Abstract
In recent years, RNA has been shown to fulfil a number of cellular functions. This has led to much interest in elucidation of the structure of functional RNA molecules, and thus, in the preparation of suitably functionalized RNAs. The chemical synthesis of RNAs allows for the site-specific modification; however, is limited to sequences of about 60–70 nucleotides in length. At the example of the flavine mononucleotide (FMN) responsive aptamer of the ypaA riboswitch from B. subtilis, we demonstrate the highly efficient preparation of site-specifically modified long-mer RNAs. Our strategy consists of the chemical synthesis of fragments followed by enzymatic or chemical ligation. Splint ligation with T4 RNA ligase turned out to be most successful among the enyzymatic protocols. Highly efficient chemical ligation was performed by azide–alkyne cycloaddition of suitably modified RNA fragments. Wild-type and 2-aminopurine (2-AP)-modified variants of the ypaA aptamer were prepared. FMN binding to all synthesized ypaA aptamer variants is demonstrated. However, dissociation of FMN from its binding site by reduction of the isoalloxazin unit as demonstrated before for a small-hairpin-derived aptazyme could not be shown. This implies that either FMN is less accessible to reduction when it is bound to its natural aptamer; that reduced FMN remains bound to the aptamer; or that FMN upon reduction indeed is released from its binding site, without the aptamer folding back in the natural ligand-free state. The results of this study are of general interest to the preparation of site-specifically modified RNAs for investigation into structure and function.
Introduction
Riboswitches are metabolite-, temperature-, or pH-sensitive RNA structures that occur in non-coding regions of mRNAs, where they control gene expression.Citation1 Metabolite responsive riboswitches are composed in a modular way consisting of an aptamer domain and an expression platform. The binding of ligands to the aptamer domain induces conformational changes in the aptamer and expression platform, resulting in modulation of transcription, translation, or splicing. Over the past 10 y, extensive studies into the structure and mechanism of riboswitches have shed light on their mode of action and biological function. More than 20 distinct classes of riboswitches have been identified, and a number of atomic-resolution models were generated for natural aptamers in complexes with their ligands.Citation2,Citation3 It has been shown that the extent of structural changes accompanied with ligand binding varies among the different aptamer classes. Some aptamers are largely pre-organized, while others undergo extensive structural rearrangements upon ligand binding, referred to as type I and type II aptamers.Citation2 Riboswitches responding to flavine mononucleotide (FMN) have been identified in bacterial genes encoding proteins that are involved in biosynthesis and transport of riboflavin and related compounds.Citation4,Citation5 The aptamer domain of various FMN responsive riboswitches is also known as RFN (Riboflavin) element.Citation4 It belongs to the type II aptamers, showing less extensive FMN-induced structural changes, consistent with a largely pre-folded tertiary RNA architecture.Citation6 Aptamer domains of riboswitches are highly conserved even among various organisms, whereas the expression platform varies in sequence, structure, and in the mechanism of genetic control. Thus, the RFN element is found in FMN responsive riboswitches being involved in genetic control by either premature transcription termination or modulation of translation initiation. As learned from the crystal structure of the RFN element from Fusobacterium nucleatum, the aptamer is centered on a six-stem junction, to which FMN binds. The overall shape is butterfly-like with opposingly directed but nearly identical peripheral domains stapled on each other. FMN binding to the junction is asymmetric and relies on interaction of the junction nucleotides with the isoalloxazin ring as well as on direct and Mg2+-mediated contacts with the phosphate. Recognition of FMN is associated with conformational transitions within the junctional binding pocket, leaving the six stems surrounding the junction virtually unaffected.Citation6
In Bacillus subtilis, FMN responsive riboswitches are found, regulating both transcription termination and translation initiation. For example, the B. subtilis ypaA riboswitch controls expression of the ypaA gene, which encodes a riboflavin transport protein. It functions as regulator of translation initiation. Based on in-line probing results, Breaker and colleagues have suggested a conformational rearrangement taking place upon FMN binding to the aptamer domain, sequestering the ribosome binding site and inhibiting translation initiation.Citation7,Citation8
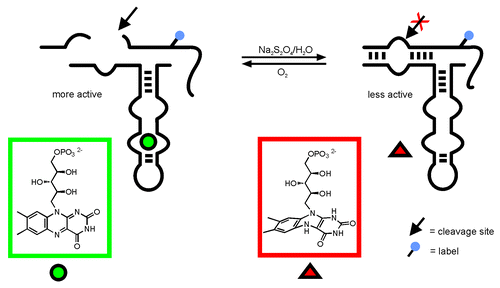
Previously, we have developed a hairpin ribozyme-derived aptazyme responding to FMN, and we have demonstrated that ribozyme activity is controlled by the FMN oxidation state ().Citation9,Citation10 FMN, in its oxidized state, is a planar compound (owing to its conjugated aromatic-ring system) and as such binds to the aptamer region of the hairpin aptazyme. Hydrogen transfer causes reduction of FMN, destroys the aromatic character, and induces a change in the molecular geometry: FMN is transformed from a planar structure to a roof-like bent shape ().Citation11,Citation12 As a result, stacking interactions of the isoalloxazin ring with nucleotides of the small FMN binding aptamerCitation13 used in that study, are interrupted, and FMN is released from its binding site. As mentioned above, also in the natural occurring FMN aptamer, FMN binding partially relies on interaction of the junction nucleotides with the isoalloxazin ring. The planar isoalloxazin ring system intercalates between two adenosines, allowing a continuous stacking alignment.Citation6 Therefore, one might assume that reduction of the isoalloxazin ring leads to disruption of stacking and in consequence to release of FMN from the aptamer. Based on our previous results on allosteric regulation of ribozyme activity by control of the FMN shape,Citation9 we sought to elucidate the possibility of redox regulation of natural FMN responsive riboswitches at the example of the ypaA aptamer domain from B. subtilis.
Figure 1. Allosteric regulation of a hairpin ribozyme derived aptazyme by FMN:Citation9 FMN binding to the aptamer leads to increased activity. Reduction changes the molecular shape of FMN such that binding is inhibited. As a result, ribozyme activity is clearly decreased.
Some of the nowadays most routinely used approaches for analysis of biomolecular conformational changes are gel shift assays or, when going into more detail, in-line probing, chemical, and enzymatic structure mapping, or SHAPE.Citation14 More sophisticated assays use spectroscopic set-ups to detect fluorescence quenching upon conformational rearrangementsCitation15 or changes of nuclearCitation16 or electron spin properties.Citation17 Binding of FMN to its natural aptamer has been shown before by using two of the above mentioned methods: in-line probing and fluorescence quenching,Citation7 as well as, in the most resolved way, by crystallization and X-ray diffraction of the aptamer–FMN complex.Citation6 In addition to choosing the appropriate method, structural analysis of RNA folding requires efficient preparation of the RNA under investigation. Moreover, in particular, the spectroscopic methods depend on the site-specific introduction of particular functionalities, as for example fluorescence dyes, isotope, or spin labels. While natural RNA can be prepared by in vitro transcription with T7 RNA polymerase, the purpose of site-specific functionalization can be fulfilled only by chemical synthesis. Either the desired functionality is available as a phosphoramidite, and thus, is directly incorporated during RNA synthesis, or alternatively, functional entities such as for example aliphatic amines, thiols, alkynes, or azides are introduced via suitable phosphoramidite building blocks and used afterwards for post-synthetic conjugation of a desired modification.Citation18 However, efficient chemical synthesis is limited to RNA fragments of about 60–70 nucleotides length. Accordingly, a number of protocols using chemical and enzymatic ligation of chemically synthesized oligonucleotidesCitation19 as well as using DNAzymes for preparation of large RNAs with site-specific labelsCitation20,Citation21 have been developed. Previously, we have synthesized a variety of amino-functionalized RNA building blocks for post-synthetic conjugation,Citation18 and applied those in RNA-structure elucidation by single-molecule FRET analysis.Citation22 This has been combined with the development of efficient protocols for joining RNA fragments by enzymatic ligation or Click chemistry to make also rather long modified RNAs amenable to analysis. Furthermore, we have developed ribozymes that support an RNA fragment exchange reaction, and thus, can be used for introduction of specific modifications into long natural RNAs.Citation23,Citation24 In the aggregate, we have the methods available to prepare site-specifically functionalized long-mer RNAs at preparative scale. Several projects addressing the question of RNA synthesis for structural investigation, i.e., structural rearrangements of RNA thermometers ROSE and fourUCitation25 as well as an extensive FRET analysis of the hairpin ribozyme four-way junction (unpublished data) are in progress. Herein, we present our strategy for efficient preparation of natural and modified versions of the 129 nt ypaA aptamer from B. subtilis and the analysis of FMN binding to these aptamer variants in response to the FMN oxidation state.
