoo3 in the regulation of oxidation-reduction processes, stress response, and metabolic pathways.Citation21 A role in housekeeping processes was suggested for sRNA–Xoo4, which affects the abundance of proteins involved in DNA replication, rRNA processing, and metabolism.Citation21 It will be interesting to determine whether the impact of Xoo sRNAs on protein abundance is due to base-pairing interactions with corresponding mRNAs.
Phylogenetic Distribution of sRNAs in Xanthomonas spp.
At present, the Rfam database (Rfam 11.0; updated Aug. 2012)Citation29 comprises 23 sRNA families, whose founding member is a Xanthomonas sRNA (). Many of the corresponding sRNAs were first described in Xcv.Citation19,Citation22 Phylogenetic analyses suggest that the majority of Xanthomonas sRNAs is exclusively conserved in members of the Xanthomonadaceae family. Exceptions include PtaRNA1 from Xcv, which exhibits an erratic phylogenetic distribution, and widely conserved sRNAs with housekeeping functions, e. g. 6S RNA, SRP RNA, tmRNA, and RNase P.Citation19,Citation22,Citation34 Homologs of some of the clade-specific sRNAs were identified by independent studies in different Xanthomonas spp. and in Smal (). While similar lengths were reported for homologs like sX13 (115 nt) and SmsR39 (113 nt), the reported lengths of asX4 (309 nt) and sRNA-Xoo4 (145 nt) differ significantly. The Xcv sRNA sX13 is highly conserved in members of the Xanthomonadaceae () and originates from a locus downstream of the DNA–polymerase I-encoding polA gene.Citation24 Most interestingly, this locus encodes the sX13-unrelated Spot42 sRNA in Escherichia coli and members of the αr7 sRNA family in α-proteobacteria.Citation35,Citation36 Thus, the polA locus appears to be a hotspot for sRNA genes in distantly related bacteria.
Figure 1. (A) Conservation of sX13. LocARNA prediction of the consensus secondary structure of all sX13 family members in xanthomonads listed in the Rfam database (Rfam 11.0, Aug. 2012).Citation29,Citation55 Compensatory mutations that retain the sX13 structure are indicated by circles. (B) Model of potential physiological functions of sX13 in Xcv according to Schmidtke et al.Citation24 sX13 accumulates in response to extracellular stimuli and indirectly promotes HrpG activation, which is induced by unknown signaling pathways.Citation56,Citation57 sX13 supposedly impacts on motility, quorum sensing (QS), and the activity of other sRNAs by repressing Hfq synthesis. Boxed arrows, wavy lines, and circles indicate genes, mRNAs, and proteins. Dashed circles mark unknown proteins or signaling pathways. Green, red, and dashed arrows denote positive, negative, and putative effects.
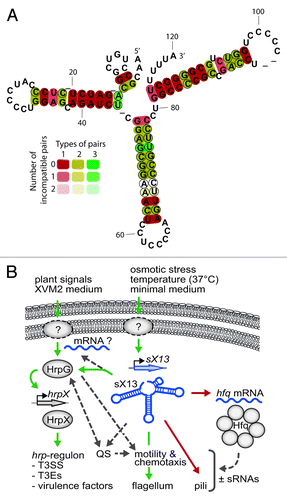
sX13 from Xcv Regulates Virulence
While the biological functions of most sRNAs identified in Xanthomonas spp. are unknown, a role in the pathogen–plant interaction was reported for sRNAs from Xcc and Xcv ().Citation22-Citation24 However, insights into underlying regulatory mechanisms are limited to sX13, which we recently discovered as novel regulator of virulence gene expression in Xcv strain 85–10.Citation24 Deletion of sX13 affected bacterial growth in complex and minimal medium and severely reduced Xcv virulence. This was unexpected, because sX13 is constitutively expressed, whereas most known Xcv virulence factors are co-expressed with the T3SS, e. g., T3Es and sRNA sX12.Citation2,Citation22,Citation24 While sX13 is not part of the hrp-regulon, it promotes mRNA accumulation of hrpX and HrpX-induced genes, which encode components and substrates of the T3SS. By contrast, the mRNA level of hrpG, the master regulator of the T3SS, was not affected. Complementation studies suggest that sX13 acts upstream of HrpG and indirectly affects HrpG activity, presumably by targeting an upstream regulator.Citation24 Although post-translational control of HrpG activity was recently reported for Xcc,Citation37 signals or proteins that activate HrpG in Xcv are unknown.