Results
Experimental design
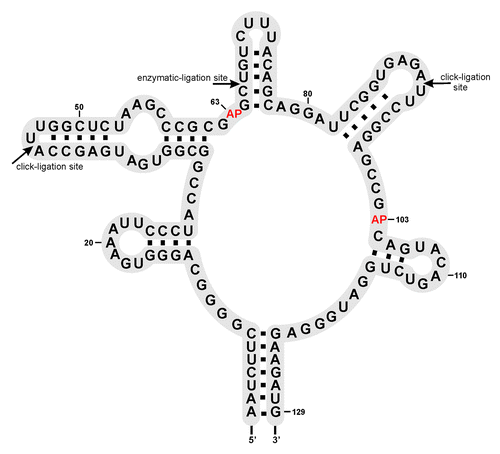
A convenient way to detect the formation of the FMN aptamer complex is following the fluorescence of the FMN chromophore that becomes quenched upon FMN binding to the RNA. This assay was used before by Winkler et al.Citation7 and being “label-free” regarding the RNA under investigation, it allows for preparation of the ypaA aptamer by enzymatic transcription in vitro. However, the fluorescence assay does not allow observing release of FMN from its binding site triggered by reduction of the isoalloxazin unit as shown for the small in vitro selected aptamer.Citation9 Reduction of FMN destroys the conjugated π-system of the chromophore rendering it non-fluorescent, and thus, invisible irrespective of its binding capability to the aptamer. Therefore, we decided to combine the FMN quenching assay with gelelectrophoretic mobility experiments, and with an alternative fluorescent assay, now using the intrinsic fluorescence of the nucleobase analog 2-aminopurine (2-AP) incorporated at suitable positions of the ypaA aptamer. The latter requires 2-AP to be placed in a part of the aptamer that undergoes structural changes upon FMN-induced folding/refolding. As mentioned in the introduction, the RFN element belonging to the type II aptamers is largely pre-organized, such that FMN binding does not induce significant conformational rearrangements. Nevertheless, potentially suitable positions for 2-AP incorporation were chosen on the basis of the crystal structure of the FMN aptamer complex from Fusobacterium nucleatum.Citation6 A suitable set of nucleobases to be replaced by 2-AP is A63 and A103 (corresponding to A48 and A85 in the F. nucleatum structure),Citation6 because both adenine residues are involved in stacking interactions with the bound FMN. The intercalation of FMN and formation of stacking interactions with the two nucleobases might be visible by a change in the fluorescence intensity when either of the adenine residues is replaced with 2-AP. Thus, aptamer RNAs carrying 2-AP at either of the two sites were required to be prepared. As mentioned above, a suitable strategy is chemical synthesis of fragments for site-specific incorporation of 2-AP followed by ligation to the full-lengths aptamer (). Accordingly, we decided dividing the aptamer into two, three, or even four parts (Fig. S1) that following their chemical preparation can be joined enzymatically, or as a promising alternative, by chemical Click ligation.Citation26
Preparation of the ypaA aptamer by enzymatic ligation of chemically synthesized fragments
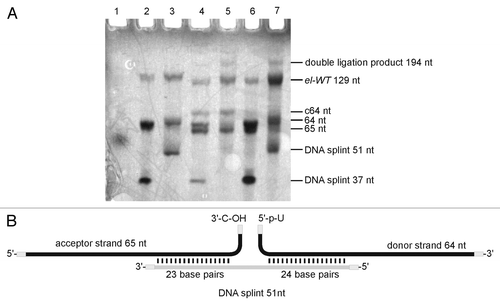
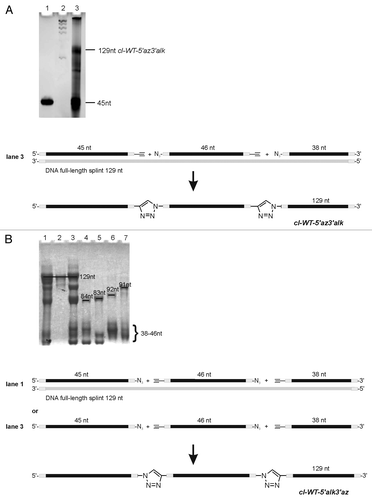
The aptamer domain was divided into fragments with lengths of 22–64 nt (Fig. S1). In order to establish a suitable protocol for enzymatic ligation, we initially used non-modified RNA fragments. After successful ligation of the chemically synthesized oligonucleotides to form the non-modified full-length aptamer, the procedure was planned to be repeated with fragments containing 2-AP as fluorescent probe. While the chemical synthesis of natural and modified fragments proceeded without problems, ligation turned out being troublesome. We screened a large number of conditions in ligation assays using T4 DNA and T4 RNA ligase from different suppliers (Fig. S1). Finally, we succeeded in ligation of two chemically prepared fragments, obtaining the full-length aptamer (, lane 7; see Supplemental Material for experimental details). The ligation site was placed between C65 and U66 of the ypaA aptamer sequence (). The 64mer donor strand was ligated to the 65mer acceptor strand by T4 RNA ligase 1 in the presence of a 51 nt DNA splint (). The splint was designed to interact with the sequence around the ligation site, but leaving the ligation site itself single-stranded. Hence, the 51 nt DNA splint formed 23 base pairs with the acceptor oligonucleotide and 24 base pairs with the donor oligonucleotide, such that three nucleotides of both donor and acceptor strand remained single stranded at the ligation junction. Encouraged by the successful preparation of the wild-type aptamer el-WT, two modified versions el-A63AP and el-A103AP, carrying 2-AP at the indicated positions were prepared following this strategy.
Figure 3. Enzymatic ligation of two fragments. (A) Preparative denaturing polyacrylamide gel of enzymatically ligated fragments. Bands were visualized by UV shadowing. Ligation reactions were conducted in a total volume of 20 µl with short (37 nt) or long (51 nt) DNA splint. The relation acceptor:splint:donor is given in brackets: Lane 1: dye-marker (~55 nt), lane 2: 5 µM acceptor, (1:1.5:2), short splint; lane 3: 5 µM acceptor, (1:1.5:2), long splint; lane 4: 10 µM acceptor, (1:0.5:1), short splint; lane 5: 10 µM acceptor, (1:0.5;1), long splint; lane 6: 10 µM acceptor, (1:1.5:2), short splint; lane 7: 10 µM acceptor, (1:1.5:2), long splint. See Supplementary Material for detail information. (B) refer to lane 7 in panel A: Ligation scheme (lane 7).
Preparation of the ypaA aptamer by Click ligation of chemically synthesized fragments
In parallel to the enzymatic ligation experiments, we have used chemical strategies for joining synthetic RNA fragments to the full-length aptamer. We decided to use a three-fragment strategy in combination with the cupper catalyzed azide-alkyne cycloaddition (in short CuAAC reaction), which originally was developed by Sharpless and coworkers as well as by Meldal and coworkers.Citation27,Citation28 More recently, the CuAAC reaction was adapted to nucleic acids, and as such termed Click ligation.Citation26 It requires the synthesis of RNA fragments carrying azide and/or alkyne functionalities at the termini and joining them by means of Click chemistry. The reaction generates a triazole as a backbone modification at the ligation site, which of course is an obvious disadvantage compared with enzymatic ligation. However, we chose ligation sites within the terminal loops of the FMN aptamer (). According to the crystal structure of the apatmer from F. nucleatum,Citation6 the terminal loops are not involved in tertiary interactions and binding of FMN, such that the triazole linkages may well be accepted at the chosen positions.