Potential functions of sX13 extend beyond the regulation of the T3SS, as implied by microarray analyses.Citation24 The sX13 regulon comprises more than 60 genes involved in virulence, quorum sensing, and motility. This suggests that sX13 coordinates the activity of different regulatory networks in response to environmental changes (). Such a role is highlighted further by the increased accumulation of sX13 under certain stress conditions.Citation24 Because the microarray analyses did not allow for the identification of mRNAs that are directly targeted by sX13, some genes are probably indirectly affected on the transcriptional level. Intriguingly, sX13 might also impact on post-transcriptional processes, because it represses the accumulation of the hfq mRNA and synthesis of Hfq (),Citation24 which encodes a conserved RBP required for sRNA activity in most bacteria (see below).
Overall, there is striking evidence that sX13 acts on certain mRNAs via antisense base-pairing: the sX13 structure consists of three stems and apical C-rich loops (), whereas 70% of the negatively regulated mRNAs carry complementary G-rich motifs in proximity of the translation start codon.Citation24 Complementation studies demonstrated that both C-rich sX13 loops and G-rich mRNA motifs are required for sX13-mediated repression of target–mRNA expression, suggesting base-pairing interactions. However, complementary mutations in sX13 loops and the G-rich mRNA motif did not restore sX13-dependency, which might be due to the fact that mutation of G-rich motifs decreased or abolished mRNA translation. Thus, potential sX13-targeting sites in mRNAs appear to serve as translational enhancers, implying that sX13 interferes with enhancer activity.Citation24 A similar case was reported for the Salmonella sRNA GcvB, which targets “CA”-rich enhancer elements.Citation32,Citation33
Despite the obvious redundancy, the C-rich sX13 loops differentially contribute to sX13 activity: while loops 2 and 3 promote Xcv virulence, only loop 2 appears to have a major role in target–mRNA repression.Citation24 So far, potential targets of loop 1 and loop 3 are elusive. It will be interesting to see whether multiple sX13 loops contribute to the regulation of some of the presumed target–mRNAs, which contain multiple G-rich motifs. Such a mechanism is employed by RNAIII of the Gram-positive pathogen S. aureus,Citation14 which resembles sX13 with respect to C-rich loops. Besides structural similarity of sX13 and RNAIII, the latter also regulates the expression of virulence genes.Citation14,Citation38
The RNA Chaperone Hfq
Hfq is a widely conserved bacterial RBP, which has been extensively studied in enterobacteria and in most cases facilitates base-pairing interactions of sRNAs and their target-mRNAs.Citation39,Citation40 Furthermore, Hfq is assumed to protect sRNAs from degradation, as the stability of Hfq-dependent sRNAs decreases in absence of Hfq. Inactivation of Hfq often results in pleiotropic phenotypes, e. g., reduced bacterial growth, altered motility, and stress responses, as well as decreased virulence of pathogenic bacteria.Citation11,Citation41
In Xanthomonas spp., the role of Hfq was investigated in Xoo and Xcv using in-frame deletion mutagenesis and a frame-shift mutation, respectively. Hfq contributes to growth of Xoo in complex medium, but does not appear to play a role in virulence.Citation21 Similarly, inactivation of hfq in Xcv did not affect virulence, which was surprising, because the sRNA sX13 contributes to plant infection and represses Hfq synthesis.Citation24 Thus, our findings demonstrate that sX13 acts Hfq-independently, which is supported by the fact that sX13-dependent expression of putative target mRNAs is not affected in the hfq mutant.Citation24 It remains to be seen whether yet-unidentified RBPs in Xanthomonas are required for the activity of sX13 and other sRNAs. Similar to sX13 in Xcv, the S. aureus RNAIII controls virulence gene expression in an Hfq-independent manner.Citation42 In other plant–pathogenic bacteria, i.e., A. tumefaciens and Erwinia spp., Hfq was shown to contribute to virulence.Citation43,Citation44 In addition, Hfq is involved in symbiotic interactions of S. meliloti with the plant.Citation45
Although the function of Hfq is obscure in Xanthomonas spp., the presumed role as RBP is supported by the Hfq-dependent accumulation of the Xcv sRNA sX14 and three Xoo sRNAs, including the sX14 homolog sRNA–Xoo3 ().Citation21,Citation24 In Smal, two Hfq-dependent sRNAs were identified, one of which, SmsR36,Citation34 is homologous to the Hfq-dependent sRNA–Xoo1 (). Hfq-independent Smal sRNAs include SmsR39, which is homologous to sX13 from Xcv. Overall, Hfq appears to have different roles in Xanthomonas spp. and the related Smal, as an hfq mutation in Smal impacts on growth, motility, biofilm formation, tolerance against antimicrobial agents, and the bacterial interaction with eukaryotic cells.Citation34 Further research is needed to address physiological functions and the RNA-binding capability of Hfq in Xanthomonas spp.
The CsrA-/RsmA-Regulatory System
Besides Hfq, proteins of the CsrA/RsmA family are well-studied bacterial RBPs, which contribute to a variety of cellular processes e.g., glycogen biosynthesis, flagellum-mediated motility, biofilm formation, and virulence.Citation46,Citation47 Members of this RBP family bind to the 5′-UTRs of target mRNAs, and thus, repress translation and stimulate mRNA decay or, in a few cases, act positively on target mRNAs.Citation48 In E. coli, the activity of CsrA is modulated by binding of the sRNAs CsrB and CsrC, which mimic target mRNAs.Citation46,Citation49 Similarly, in P. fluorescens, the RsmA activity is controlled by corresponding sRNAs, i. e., RsmX, RsmY, and RsmZ.Citation50
In Xanthomonas spp., the role of RsmA has been studied in Xcc and Xoo. In both strains, deletion of rsmA led to a loss of pathogenicity, which was associated with an altered expression of hrp- and rpf-genes.Citation51-Citation53 These mutants also showed pleiotropic phenotypes, e.g., higher levels of intracellular glycogen, increased bacterial adhesion and aggregation, and reduced production of extracellular enzymes.Citation51-Citation53 RsmA in Xanthomonas spp. appears to control quorum sensing and pathogenicity. The underlying regulatory mechanism is still elusive, because Xanthomonas spp. lack homologs of the known CsrA/RsmA-binding sRNAs of other bacteria.
Conclusions
Since the discovery of the first sRNAs in Xanthomonas spp. four years ago, 44 sRNAs were experimentally validated in members of the genus, which was substantially fostered by high-throughput transcriptome-sequencing approaches. Since these methods generate huge data sets and reveal many sRNA candidates, careful evaluation by independent methods is needed to confirm sRNA expression, i.e., northern blots. Xcv transcriptomics demonstrated the benefits of dRNA–seq for the improvement of genome annotations and the identification of TSSs and laid an excellent foundation for more in-depth studies. Besides sX13 in Xcv, a role in virulence was reported for other Xanthomonas sRNAs, although their precise function and targets remain to be identified. Future studies will lead to exciting new insights into post-transcriptional regulation of gene expression and virulence of bacterial plant pathogens.