Two different Click ligation strategies were applied: ligation of a 5′-azide with a 3′-alkyne (strategy I), or vice versa of a 5′-alkyne with a 3′-azide (strategy II) ( and ). For the synthesis of 3′-alkyne modified fragments, commercially available CPG was used, to which N-phenoxyacetyl-5′-O-dimethoxytrityl-3′-O-propargyl-adenosine was attached as starting nucleoside (). The 5′-azide was generated post-synthetically with the RNA still being bound to the solid support according to a protocol provided in the literature ().Citation29 For the opposite strategy we applied a hexynyl phosphoramidite for 5′-terminal coupling () and a 3′-azido nucleoside linked to the solid support. The latter is not commercially available and therefore was synthesized starting from arabinoadenosine,Citation30 followed by immobilization as described ().Citation31 At reaction conditions reported in the literature, we observed decomposition of the azide to an amine, very likely owing to the known acid and light sensitivity of azides.Citation32 Therefore, the original protocol was slightly changed; reactions were performed in the dark with strict control of the pH during all synthesis and purification steps, thus preventing azide decomposition. After synthesis was successfully completed, the 3′-azide derivative was converted to 3′-azido-N-(di-n-butylamino)methylene-5′-O-dimethoxytrityl-2′-O-[1,6-dioxo-6-(pentafluorophenyl-oxy)hexyl]-adenosine, followed by immobilization as described in reference Citation31. It has been discussed in the literature that the azide is not stable at RNA synthesis conditions, in particular, suffering from the applied P(III) chemistry.Citation33,Citation34 Therefore, in order to evaluate the applicability of our 3′-azido functionalized support, we used it for the synthesis of a short model oligonucleotide bearing a 3′-terminal azide. The 3′-functionalized oligomer was successfully Click-ligated to a short 5′-alkyne modified oligonucleotide, demonstrating that the 3′-azido functionality was preserved under the conditions of chain assembly. This result is in agreement with a very recent report of Micura and co-workers, showing that an azide, present as 2'-azidoethyl group at the 3′-terminal nucleotide, survives RNA chain assembly by solid phase synthesis, and can be used for post-synthetic conjugation of dye labels.Citation35 Thus, we felt confident enough to apply the 3′-azide support also for the synthesis of the longer fragments, namely 45mer fragment 1 and 46mer fragment 2, such that both carry an azide at their 3′-ends (). Fragment 2 was additionally functionalized with a 5′-terminal alkyne by coupling of a hexynyl phosphoramidite. The same phosphoramidite was used for 5′-terminal alkynylation of 38mer fragment 3. Thus, the three fragments were suitably functionalized to be joined by Click ligation to the full-length aptamer ( and ). Click ligation was performed basically following the protocols of El-Sagheer and BrownCitation29 and Paredes and Das,Citation34 although we varied conditions in terms of reaction with or without DNA splint, and changed buffer composition, more appropriate for DNA splint digestion right after successful ligation (see Materials and Methods). Particular care was taken to work oxygen-free in order to allow Cu(I) as catalyst to be formed. Furthermore, the water-soluble ligand tris-hydroxypropyltriazolylamine (THPTA)Citation36 was used for chelating the reactive Cu(I) species to prevent RNA degradation. Ligation was successful with both strategies in the presence and, as tried for strategy II, also in the absence of a DNA splint (, lane 3; , lanes 1 and 3). Obviously, the three fragments are capable of assembling in solution to the required structural fold, such that ligation sites are positioned in the right orientation to form the triazole linkages. In addition to the full-lengths product, a number of side products resulting from ligation of only two fragments were detected, and their identity was confirmed by using all possible combinations of two out of the three fragments for single site ligation (, lanes 4–7). We did not observe significant differences between the two strategies. Ligation proceeded with good yields irrespective of the azide being at the 5′-terminus of the oligonucleotide and the alkyne at the 3′-terminus or vice versa.
Figure 4. Synthesis of RNA-fragments for Click ligation. Preparation of 3′-alkyne building block (A); 5′-azid building block (B); 5′-alkyne building block (C); 3′-azid building block (D) See Supplementary Material for details.
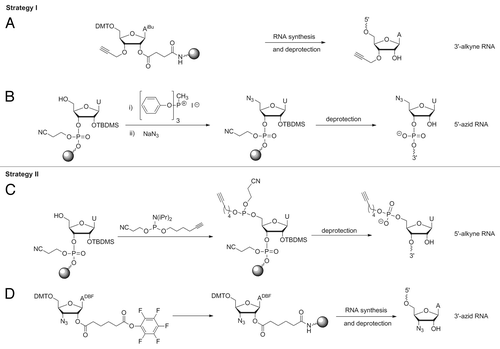
Figure 5. Strategies for Click ligation: (A) Click reaction of 3′-terminal 3′-alkyne with 5′-terminal azide resulting in 1,4-dimethylen tetrazol moiety; (B) Click reaction of 3′-terminal azide with 5′-terminal alkyne resulting in a more flexible tatrazol-linkage with n-butyl-bridge and phosphate moiety.
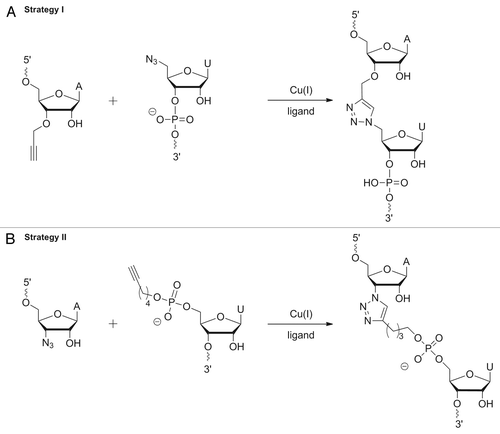
Figure 6. Preparation of the ypaA aptamer by Click ligation. Preparative denaturing PAA gels. Bands were visualized by UV shadowing: (A) Strategy I: 5′-terminal 45nt fragment; lane 2: RNA ladder, lane 3: ligation reaction using the three fragments and a full-length DNA splint as shown on the right. (B) Strategy II: ligation reaction using three fragments and a full-length DNA splint as shown on the right, lane 2: full-length aptamer wt-tr resulting from in vitro transcription, lane 3: ligation reaction using three fragments without splint as shown on the right, lane 4–7: single site ligations of two fragments out of the three (see Fig. S5 for details).
In summary, as a result of transcription, chemical synthesis and enzymatic and chemical ligation, six versions of the ypaA aptamer were produced: (1) tr-WT resulting from in vitro transcription and carrying a six nucleotide overhang (introduced to enhance transcription efficiency) at the 5′-end, (2) el-WT resulting from enzymatic ligation of chemically synthesized fragments, (3) el-A63AP, and (4) el-A103AP resulting from enzymatic ligation of chemically synthesized fragments carrying 2-AP, (5) cl-WT-5′az3′alk, and (6) cl-WT-5′alk3′az, resulting from Click ligation, and thus, having two triazole linkages in the backbone, which differ in their flexibility. The sequence of all variants is identical, corresponding to the structure shown in .
FMN fluorescence quenching assay
We made use of the characteristic feature of quenching of the FMN fluorescence upon FMN binding to first check if all synthesized versions of the ypaA aptamer are functional. As shown in , all six aptamer versions, even those having triazole linkages in the backbone, bind FMN as can be concluded from the observed quenching of the FMN fluorescence upon addition of the individual aptamers. However, quenching is not equally efficient for all apatmer variants, implying that the folding behavior is slightly influenced by the 2-AP modification as well as by the backbone triazole (; ). Looking at the 2-AP-modified aptamer variants (), it has to be noticed that quenching by el-A103AP is less efficient, indicating that the substitution of A103 by 2-AP might somewhat interfere with formation of a competent aptamer conformation. On the contrary, el-A63AP induces very strong quenching of the FMN fluorescence (). The most obvious deviation however, is observed for the chemically ligated aptamer cl-WT-5′alk3′az, which does not at all induce FMN quenching in the initial experiment (). We hypothesized that the alkyl spacers at the two triazole linkages in the aptamer backbone () induce too much flexibility, making the aptamer structure rather floppy. Therefore, we changed the experimental set-up and let the aptamer fold in the presence of FMN instead of adding the readily folded aptamer to the FMN solution. Now, quenching of the FMN fluorescence is equally efficient as observed for the other aptamer variants (). Probably, in this set-up, FMN acts as a sort of chaperone, providing a scaffold around which the aptamer can fold. It is noteworthy that with this assay the functionality for FMN binding was demonstrated for both backbone triazole-modified aptamer variants, cl-WT5′alk3′az and cl-WT5′az3′, demonstrating that Click ligation indeed is a suitable method allowing for the synthesis of long-mer RNAs with a complex functional fold.
Figure 7. FMN fluorescence quenching curves: (A) Blue curve: FMN [0.1 µM]. FMN [0.1 µM] was incubated with cl-WT-5′alk3′az [0.15 µM] (red), cl-WT-5′az3′alk [0.15 µM] (purple), el-WT [0.15 µM] (yellow), tr-WT [0.15 µM] (green). (B) Blue curve: FMN [0.1 µM], red: cl-WT-5′alk3′az [0.15 µM] upon folding in the presence of FMN as folding mediator; (C) Blue curve: FMN [0.15 µM], red: upon incubation with el-A63AP [0.3 µM]; (D) blue curve: FMN [0.15 µM], red: upon incubation with el-A103AP [0.3 µM].
![Figure 7. FMN fluorescence quenching curves: (A) Blue curve: FMN [0.1 µM]. FMN [0.1 µM] was incubated with cl-WT-5′alk3′az [0.15 µM] (red), cl-WT-5′az3′alk [0.15 µM] (purple), el-WT [0.15 µM] (yellow), tr-WT [0.15 µM] (green). (B) Blue curve: FMN [0.1 µM], red: cl-WT-5′alk3′az [0.15 µM] upon folding in the presence of FMN as folding mediator; (C) Blue curve: FMN [0.15 µM], red: upon incubation with el-A63AP [0.3 µM]; (D) blue curve: FMN [0.15 µM], red: upon incubation with el-A103AP [0.3 µM].](/cms/asset/27003067-4e28-49b4-b86f-7aa22f7634a4/krnb_a_10928526_f0007.gif)
Table 1. FMN quenching assay, normalized fluorescence
With the binding assay, we also addressed the question of the contribution of individual structural elements of FMN to binding. For this purpose we studied the binding of a set of FMN analogs containing a simple hexyl or propyl chain instead of the ribitol, as well as of riboflavine lacking the phosphate moiety (Fig. S9). Neither of the analogs showed fluorescence quenching upon addition of the aptamer, indicating loss of binding capacity. This is consistent with results obtained by others, showing that the terminal phosphate of FMN is essential for binding to the aptamer.Citation6
In summary, all synthesized ypaA aptamer variants were shown to bind FMN, although with apparently slightly differing affinities. The specificity for FMN is high; loss of the terminal FMN phosphate leads to loss of binding.
Electrophoretic mobility shift assay
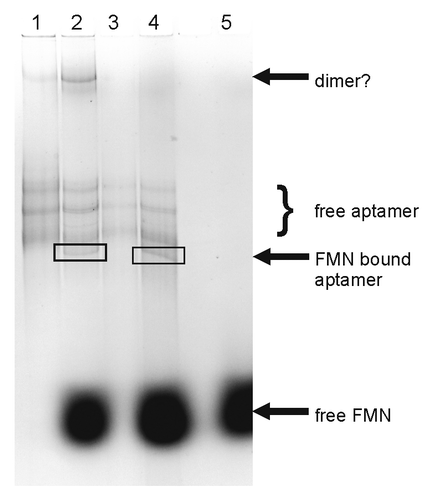
As already mentioned, the FMN quenching assay is not suitable to observing release of FMN from its binding site triggered by reduction of the isoalloxazin unit. Therefore, we decided to use a gelelectrophoretic mobility assay to follow the conformational changes of the aptamer RNA associated with FMN binding. For this study, a particular probe was not necessary, such that the in vitro transcribed aptamer tr-WT could be used. First trials with a 10 000-fold excess of FMN over RNA as used previously in in-line probing experiments,Citation7 were not successful, since the all overlaying FMN fluorescence hampered detection of RNA bands in the gel. At a 1000-fold excess of FMN over the aptamer, RNA bands were visible, and indeed we could observe a difference in the band pattern between the free and the FMN-bound aptamer (). Interestingly, the FMN-free state of the aptamer seems to be conformationally heterogeneous as indicated by several bands in the gel (lane 1). The pattern changes when the aptamer RNA is denatured before subjecting it onto the gel: the upper band with the lowest mobility disappears (lane 3), implying that it represents a less stable secondary structure, possibly even a dimer of the aptamer RNA. Since the structure of the FMN aptamer has been shown to be largely pre-organized,Citation6 it is well possible that the remaining individual bands represent members of an ensemble of possibly interconverting structures with different interhelical angles as observed for the adenine riboswitch.Citation37 A new band corresponding to a species with higher mobility appears in the presence of FMN (lane 2 and 4), possibly indicating the FMN-bound state of the aptamer. Unfortunately, it was not possible to observe the band pattern after reduction of FMN with dithionite. The high excess of dithionite that had to be used for quantitative FMN reduction hampered gelelectrophoretic mobility analysis; no bands at all were detectable.
Figure 8. Electrophoretic mobility gel-shift analysis of FMN binding to the ypaA aptamer. Bands were visualized by ethidium bromide staining: lane 1: free aptamer, lane 2: aptamer incubated with 1000-fold excess of FMN, lane 3: free aptamer, denatured prior to subjecting onto the gel, lane 4: aptamer, denatured prior to addition of a 1000-fold excess of FMN and subjecting onto the gel, lane 5: free FMN.
2AP quenching assay
Further analysis of the effect of FMN reduction on aptamer binding was performed by another fluorescence quenching assay, now following the changes of the aptamer's intrinsic 2-AP fluorescence upon FMN binding (). In a first row of experiments, the change of the intensity of the fluorescence signal of 2-AP at 370 nm upon irradiation at 305 nm was used as readout for structural changes upon ligand binding. We have recorded fluorescence spectra of both variants, el-A63AP und el-A103AP, in the absence of FMN, in the presence of FMN, and in the presence of FMN at reducing conditions. Due to the incompatibility of dithionite with the assay,Citation38 dithiothreitol (DTT) and irradiation with light was chosen as suitable alternative for reduction of the planar isoalloxazin ring.Citation39 First, we defined conditions for photochemical reduction of FMN that leave the 2-AP fluorescence intact. Reduction was successful by irradiation of the sample at 406 nm over 15 min without observable changes of the 2-AP fluorescence signal, and was completely reversible by bubbling air through the sample in the cuvette (). Thus, fluorescence spectra of the two 2-AP-containing aptamer variants with and without FMN were recorded (), followed by addition of DTT and irradiation, and recording of another fluorescence spectrum (). Finally, FMN was allowed to re-oxidize by atmospheric oxygen before taking another set of spectra (). For el-A63AP, only a very weak change of the 2-AP fluorescence upon addition of FMN is observed (, red curve), and the following reduction and re-oxidation of FMN has also no significant effect on the fluorescence spectra (, yellow and purple curves). This does not allow for a viable conclusion on the effects of FMN binding or dissociation on the aptamer conformation. The situation is clearer for el-A103AP. Here, 2-AP fluorescence is clearly quenched upon addition of FMN (, red curve). Reduction leads to further decrease of the 2-AP fluorescence signal (, yellow curve), and there is only a very weak recovery of 2-AP fluorescence upon back-oxidation of FMN (, purple curve). This result is surprising, because one would expect that 2-AP fluorescence is recovered to the level of the free aptamer when reduced FMN due to its changed geometry dissociates from the aptamer binding site. On the contrary, a further fluorescence decrease is observed (), implying that either the reduced form of FMN is not completely released from its binding site and just induces another conformational change in the aptamer. Alternatively, folding of the ypaA aptamer may be not fully reversible, such that the aptamer after initial folding and FMN binding does not refold in the conformation of the “ligand-free” state, even though FMN is reduced and released from its binding site. Furthermore, it is noticeable that reduction and re-oxidation of FMN at the conditions tested in the control experiment with free FMN () were less efficient in the presence of the aptamers, as can be seen from the very similar fluorescence intensities at 520 nm upon reduction and oxidation (, yellow and purple curves). Control experiments confirmed the intactness of DTT as reducing agent, indicating that FMN may be less accessible for reduction/re-oxidation when sitting in its binding pocket, and thus, being sequestered in the interior of the aptamer structure.
Figure 9. 2-AP fluorescence curves: (A) blue curve: FMN [20 µM], DTT [1 mM]; red: FMN [20 µM], DTT [1 mM], irridation with light (405 nm for 15 min); purple: FMN [20 µM], DTT [1 mM], irridation with light (405 nm for 15 min), oxygen. (B) blue curve: RNA [0.3 µM], DTT [1 mM]; red: RNA [0.3 µM], DTT [1 mM], irridation with light (405 nm for 15 min). (C) blue curve: el-A63AP [0.3 µM]; red: el-A63AP [0.3 µM] and FMN [0.6 µM]; (D) blue curve: el-A63AP [0.3 µM]; red: el-A63AP [0.3 µM] and FMN [0.6 µM]; yellow: el-A63AP [0.3 µM], FMN [0.6 µM] and DTT; purple: el-A63AP [0.3 µM], FMN [0.6 µM] and DTT, oxygen. (E) blue curve: el-A103AP [0.3 µM]; red: el-A103AP [0.3 µM] and FMN [0.6 µM]; (F) blue curve: el-A103AP [0.3 µM]; red: el-A103AP [0.3 µM] and FMN [0.6 µM]; yellow: el-A103AP [0.3 µM], FMN [0.6 µM], DTT and light (405 nm, 15 min); purple: el-A103AP [0.3 µM], FMN [0.6 µM] and DTT, oxygen.
![Figure 9. 2-AP fluorescence curves: (A) blue curve: FMN [20 µM], DTT [1 mM]; red: FMN [20 µM], DTT [1 mM], irridation with light (405 nm for 15 min); purple: FMN [20 µM], DTT [1 mM], irridation with light (405 nm for 15 min), oxygen. (B) blue curve: RNA [0.3 µM], DTT [1 mM]; red: RNA [0.3 µM], DTT [1 mM], irridation with light (405 nm for 15 min). (C) blue curve: el-A63AP [0.3 µM]; red: el-A63AP [0.3 µM] and FMN [0.6 µM]; (D) blue curve: el-A63AP [0.3 µM]; red: el-A63AP [0.3 µM] and FMN [0.6 µM]; yellow: el-A63AP [0.3 µM], FMN [0.6 µM] and DTT; purple: el-A63AP [0.3 µM], FMN [0.6 µM] and DTT, oxygen. (E) blue curve: el-A103AP [0.3 µM]; red: el-A103AP [0.3 µM] and FMN [0.6 µM]; (F) blue curve: el-A103AP [0.3 µM]; red: el-A103AP [0.3 µM] and FMN [0.6 µM]; yellow: el-A103AP [0.3 µM], FMN [0.6 µM], DTT and light (405 nm, 15 min); purple: el-A103AP [0.3 µM], FMN [0.6 µM] and DTT, oxygen.](/cms/asset/efacd229-e47c-4c41-bfdb-851e989cbb1e/krnb_a_10928526_f0009.gif)
Taken together, the results of the 2-AP fluorescence assay demonstrate that 2-AP at nucleotide position 103 is more sensitive than at position 63 toward structural and/or conditional changes induced by FMN binding. Quantitative reduction of free FMN is achieved by irradiation with light at 406 nm in combination with DTT. However, in the presence of the aptamer, efficient reduction seems to be hindered. Furthermore, we have no indication of reversible aptamer folding. 2-AP fluorescence was quenched upon addition of FMN, but it was not recovered to the level of the free aptamer when FMN was reduced.