| Abbreviations: | ||
| bp | = | base pair |
| CDS | = | coding sequence |
| dRNA-seq | = | differential RNA-seq |
| HR | = | hypersensitive response |
| hrp | = | hypersensitive response and pathogenicity |
| nt | = | nucleotide |
| ncRNA | = | non-coding RNA |
| pv. | = | pathovar |
| RBP | = | RNA-binding protein |
| Smal | = | Stenotrophomonas maltophilia |
| sRNA | = | small RNA |
| T3E | = | type III effector protein |
| T3SS | = | type III secretion system |
| TSS | = | transcription start site |
| UTR | = | untranslated region |
| Xac | = | Xanthomonas axonopodis pv. citri |
| Xcc | = | Xanthomonas campestris pv. campestris |
| Xcv | = | Xanthomonas campestris pv. vesicatoria |
| Xoc | = | Xanthomonas oryzae pv. oryzicola |
| Xoo | = | Xanthomonas oryzae pv. oryzae |
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Funding
This work was supported by grants of the Deutsche Forschungsgemeinschaft (DFG, Priority program “Sensory and Regulatory RNAs in Prokaryotes,” SPP 1258) to U. Bonas.
References
- Leyns F, De Cleene M, Swings J-G, De Ley J. The host range of the genus Xanthomonas.. Bot Rev 1984; 50:308 - 56; http://dx.doi.org/10.1007/BF02862635
- Büttner D, Bonas U. Regulation and secretion of Xanthomonas virulence factors. FEMS Microbiol Rev 2010; 34:107 - 33; http://dx.doi.org/10.1111/j.1574-6976.2009.00192.x; PMID: 19925633
- Ryan RP, Monchy S, Cardinale M, Taghavi S, Crossman L, Avison MB, Berg G, van der Lelie D, Dow JM. The versatility and adaptation of bacteria from the genus Stenotrophomonas.. Nat Rev Microbiol 2009; 7:514 - 25; http://dx.doi.org/10.1038/nrmicro2163; PMID: 19528958
- Bonas U, Schulte R, Fenselau S, Minsavage GV, Staskawicz BJ, Stall RE. Isolation of a gene cluster from Xanthomonas campestris pv. vesicatoria that determines pathogenicity and the hypersensitive response on pepper and tomato. Mol Plant Microbe Interact 1991; 4:81 - 8; http://dx.doi.org/10.1094/MPMI-4-081
- Ryan RP, Vorhölter FJ, Potnis N, Jones JB, Van Sluys MA, Bogdanove AJ, Dow JM. Pathogenomics of Xanthomonas: understanding bacterium-plant interactions. Nat Rev Microbiol 2011; 9:344 - 55; http://dx.doi.org/10.1038/nrmicro2558; PMID: 21478901
- Thieme F, Koebnik R, Bekel T, Berger C, Boch J, Büttner D, Caldana C, Gaigalat L, Goesmann A, Kay S, et al. Insights into genome plasticity and pathogenicity of the plant pathogenic bacterium Xanthomonas campestris pv. vesicatoria revealed by the complete genome sequence. J Bacteriol 2005; 187:7254 - 66; http://dx.doi.org/10.1128/JB.187.21.7254-7266.2005; PMID: 16237009
- Klement Z, Goodman R. The hypersensitive reaction to infection by bacterial plant pathogens. Annu Rev Phytopathol 1967; 5:17 - 44; http://dx.doi.org/10.1146/annurev.py.05.090167.000313
- Waters LS, Storz G. Regulatory RNAs in bacteria. Cell 2009; 136:615 - 28; http://dx.doi.org/10.1016/j.cell.2009.01.043; PMID: 19239884
- Gottesman S, Storz G. Bacterial small RNA regulators: versatile roles and rapidly evolving variations. Cold Spring Harb Perspect Biol 2011; 3:a003798; http://dx.doi.org/10.1101/cshperspect.a003798; PMID: 20980440