Discussion
The synthesis of site-specifically modified long-mer RNAs is an ever growing task in the field of RNA research. In addition to enzymatic protocols for ligation of chemically synthesized fragments, chemical protocols were developed,Citation40-Citation43 whereby the cupper-catalyzed azide-alkyne coupling (Click ligation)Citation26,Citation29,Citation34,Citation44 stands out as most promising. A drawback of Click ligation is the need for terminal functionalization of fragments to be ligated (5′-azide and 3′-alkyne or vice versa), and the generation of a triazole linkage in the nucleic acid backbone. Nevertheless, Click ligation has been used for fragment joining in a number of applications.Citation26,Citation29,Citation34,Citation40-Citation44 Among those, also functional RNAs, such as hairpin and hammerhead ribozymes, were prepared.Citation29 However, to the best of our knowledge, we here show the first example of a Click ligated natural aptamer as long as 129 nucleotides that retains full functionality in binding of its cognate ligand, in spite of the two triazole backbone linkages. This is a promising result paving the way to using Click ligation more routinely for the preparation of site-specifically modified long-mer RNAs, such as, for example, other natural aptamers and entire riboswitchesCitation45-Citation47 or RNA thermometers.Citation25
In addition to Click ligation, we have also used enzymatic ligation to join chemically synthesized fragments to the full-length ypaA aptamer. After extensive screening and optimization of conditions, we were successful with a two-fragment strategy and RNA ligase-mediated splint-ligation. However, in our hands, the tedious way and the huge effort put into finding suitable conditions for enzymatic ligation of the ypaA aptamer was disproportionate to the rather quick set-up of the Click ligation protocol. Moreover, we succeeded in Click-ligating three fragments, whereas enzymatically only two fragments could be satisfactorily linked. The two-fragment strategy requires chemical synthesis of rather long RNA fragments (64- and 65mers), and provides little combinatorial space for the preparation of aptamer variants. On the contrary, by Click ligation of three fragments, natural and differently modified aptamer versions can be synthesized with less effort by variable fragment combination, and the individual fragments, being shorter, can be efficiently prepared by chemical synthesis. The development of efficient protocols for enzymatic and chemical ligation of the FMN responsive ypaA aptmer allowed us to synthesize a number of natural and modified aptamer variants to be used for FMN binding studies. FMN binding to all synthesized variants was confirmed by FMN fluorescence quenching, showing that functional folding of the aptamer was not significantly hampered by 2-AP or triazole backbone modifications. Yet, binding of FMN was not equally efficient with all aptamers. Interestingly, one of the 2-AP-modified versions, el-A63AP, induced stronger quenching of FMN than the wild-type aptamers, tr-WT and el-WT (; ), whereas the other, el-A103A, was clearly less efficient in FMN quenching (; ). According to the assay, stronger quenching may be interpreted as stronger binding, less efficient quenching as weaker binding. Nevertheless, it has to be considered that in both 2-AP-modified aptamers, FMN is stacked upon 2-AP. This may additionally influence the fluorescence properties of FMN in terms of fluorescence enhancement instead of quenching, or alternatively, increased quenching compared with FMN binding to the wild-type sequence aptamer versions.
The aptamer el-A63AP shows a strong effect in the FMN fluorescence quenching assay (); however, it exhibits rather low sensitivity of the intrinsic 2-AP fluorescence toward FMN addition, reduction, and re-oxidation (). This result may be interpreted in terms of weak FMN binding to the aptamer and/or no or little conformational changes induced by FMN binding. Taking into account the result of the FMN quenching assay, the latter interpretation seems more appropriate. This leads to the conclusion that nucleotide position 63 is less suitable for substitution with 2-AP as fluorescent probe, because local structural changes upon FMN binding/dissociation are too small to be mirrored in the 2-AP fluorescent properties. This trend is seen also with the second 2-AP-containing aptamer el-A103AP, although here initial FMN addition leads to considerable quenching of the 2-AP fluorescence (). Nevertheless, reduction of FMN turned out to be inefficient, although proper working of the reduction protocol was demonstrated in control experiments (). Thus, the inefficient reduction of FMN may be attributed to hampered accessibility of FMN when it is bound to the aptamer. Nevertheless, reduction of FMN is associated with additional quenching of the 2-AP fluorescence, indicative of local structural changes. Interestingly, 2-AP fluorescence is not restored to the level of the free aptamer, what one would expect, if aptamer folding upon FMN binding is fully reversible. Reversible folding of natural aptamers and riboswitches remains an open question, and it is well possible that RNA decay and de novo synthesis is easier accomplished by and more attractive to the cell as ligand-dependent reversible RNA folding. The results of our study might imply that reversible folding of the ypaA aptamer in response to its cognate ligand FMN is not the mode of choice, because 2-AP fluorescence remains quenched, instead being restored to the starting level. Strikingly, there is even additional quenching upon FMN reduction. FMN has a high affinity to the aptamer (KD ~5 nM),Citation7 and therefore, it might be more difficult to induce dissociation of FMN by simple destruction of planarity. Assuming that FMN remains bound to the aptamer, a possible scenario is that destruction of the planar isoalloxazin ring upon reduction, and thus, disruption of stacking interactions indeed is not sufficient for release of FMN from its binding site, because other interactions (i.e. the phosphate that was shown being essential for binding; see Fig. S9 and Serganov et al.Citation6) are sufficiently strong to hold the ligand in place. Nevertheless, due to at least partial reduction, the position of the flavine unit may be varied, leading to local structural rearrangements and an even stronger quenching of the 2-AP fluorescence (). Taken additionally into account the indicated insufficient reduction (), one might even speculate that the isoalloxazin unit is reduced only to the semiquinone, thus retaining fluorescence at 520 nm as seen in the spectra, and remaining bound to the aptamer, although with slightly varied structural parameters.
In conclusion, the data are indicative of FMN binding to a number of ypaA aptamer variants, but forced dissociation of FMN from its binding site by reduction could not be demonstrated. The results rather imply that (1) FMN is less accessible to reduction when it is bound to its natural aptamer, (2) FMN upon reduction possibly remains bound to the aptamer; however, reduction is associated with local structural changes mirrored in another change of the 2-AP fluorescence, and (3) alternatively, FMN upon reduction indeed may be released from its binding site, but the aptamer does not fold back in the natural ligand-free state, meaning that aptamer folding is not fully reversible. In order to draw a more precise picture of the ypaA folding dynamics in response to FMN, the fluorescent assay described herein is thought to be complemented by other methods requiring no or applying different kinds of reporter probes. Thus, we have started setting up an electrochemical assay to follow binding and re-binding of the aptamer in response to the FMN oxidation state.Citation10,Citation48 Furthermore, we intend to use Terahertz spectroscopy to observe local changes in the water network of the aptamer upon folding, as well as EPR and/or NMR spectroscopy with nitroxide spin labels or fluorine nuclei as folding reporters to follow structural rearrangements of the aptamer in response to FMN.
Materials and Methods
Materials
Chemicals for RNA synthesis were obtained from commercial suppliers (Roth, Sigma Aldrich, and Acros). NTP's, dNTP's, Klenow fragment exo-, T4 PNK, T4 PNK buffer, and DNase I were purchased from Fermentas. Core buffers were purchased from Promega. Taq DNA polymerase, ThermoPol buffer, T4 RNA ligase was purchased from Fermentas. DNA ligase, T4 RNA ligase 1, and T4 RNA ligase 2 were purchased from New England Biolabs. T7 RNA polymerase was a non-commercial product with a concentration of 76 mg/ml. DNA primer were purchased from Biomers. Standard PAC-phosphoramidites as well as CPG supports were obtained from ChemGenes or Link Technologies. Buffers used for RNA purification were prepared from autoclaved micro-pore water with a resistivity of 18.2 MΩ-cm and were filtered through a 0.20 µm membrane.
tr-WT—Generation of the aptamer domain by transcription
The transcribed aptamer domain tr-WT was prepared by in vitro transcription with T7 RNA polymerase. Double-stranded DNA template was prepared by extension reaction of two primers and amplified by PCR. The primer (2 µM) ypaA1 and ypaA2 with an overlapping region of 20 bp (forward primer 5′-TAATACGACT CACTAGGGAG AAATCTTCGG GGCAGGGTGA AATTCCCTAC CGGCGGCGGT GATGAGCCAT TGGCTCTAAG CCCGCGAG-3′, reverse primer: 5′-CATCTTCTCC CATCCAGACT GTACTGTCGG CTCCGGAATC TCACCGAATC CTGCTGTAAA GACAGCTCGC GGGCTTAGAG CCAA T-3′, bold letters represents the T7 promotor region, italic part indicates T7 Polymerase starting sequence) were solved in 1x Core buffer (50 mM Tris, 10 mM MgCl2, 50 mM NaCl, pH 7.6), denatured for 5 min at 92 °C, cooled-down to rt in 15 min, and filled up to dsDNA with Klenow exo- by adding 500 µM dNTPs and 0.04 U/µl Klenow exo- in a total reaction volume of 500 µl. Reaction was left to proceed for 30 min at 37 °C. PCR amplification was performed as follows: To 50 µl of the primer extension product, PCR primers (PCR Primer ypaA1: 5′-TCTGGATGGG AGAAGATG-3′ and PCR primer ypaA2: 5′-CTCCCTAGTG AGTCGTATTA-3′, final concentration: 1.0 µM each), 1x ThermoPol buffer (20 mM TRIS-HCl, 10 mM (NH4)2SO4, 10 mM KCl, 2 mM MgSO4, 0.1% Triton X-100, pH 8.8), 2 mM MgCl2, 250 µM dNTPs, 0.025 U/µl Taq polymerase, and 0.01 U/µl of Vent polymerase were added in a total reaction volume of 100 µl. After 2 min incubation at 95 °C, 25 cycles (94 °C 30 s, 55 °C 30 s, 72 °C 1 min) were run. Finale elongation time was 7 min at 72 °C. Purification of PCR product was conducted with QIAquick PCR Purification Kit (Qiagen). For in vitro transcription, 0.5 µM PCR product was incubated with 80 mM HEPES, pH 7.5, 12 mM MgCl2, 2 mM Spermidin, 40 mM DTT, 2 mM of each NTP and T7 RNA polymerase (35 ng/µl) for 6 h at 37 °C in a total reaction volume of 100 µl. After treatment with 2 µl DNase I for 30 min at 37 °C, the RNA was precipitated, re-solved in gel loading buffer (98% formamid, 2% 0.5 M aqueous EDTA [v/v]), and purified by denaturing PAGE. Elution from the gel was performed using 0.3 M NaOAc (pH 5.0) followed by ethanol precipitation.
Chemical synthesis of oligonucleotides
All RNA fragments for enzymatic and Click ligation were chemically synthesized by the phosphoramidite method on a Pharmacia Gene Assembler Plus at 1 µmol scale as described elsewhere.Citation18 Benzylmercaptotatrazol (BMT, emp Biotech) was used as activator.Citation49 The solutions of phosphoramidites, BMT, and acetonitrile were kept over molecular sieve 0.3 nm (Roth). The coupling time for all phosphoramidites was 5 min. All syntheses were performed “trityl-off.” The obtained RNAs were cleaved from the support and deprotected using NH3 (30%) in a 1:1 mixture with 8 M ethanolic amine, followed by treatment with TEAx3HF as described.Citation50 Deprotected crude RNAs were purified by gel electrophoresis using 10 or 15% denaturing polyacrylamide gels. Elution was performed using a 2 M solution of lithium perchlorate or 0.3 M NaOAc (pH 5.0) followed by EtOH precipitation.
Phosphorylation of donor strands for enzymatic ligation
Fragments (40 µM) were solved in 1x T4 PNK buffer A (Fermentas). ATP (5 mM), T4 PNK (0.4 U/µl, Fermentas), and water were added to give a final reaction volume of 50 µl. Phosphorylation reaction was performed at 37 °C for 2 h followed by adding again 2.5 µl ATP and 2.5 µl T4 PNK and incubation for 1 h. The phosphorylated donor strands were purified using the RNA Clean and Concentrator Kit (Zymo Research).
el-WT, el-A63AP, and el-A103AP—Enzymatic ligation with T4 RNA ligase I
The 64 nt and 65 nt fragments of the aptamer el-WT were most successfully ligated at the following conditions: hybridization of the fragments to the full-length DNA splint (acceptor:splint:donor 1:1.5:2) at a temperature of 90 °C for 5 min and cooling to 22 °C over 15 min, addition of T4 RNA Ligase I (NEB), cooling from 22 °C to 16 °C in 2 °C steps over 3h, ligation over night at 16 °C in T4 RNA ligase buffer (NEB), additionally containing 10% DMSO (Fig. S2). With this assay we obtained the full-lengths aptamer with 42% yield.Citation19 2-AP modified ypaA aptamers (el-A63AP and el-A103AP) were prepared following this protocol.
Preparation of 3′-Azido-N-(di-n-butylamino)methylene-5′-O-dimethoxytrityl-2'-O-[1,6-dioxo-6-(pentafluorophenyloxy)hexyl]-adenosine
(See Fig. S5 for details.) Starting from arabinofuranosyladenosine synthesis was conducted according to the protocol by Chen et al.,Citation51 to first deliver 3′-Azido-N-[(di-n-butylamino)methylene]-5′-O-dimethoxytrityl-adenosine. Once the 3′-azide was generated, standard protecting groups of RNA synthesis were introduced at the 5′-hydroxyl group and the exocyclic amino function. Epimerization of the arabinose to ribose was performed using the protocol described by Micura and co-workers.Citation30 The protected 3′-azido-adenosine was linked to the solid support as described.Citation30 Briefly, the 2′-hydroxyl group was converted to an adipinic-acid-ester providing a second functionalized pentafluorophenylester to be coupled onto amino functionalized polysterene (Fig. S5).
cl-WT-5′az3′alk—Preparation of the 5′ azide modification
The transformation of the 5′-hydroxyl group into an azide was conducted following the protocol by El-Sagheer and Brown.Citation29 After 1.0 µmol scale trityl-off RNA synthesis the fully protected RNA (still on the solid-support) was incubated in 1 ml of methyltriphenoxyphosphoniun iodide solution (0.5 M) in dry DMF for 15 min at room temperature. The solid was washed with dry DMF. Sodium azide (50 mg) was suspended in 1 ml dry DMF, heated to 70 °C for 10 min and cooled down to room temperature. From the resulting supernatant 1 ml was used for incubation of the RNA-loaded solid support over night at room temperature. After reaction was completed, the solid was washed with dry DMF followed by dry acetonitrile, and subsequently dried with the help of argon. The produced 5′-azido modified RNA was cleaved from the support and treated according to the standard protocol for removal of protecting groups and purification.
cl-WT-5′az3′alk—Click ligation with full-length DNA splint—Strategy I
Ligation was performed under inert gas atmosphere following the protocol by El-Sagheer and BrownCitation29 with slight changes in the concentration of the reagents. The fragments and the full-length DNA splint were solved in 0.2 M NaCl solution, incubated at 85 °C and cooled down slowly to room temperature. THPTA, sodium ascorbate and CuSO4 were solved in 0.2 M NaCl solution and were added to the fragment solution. The final concentration of each fragment and the DNA splint was 1.5 µM; furthermore, the reaction mix contained 9 mM THPTA, 13 mM sodium ascorbate, and 1.3 mM CuSO4. The reaction mixture was stirred for 3 h at room temperature. Reaction was stopped by addition of ethanol and the product was isolated by PAGE.
cl-WT-5′alk3′az—Click ligation with full-length DNA splint—Strategy II
For the splint-mediated reaction, 0.8 nmol of a full-length DNA splint was added to the reaction mixture from above. Additionally, the sodium ascorbate buffer was replaced with 100 mM TRIS-HCl-buffer (pH 8.0) containing 10 mM MgCl2 and 1 mM CaCl2, being more appropriate for the following DNA digestion with DNase1. After reaction was finished, 3 µl of DNase I (1 u/µl) was added to the mixture, followed by incubation for 45 min by 37 °C. Reaction was stopped by addition of ethanol and the product was isolated by PAGE.
cl-WT-5′alk3′az—Click ligation without DNA splint—Strategy II
The click protocol was adapted from El-Sagheer and Brown.Citation29 RNA fragments (1 nmol each) were dissolved in 95 µl of a 0.2 M solution of NaCl (pH 7.0). The three fragments were annealed by heating at 90 °C for 2 min, followed by slow cooling to room temperature. Under inert atmosphere 5 µl of THPTA (0.35 µmol) and 0.5 µl of CuSO4 (0.05 µmol), both dissolved in sodium chloride solution (0.2 M), were added. Reaction was started by addition of 5 µl sodium ascorbate (5 µmol in 0.2 M NaCl-solution) followed by shaking for two hours at room temperature. The whole reaction was stopped by ethanol precipitation. The ligated RNA was purified by denaturing PAGE.