- Bejerano-Sagie M, Xavier KB. The role of small RNAs in quorum sensing. Curr Opin Microbiol 2007; 10:189 - 98; http://dx.doi.org/10.1016/j.mib.2007.03.009; PMID: 17387037
- Papenfort K, Vogel J. Regulatory RNA in bacterial pathogens. Cell Host Microbe 2010; 8:116 - 27; http://dx.doi.org/10.1016/j.chom.2010.06.008; PMID: 20638647
- Novick RP, Ross HF, Projan SJ, Kornblum J, Kreiswirth B, Moghazeh S. Synthesis of staphylococcal virulence factors is controlled by a regulatory RNA molecule. EMBO J 1993; 12:3967 - 75; PMID: 7691599
- Bardill JP, Hammer BK. Non-coding sRNAs regulate virulence in the bacterial pathogen Vibrio cholerae.. RNA Biol 2012; 9:392 - 401; http://dx.doi.org/10.4161/rna.19975; PMID: 22546941
- Boisset S, Geissmann T, Huntzinger E, Fechter P, Bendridi N, Possedko M, Chevalier C, Helfer AC, Benito Y, Jacquier A, et al. Staphylococcus aureus RNAIII coordinately represses the synthesis of virulence factors and the transcription regulator Rot by an antisense mechanism. Genes Dev 2007; 21:1353 - 66; http://dx.doi.org/10.1101/gad.423507; PMID: 17545468
- Wilms I, Overlöper A, Nowrousian M, Sharma CM, Narberhaus F. Deep sequencing uncovers numerous small RNAs on all four replicons of the plant pathogen Agrobacterium tumefaciens.. RNA Biol 2012; 9:446 - 57; http://dx.doi.org/10.4161/rna.17212; PMID: 22336765
- Filiatrault MJ, Stodghill PV, Bronstein PA, Moll S, Lindeberg M, Grills G, Schweitzer P, Wang W, Schroth GP, Luo S, et al. Transcriptome analysis of Pseudomonas syringae identifies new genes, noncoding RNAs, and antisense activity. J Bacteriol 2010; 192:2359 - 72; http://dx.doi.org/10.1128/JB.01445-09; PMID: 20190049
- Liu Y, Cui Y, Mukherjee A, Chatterjee AK. Characterization of a novel RNA regulator of Erwinia carotovora ssp. carotovora that controls production of extracellular enzymes and secondary metabolites. Mol Microbiol 1998; 29:219 - 34; http://dx.doi.org/10.1046/j.1365-2958.1998.00924.x; PMID: 9701816
- Zeng Q, McNally RR, Sundin GW. Global small RNA chaperone Hfq and regulatory small RNAs are important virulence regulators in Erwinia amylovora.. J Bacteriol 2013; 195:1706 - 17; http://dx.doi.org/10.1128/JB.02056-12; PMID: 23378513
- Findeiss S, Schmidtke C, Stadler PF, Bonas U. A novel family of plasmid-transferred anti-sense ncRNAs. RNA Biol 2010; 7:120 - 4; http://dx.doi.org/10.4161/rna.7.2.11184; PMID: 20220307
- Jiang RP, Tang DJ, Chen XL, He YQ, Feng JX, Jiang BL, Lu GT, Lin M, Tang JL. Identification of four novel small non-coding RNAs from Xanthomonas campestris pathovar campestris.. BMC Genomics 2010; 11:316; http://dx.doi.org/10.1186/1471-2164-11-316; PMID: 20482898
- Liang H, Zhao Y-T, Zhang J-Q, Wang X-J, Fang R-X, Jia Y-T. Identification and functional characterization of small non-coding RNAs in Xanthomonas oryzae pathovar oryzae.. BMC Genomics 2011; 12:87; http://dx.doi.org/10.1186/1471-2164-12-87; PMID: 21276262
- Schmidtke C, Findeiss S, Sharma CM, Kuhfuss J, Hoffmann S, Vogel J, Stadler PF, Bonas U. Genome-wide transcriptome analysis of the plant pathogen Xanthomonas identifies sRNAs with putative virulence functions. Nucleic Acids Res 2012; 40:2020 - 31; http://dx.doi.org/10.1093/nar/gkr904; PMID: 22080557
- An SQ, Febrer M, McCarthy Y, Tang DJ, Clissold L, Kaithakottil G, Swarbreck D, Tang JL, Rogers J, Dow JM, et al. High-resolution transcriptional analysis of the regulatory influence of cell-to-cell signalling reveals novel genes that contribute to Xanthomonas phytopathogenesis. Mol Microbiol 2013; 88:1058 - 69; http://dx.doi.org/10.1111/mmi.12229; PMID: 23617851
- Schmidtke C, Abendroth U, Brock J, Serrania J, Becker A, Bonas U. Small RNA sX13: a multifaceted regulator of virulence in the plant pathogen Xanthomonas.. PLoS Pathog 2013; 9:e1003626; http://dx.doi.org/10.1371/journal.ppat.1003626; PMID: 24068933
- Canonne J, Marino D, Noël LD, Arechaga I, Pichereaux C, Rossignol M, Roby D, Rivas S. Detection and functional characterization of a 215 amino acid N-terminal extension in the Xanthomonas type III effector XopD. PLoS One 2010; 5:e15773; http://dx.doi.org/10.1371/journal.pone.0015773; PMID: 21203472
- Rossier O, Wengelnik K, Hahn K, Bonas U. The Xanthomonas Hrp type III system secretes proteins from plant and mammalian bacterial pathogens. Proc Natl Acad Sci U S A 1999; 96:9368 - 73; http://dx.doi.org/10.1073/pnas.96.16.9368; PMID: 10430949
- Georg J, Hess WR. cis-antisense RNA, another level of gene regulation in bacteria. Microbiol Mol Biol Rev 2011; 75:286 - 300; http://dx.doi.org/10.1128/MMBR.00032-10; PMID: 21646430
- Schuster CF, Bertram R. Toxin-antitoxin systems are ubiquitous and versatile modulators of prokaryotic cell fate. FEMS Microbiol Lett 2013; 340:73 - 85; http://dx.doi.org/10.1111/1574-6968.12074; PMID: 23289536
- Burge SW, Daub J, Eberhardt R, Tate J, Barquist L, Nawrocki EP, Eddy SR, Gardner PP, Bateman A. Rfam 11.0: 10 years of RNA families. Nucleic Acids Res 2013; 41:D226 - 32; http://dx.doi.org/10.1093/nar/gks1005; PMID: 23125362
- Barber CE, Tang JL, Feng JX, Pan MQ, Wilson TJ, Slater H, Dow JM, Williams P, Daniels MJ. A novel regulatory system required for pathogenicity of Xanthomonas campestris is mediated by a small diffusible signal molecule. Mol Microbiol 1997; 24:555 - 66; http://dx.doi.org/10.1046/j.1365-2958.1997.3721736.x; PMID: 9179849
- Tang JL, Liu YN, Barber CE, Dow JM, Wootton JC, Daniels MJ. Genetic and molecular analysis of a cluster of rpf genes involved in positive regulation of synthesis of extracellular enzymes and polysaccharide in Xanthomonas campestris pathovar campestris.. Mol Gen Genet 1991; 226:409 - 17; http://dx.doi.org/10.1007/BF00260653; PMID: 1645442
- Sharma CM, Darfeuille F, Plantinga TH, Vogel J. A small RNA regulates multiple ABC transporter mRNAs by targeting C/A-rich elements inside and upstream of ribosome-binding sites. Genes Dev 2007; 21:2804 - 17; http://dx.doi.org/10.1101/gad.447207; PMID: 17974919