FMN quenching assay
A solution of fresh FMN (purchased from Alfa Aesar) was prepared in TRIS-HCl-buffer (50 mM TRIS-HCl, 2 mM MgCl2, 100 mM KCl, pH 8.0). Before measurements all oligonucleotides were heated at 90 °C for 2 min for denaturation and cooled down to room temperature for 15 min. 0.15 µM of the respective oligonucleotide was diluted in 100 µl TRIS-HCl-buffer, followed by addition of 0.1 µM FMN. The solution was held at 37 °C for 15 min for the purpose of incubation. Fluorescence measurements were performed in a 15 mm × 3 mm quarz cuvette using a Jasco FP-6500 Spectrofluorimeter. For irradiation a wavelength of 445 nm was chosen and the spectra were recorded from 455–660 nm. All FMN samples were handled under exclusion of light as much as possible.
Electrophoretic mobility shift assay
tr-WT (1 µM) was solved in buffer (0.02 M MgCl2, 0.05 M TRIS-HCl, 0.1 M KCl, pH 8.3) according to the protocol by Winkler et al.Citation7 The solution was incubated at 90 °C for 10 min. FMN (1 mM) and water were added to a final volume of 50 µl and the sample was incubated at rt for 5 h. Samples were loaded onto an 15% native PAA gel (500 V, 16 h).
2-AP fluorescence assay
Fluorescence data were collected using a Jasco FP-6500 Spectrofluorimeter with a 15 mm × 3 mm quarz cuvette. To 0.3 µM of the 2AP-modified oligonucleotide, dissolved in 100 µl TRIS-HCl-buffer (50 mM TRIS-HCl, 2 mM MgCl2, 100 mM KCl, pH 8.0), fresh FMN was added to a final concentration of 0.6 µM. The sample was mixed via repetitive pipetting for 10 s and remained at 37 °C for 30 min before the measurement was started. Irradiation of the samples occurred at 305 nm, and emission was recorded from 315–600 nm.
Reduction of FMN
The reduction experiment was connected with the 2-AP fluorescence assay. Therefore all solvents had to be free of oxygen beforehand in order to decrease the re-oxidation of FMN as much as possible (using degasing via ultrasonic and purging with argon). DTT-solution (1 M, 1 µl) was added to the FMN-aptamer complex containing mixture from above, followed by constant irradiation at 406 nm for 15 min with a 150 W high-pressure xenon lamp. The irradiation wavelength was switched back to 305 nm, and the spectrum was recorded immediately from 315–600 nm. For FMN re-oxidation, 1 cm3 of air was bubbled through the sample solution. After a time slot of about 10 min another spectrum was recorded.
Additional material
Download Zip (2.8 MB)Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
Financial support by the DFG (SPP 1258 “Sensory and regulatory RNAs in procaryotes”) is gratefully acknowledged. We thank Beatrice Karg and Klaus Weisz for valuable help with fluorescence spectroscopy.
References
- Bastet L, Dubé A, Massé E, Lafontaine DA. New insights into riboswitch regulation mechanisms. Mol Microbiol 2011; 80:1148 - 54; http://dx.doi.org/10.1111/j.1365-2958.2011.07654.x; PMID: 21477128
- Montange RK, Batey RT. Riboswitches: emerging themes in RNA structure and function. Annu Rev Biophys 2008; 37:117 - 33; http://dx.doi.org/10.1146/annurev.biophys.37.032807.130000; PMID: 18573075
- Serganov A, Patel DJ. Molecular recognition and function of riboswitches. Curr Opin Struct Biol 2012; 22:279 - 86; http://dx.doi.org/10.1016/j.sbi.2012.04.005; PMID: 22579413
- Gelfand MS, Mironov AA, Jomantas J, Kozlov YI, Perumov DA. A conserved RNA structure element involved in the regulation of bacterial riboflavin synthesis genes. Trends Genet 1999; 15:439 - 42; http://dx.doi.org/10.1016/S0168-9525(99)01856-9; PMID: 10529804
- Vitreschak AG, Rodionov DA, Mironov AA, Gelfand MS. Regulation of riboflavin biosynthesis and transport genes in bacteria by transcriptional and translational attenuation. Nucleic Acids Res 2002; 30:3141 - 51; http://dx.doi.org/10.1093/nar/gkf433; PMID: 12136096
- Serganov A, Huang L, Patel DJ. Coenzyme recognition and gene regulation by a flavin mononucleotide riboswitch. Nature 2009; 458:233 - 7; http://dx.doi.org/10.1038/nature07642; PMID: 19169240
- Winkler WC, Cohen-Chalamish S, Breaker RR. An mRNA structure that controls gene expression by binding FMN. Proc Natl Acad Sci U S A 2002; 99:15908 - 13; http://dx.doi.org/10.1073/pnas.212628899; PMID: 12456892
- Mironov AS, Gusarov I, Rafikov R, Lopez LE, Shatalin K, Kreneva RA, Perumov DA, Nudler E. Sensing small molecules by nascent RNA: a mechanism to control transcription in bacteria. Cell 2002; 111:747 - 56; http://dx.doi.org/10.1016/S0092-8674(02)01134-0; PMID: 12464185
- Strohbach D, Novak N, Müller S. Redox-active riboswitching: allosteric regulation of ribozyme activity by ligand-shape control. Angew Chem Int Ed Engl 2006; 45:2127 - 9; http://dx.doi.org/10.1002/anie.200503820; PMID: 16502442
- Strohbach D, Turcu F, Schuhmann W, Muller S. Electrochemically induced modulation of the catalytic activity of a reversible redoxsensitive riboswitch. Electroanalysis 2008; 20:935 - 40; http://dx.doi.org/10.1002/elan.200704173
- Moonen CT, Vervoort J, Müller F. Reinvestigation of the structure of oxidized and reduced flavin: carbon-13 and nitrogen-15 nuclear magnetic resonance study. Biochemistry 1984; 23:4859 - 67; http://dx.doi.org/10.1021/bi00316a007; PMID: 6498164
- Moonen CT, Vervoort J, Müller F. Carbon-13 nuclear magnetic resonance study on the dynamics of the conformation of reduced flavin. Biochemistry 1984; 23:4868 - 72; http://dx.doi.org/10.1021/bi00316a008; PMID: 6541948
- Burgstaller P, Famulok M. Isolation of Rna Aptamers for Biological Cofactors by in-Vitro Selection. Angew Chem Int Ed Engl 1994; 33:1084 - 7; http://dx.doi.org/10.1002/anie.199410841
- Weeks KM. Advances in RNA structure analysis by chemical probing. Curr Opin Struct Biol 2010; 20:295 - 304; http://dx.doi.org/10.1016/j.sbi.2010.04.001; PMID: 20447823
- Soulière MF, Altman RB, Schwarz V, Haller A, Blanchard SC, Micura R. Tuning a riboswitch response through structural extension of a pseudoknot. Proc Natl Acad Sci U S A 2013; 110:E3256 - 64; http://dx.doi.org/10.1073/pnas.1304585110; PMID: 23940363
- Reining A, Nozinovic S, Schlepckow K, Buhr F, Fürtig B, Schwalbe H. Three-state mechanism couples ligand and temperature sensing in riboswitches. Nature 2013; 499:355 - 9; http://dx.doi.org/10.1038/nature12378; PMID: 23842498
- Krstić I, Endeward B, Margraf D, Marko A, Prisner TF. Structure and dynamics of nucleic acids. Top Curr Chem 2012; 321:159 - 98; http://dx.doi.org/10.1007/128_2011_300; PMID: 22160388
- Rublack N, Nguyen H, Appel B, Springstubbe D, Strohbach D, Müller S. Synthesis of specifically modified oligonucleotides for application in structural and functional analysis of RNA. J Nucleic Acids 2011; 2011:805253; http://dx.doi.org/10.4061/2011/805253; PMID: 22013508
- Turunen JJ, Pavlova LV, Hengesbach M, Helm M, Müller S, Hartmann RK, Frilander MJ. (2013) RNA Ligation. In: Hartmann R, Bindereif A, Schön A, Westhof E (eds) Handbook of RNA Biochemistry, 2nd edn. Wiley, Weinheim (in press).