- Sharma CM, Papenfort K, Pernitzsch SR, Mollenkopf HJ, Hinton JC, Vogel J. Pervasive post-transcriptional control of genes involved in amino acid metabolism by the Hfq-dependent GcvB small RNA. Mol Microbiol 2011; 81:1144 - 65; http://dx.doi.org/10.1111/j.1365-2958.2011.07751.x; PMID: 21696468
- Roscetto E, Angrisano T, Costa V, Casalino M, Förstner KU, Sharma CM, Di Nocera PP, De Gregorio E. Functional characterization of the RNA chaperone Hfq in the opportunistic human pathogen Stenotrophomonas maltophilia.. J Bacteriol 2012; 194:5864 - 74; http://dx.doi.org/10.1128/JB.00746-12; PMID: 22923593
- Rice PW, Dahlberg JE. A gene between polA and glnA retards growth of Escherichia coli when present in multiple copies: physiological effects of the gene for spot 42 RNA. J Bacteriol 1982; 152:1196 - 210; PMID: 6183252
- del Val C, Romero-Zaliz R, Torres-Quesada O, Peregrina A, Toro N, Jiménez-Zurdo JI. A survey of sRNA families in α-proteobacteria. RNA Biol 2012; 9:119 - 29; http://dx.doi.org/10.4161/rna.18643; PMID: 22418845
- Li RF, Lu GT, Li L, Su HZ, Feng GF, Chen Y, He YQ, Jiang BL, Tang DJ, Tang JL. Identification of a putative cognate sensor kinase for the two-component response regulator HrpG, a key regulator controlling the expression of the hrp genes in Xanthomonas campestris pv. campestris.. Environ Microbiol 2013; Forthcoming http://dx.doi.org/10.1111/1462-2920.12207; PMID: 23906314
- Morfeldt E, Taylor D, von Gabain A, Arvidson S. Activation of alpha-toxin translation in Staphylococcus aureus by the trans-encoded antisense RNA, RNAIII. EMBO J 1995; 14:4569 - 77; PMID: 7556100
- Vogel J, Luisi BF. Hfq and its constellation of RNA. Nat Rev Microbiol 2011; 9:578 - 89; http://dx.doi.org/10.1038/nrmicro2615; PMID: 21760622
- De Lay N, Schu DJ, Gottesman S. Bacterial small RNA-based negative regulation: Hfq and its accomplices. J Biol Chem 2013; 288:7996 - 8003; http://dx.doi.org/10.1074/jbc.R112.441386; PMID: 23362267
- Chao Y, Vogel J. The role of Hfq in bacterial pathogens. Curr Opin Microbiol 2010; 13:24 - 33; http://dx.doi.org/10.1016/j.mib.2010.01.001; PMID: 20080057
- Bohn C, Rigoulay C, Bouloc P. No detectable effect of RNA-binding protein Hfq absence in Staphylococcus aureus.. BMC Microbiol 2007; 7:10; http://dx.doi.org/10.1186/1471-2180-7-10; PMID: 17291347
- Zeng Q, McNally RR, Sundin GW. Global small RNA chaperone Hfq and regulatory small RNAs are important virulence regulators in Erwinia amylovora.. J Bacteriol 2013; 195:1706 - 17; http://dx.doi.org/10.1128/JB.02056-12; PMID: 23378513
- Wilms I, Möller P, Stock A-M, Gurski R, Lai E-M, Narberhaus F. Hfq influences multiple transport systems and virulence in the plant pathogen Agrobacterium tumefaciens.. J Bacteriol 2012; 194:5209 - 17; http://dx.doi.org/10.1128/JB.00510-12; PMID: 22821981
- Barra-Bily L, Pandey SP, Trautwetter A, Blanco C, Walker GC. The Sinorhizobium meliloti RNA chaperone Hfq mediates symbiosis of S. meliloti and alfalfa. J Bacteriol 2010; 192:1710 - 8; http://dx.doi.org/10.1128/JB.01427-09; PMID: 20081033
- Timmermans J, Van Melderen L. Post-transcriptional global regulation by CsrA in bacteria. Cell Mol Life Sci 2010; 67:2897 - 908; http://dx.doi.org/10.1007/s00018-010-0381-z; PMID: 20446015
- Lucchetti-Miganeh C, Burrowes E, Baysse C, Ermel G. The post-transcriptional regulator CsrA plays a central role in the adaptation of bacterial pathogens to different stages of infection in animal hosts. Microbiology 2008; 154:16 - 29; http://dx.doi.org/10.1099/mic.0.2007/012286-0; PMID: 18174122
- Yakhnin AV, Baker CS, Vakulskas CA, Yakhnin H, Berezin I, Romeo T, Babitzke P. CsrA activates flhDC expression by protecting flhDC mRNA from RNase E-mediated cleavage. Mol Microbiol 2013; 87:851 - 66; http://dx.doi.org/10.1111/mmi.12136; PMID: 23305111
- Babitzke P, Romeo T. CsrB sRNA family: sequestration of RNA-binding regulatory proteins. Curr Opin Microbiol 2007; 10:156 - 63; http://dx.doi.org/10.1016/j.mib.2007.03.007; PMID: 17383221
- Kay E, Dubuis C, Haas D. Three small RNAs jointly ensure secondary metabolism and biocontrol in Pseudomonas fluorescens CHA0. Proc Natl Acad Sci U S A 2005; 102:17136 - 41; http://dx.doi.org/10.1073/pnas.0505673102; PMID: 16286659
- Lu X-H, An S-Q, Tang D-J, McCarthy Y, Tang J-L, Dow JM, Ryan RP. RsmA regulates biofilm formation in Xanthomonas campestris through a regulatory network involving cyclic di-GMP and the Clp transcription factor. PLoS One 2012; 7:e52646; http://dx.doi.org/10.1371/journal.pone.0052646; PMID: 23285129
- Zhu PL, Zhao S, Tang JL, Feng JX. The rsmA-like gene rsmA(Xoo) of Xanthomonas oryzae pv. oryzae regulates bacterial virulence and production of diffusible signal factor. Mol Plant Pathol 2011; 12:227 - 37; http://dx.doi.org/10.1111/j.1364-3703.2010.00661.x; PMID: 21355995
- Chao N-X, Wei K, Chen Q, Meng Q-L, Tang D-J, He Y-Q, Lu G-T, Jiang B-L, Liang X-X, Feng J-X, et al. The rsmA-like gene rsmA(Xcc) of Xanthomonas campestris pv. campestris is involved in the control of various cellular processes, including pathogenesis. Mol Plant Microbe Interact 2008; 21:411 - 23; http://dx.doi.org/10.1094/MPMI-21-4-0411; PMID: 18321187
- Chen X-L, Tang D-J, Jiang R-P, He Y-Q, Jiang B-L, Lu G-T, Tang J-L. sRNA-Xcc1, an integron-encoded transposon- and plasmid-transferred trans-acting sRNA, is under the positive control of the key virulence regulators HrpG and HrpX of Xanthomonas campestris pathovar campestris.. RNA Biol 2011; 8:947 - 53; http://dx.doi.org/10.4161/rna.8.6.16690; PMID: 21941121
- Will S, Joshi T, Hofacker IL, Stadler PF, Backofen R. LocARNA-P: accurate boundary prediction and improved detection of structural RNAs. RNA 2012; 18:900 - 14; http://dx.doi.org/10.1261/rna.029041.111; PMID: 22450757
- Wengelnik K, Van den Ackerveken G, Bonas U. HrpG, a key hrp regulatory protein of Xanthomonas campestris pv. vesicatoria is homologous to two-component response regulators. Mol Plant Microbe Interact 1996; 9:704 - 12; http://dx.doi.org/10.1094/MPMI-9-0704; PMID: 8870269
- Wengelnik K, Rossier O, Bonas U. Mutations in the regulatory gene hrpG of Xanthomonas campestris pv. vesicatoria result in constitutive expression of all hrp genes. J Bacteriol 1999; 181:6828 - 31; PMID: 10542187