- Silverman SK. Deoxyribozymes: selection design and serendipity in the development of DNA catalysts. Acc Chem Res 2009; 42:1521 - 31; http://dx.doi.org/10.1021/ar900052y; PMID: 19572701
- Büttner L, Seikowski J, Wawrzyniak K, Ochmann A, Höbartner C. Synthesis of spin-labeled riboswitch RNAs using convertible nucleosides and DNA-catalyzed RNA ligation. Bioorg Med Chem 2013; 21:6171 - 80; http://dx.doi.org/10.1016/j.bmc.2013.04.007; PMID: 23664496
- Sindbert S, Kalinin S, Nguyen H, Kienzler A, Clima L, Bannwarth W, Appel B, Müller S, Seidel CA. Accurate distance determination of nucleic acids via Förster resonance energy transfer: implications of dye linker length and rigidity. J Am Chem Soc 2011; 133:2463 - 80; http://dx.doi.org/10.1021/ja105725e; PMID: 21291253
- Vauléon S, Ivanov SA, Gwiazda S, Müller S. Site-specific fluorescent and affinity labelling of RNA by using a small engineered twin ribozyme. Chembiochem 2005; 6:2158 - 62; http://dx.doi.org/10.1002/cbic.200500215; PMID: 16276501
- Welz R, Bossmann K, Klug C, Schmidt C, Fritz HJ, Müller S. Site-directed alteration of RNA sequence mediated by an engineered twin ribozyme. Angew Chem Int Ed Engl 2003; 42:2424 - 7; http://dx.doi.org/10.1002/anie.200250611; PMID: 12783515
- Kortmann J, Narberhaus F. Bacterial RNA thermometers: molecular zippers and switches. Nat Rev Microbiol 2012; 10:255 - 65; http://dx.doi.org/10.1038/nrmicro2730; PMID: 22421878
- El-Sagheer AH, Brown T. Synthesis and polymerase chain reaction amplification of DNA strands containing an unnatural triazole linkage. J Am Chem Soc 2009; 131:3958 - 64; http://dx.doi.org/10.1021/ja8065896; PMID: 19292490
- Tornøe CW, Christensen C, Meldal M. Peptidotriazoles on solid phase: [1,2,3]-triazoles by regiospecific copper(i)-catalyzed 1,3-dipolar cycloadditions of terminal alkynes to azides. J Org Chem 2002; 67:3057 - 64; http://dx.doi.org/10.1021/jo011148j; PMID: 11975567
- Rostovtsev VV, Green LG, Fokin VV, Sharpless KB. A stepwise huisgen cycloaddition process: copper(I)-catalyzed regioselective “ligation” of azides and terminal alkynes. Angew Chem Int Ed Engl 2002; 41:2596 - 9; http://dx.doi.org/10.1002/1521-3773(20020715)41:14<2596::AID-ANIE2596>3.0.CO;2-4; PMID: 12203546
- El-Sagheer AH, Brown T. New strategy for the synthesis of chemically modified RNA constructs exemplified by hairpin and hammerhead ribozymes. Proc Natl Acad Sci U S A 2010; 107:15329 - 34; http://dx.doi.org/10.1073/pnas.1006447107; PMID: 20713730
- Moroder H, Steger J, Graber D, Fauster K, Trappl K, Marquez V, Polacek N, Wilson DN, Micura R. Non-hydrolyzable RNA-peptide conjugates: a powerful advance in the synthesis of mimics for 3′-peptidyl tRNA termini. Angew Chem Int Ed Engl 2009; 48:4056 - 60; http://dx.doi.org/10.1002/anie.200900939; PMID: 19396850
- Steger J, Graber D, Moroder H, Geiermann AS, Aigner M, Micura R. Efficient access to nonhydrolyzable initiator tRNA based on the synthesis of 3′-azido-3′-deoxyadenosine RNA. Angew Chem Int Ed Engl 2010; 49:7470 - 2; http://dx.doi.org/10.1002/anie.201003424; PMID: 21038451
- Scriven EFV, Turnbull K. Azides - Their Preparation and Synthetic Uses. Chem Rev 1988; 88:297 - 368; http://dx.doi.org/10.1021/cr00084a001
- El-Sagheer AH, Brown T. Click chemistry with DNA. Chem Soc Rev 2010; 39:1388 - 405; http://dx.doi.org/10.1039/b901971p; PMID: 20309492
- Paredes E, Das SR. Click chemistry for rapid labeling and ligation of RNA. Chembiochem 2011; 12:125 - 31; http://dx.doi.org/10.1002/cbic.201000466; PMID: 21132831
- Santner T, Hartl M, Bister K, Micura R. Efficient Access to 3′-Terminal Azide-Modified RNA for Inverse Click-Labeling Patterns. Bioconjug Chem 2013; 25:188 - 95; http://dx.doi.org/10.1021/bc400513z; PMID: 24358989
- Chan TR, Hilgraf R, Sharpless KB, Fokin VV. Polytriazoles as copper(I)-stabilizing ligands in catalysis. Org Lett 2004; 6:2853 - 5; http://dx.doi.org/10.1021/ol0493094; PMID: 15330631
- Noeske J, Schwalbe H, Wöhnert J. Metal-ion binding and metal-ion induced folding of the adenine-sensing riboswitch aptamer domain. Nucleic Acids Res 2007; 35:5262 - 73; http://dx.doi.org/10.1093/nar/gkm565; PMID: 17686787
- Amonette JE, Szecsody JE, Schaef HT, Templeton JC, Gorby YA, Fruchter JS. Abiotic Reduction of Aquifer Materials by Dithionite - a Promising Insitu Remediation Technology. In Situ Remediation: Scientific Basis for Current and Future Technologies, Pts 1 and 2 1994:851-81.
- Song SH, Dick B, Penzkofer A, Hegemann P. Photo-reduction of flavin mononucleotide to semiquinone form in LOV domain mutants of blue-light receptor phot from Chlamydomonas reinhardtii. J Photochem Photobiol B 2007; 87:37 - 48; http://dx.doi.org/10.1016/j.jphotobiol.2006.12.007; PMID: 17292618
- Dolinnaya NG, Sokolova NI, Gryaznova OI, Shabarova ZA. Site-directed modification of DNA duplexes by chemical ligation. Nucleic Acids Res 1988; 16:3721 - 38; http://dx.doi.org/10.1093/nar/16.9.3721; PMID: 3375071
- Herrlein MK, Nelson JS, Letsinger RL. A Covalent Lock for Self-Assembled Oligonucleotide Conjugates. J Am Chem Soc 1995; 117:10151 - 2; http://dx.doi.org/10.1021/ja00145a042
- Xu Y, Kool ET. A Novel 5′-Iodonucleoside Allows Efficient Nonenzymatic Ligation of Single-stranded and Duplex DNAs. Tetrahedron Lett 1997; 38:5595 - 8; http://dx.doi.org/10.1016/S0040-4039(97)01266-5; PMID: 19924262
- Sokolova NI, Ashirbekova DT, Dolinnaya NG, Shabarova ZA. Chemical reactions within DNA duplexes. Cyanogen bromide as an effective oligodeoxyribonucleotide coupling agent. FEBS Lett 1988; 232:153 - 5; http://dx.doi.org/10.1016/0014-5793(88)80406-X; PMID: 2835262
- Kumar R, El-Sagheer A, Tumpane J, Lincoln P, Wilhelmsson LM, Brown T. Template-directed oligonucleotide strand ligation, covalent intramolecular DNA circularization and catenation using click chemistry. J Am Chem Soc 2007; 129:6859 - 64; http://dx.doi.org/10.1021/ja070273v; PMID: 17488075
- Breaker RR. Riboswitches and the RNA world. Cold Spring Harb Perspect Biol 2012; 4:a003566; http://dx.doi.org/10.1101/cshperspect.a003566; PMID: 21106649
- Serganov A, Nudler E. A decade of riboswitches. Cell 2013; 152:17 - 24; http://dx.doi.org/10.1016/j.cell.2012.12.024; PMID: 23332744
- Liberman JA, Wedekind JE. Riboswitch structure in the ligand-free state. Wiley Interdiscip Rev RNA 2012; 3:369 - 84; http://dx.doi.org/10.1002/wrna.114; PMID: 21957061
- Piekielska K, Gebala M, Gwiazda S, Muller S, Schuhmann W. Impedimetric Detection of Hairpin Ribozyme Activity. Electroanalysis 2011; 23:37 - 42; http://dx.doi.org/10.1002/elan.201000640
- Welz R, Müller S. 5-(Benzylmercapto)-1H-tetrazole as activator for 2'-O-TBDMS phosphoramidite building blocks in RNA synthesis. Tetrahedron Lett 2002; 43:795 - 7; http://dx.doi.org/10.1016/S0040-4039(01)02274-2
- Appel B, Marschall T, Strahl A, Müller S. Kinetic characterization of hairpin ribozyme variants. Methods Mol Biol 2012; 848:41 - 59; http://dx.doi.org/10.1007/978-1-61779-545-9_4; PMID: 22315062
- Chen YCJ, Hansske F, Janda KD, Robins MJ. Nucleic-Acid Related-Compounds. 64. Synthesis of 2',3′-Diazido-2',3′-Dideoxyadenosine and 2',3′-Diamino-2',3′-Dideoxyadenosine from 9-(Beta-D-Arabinofuranosyl)Adenine. J Org Chem 1991; 56:3410 - 3; http://dx.doi.org/10.1021/jo00010a042