Abstract
The eukaryotic Sm and the Sm-like (LSm) proteins form a large family that includes LSm proteins in archaea and the Hfq proteins in bacteria. Commonly referred to as the (L)Sm protein family, the various members play important roles in RNA processing, decay, and riboregulation. Particularly interesting from a structural point of view is their ability to assemble into doughnut-shaped rings, which allows them to bind preferentially the uridine-rich 3′-end of RNA oligonucleotides. With an emphasis on Hfq, this review compares the RNA-binding properties of the various (L)Sm rings that were recently co-crystallized with RNA substrates, and it discusses how these properties relate to physiological function.
Heptamer-forming (L)Sm Proteins Have a Conserved Nucleotide Binding Pocket
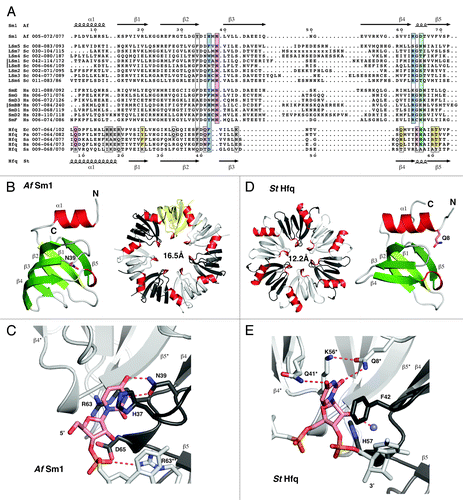
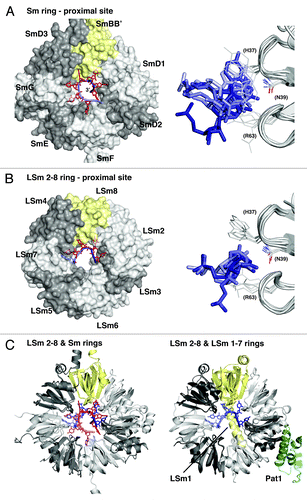
Sm proteins were originally identified as the targets of autoantibodies from the Sm (Stephanie Smith) serotype of patients with systemic lupus erythematosus.Citation1 Soon thereafter they were found to be part of the spliceosomal U snRNPsCitation2,Citation3 and to bind to internal, uridine-rich RNA sequences of the U snRNAsCitation4 around which they form heteroheptameric Sm rings.Citation5 A series of other Sm-like proteins were identified in eukaryotic species and were suggested to assemble into at least two additional heteroheptameric rings, the cytoplasmic LSm1–7 ring involved in mRNA decay, and the nuclear LSm2–8 ring that binds to the 3′-end of the U6 snRNA ().Citation6-Citation8 However, the first fully assembled LSm ring to be crystallized was the homoheptameric ring formed by the LSm protein AfSm1 from the archaeon Archaeoglobus fulgidus.Citation9 AfSm1 can be regarded as a simplified prototype for the Sm fold (), which consists of an N-terminal α-helix that packs against a five-stranded half-open β-barrel.Citation5 The structure of the AfSm1 ring confirmed that the suggested assembly of (L)Sm proteins into heptamersCitation5 via the outer β-strands (β4 and β5) was correct (). Furthermore, the structure demonstrates how one uridine per LSm monomer is recognized in a specific nucleotide binding pocket (). The uridine base is sandwiched between H37 and R63 as the upper and lower π-stacking partners. Its Watson-Crick edge is read out by hydrogen bonds to N39 on β-strand β3, explaining why uridines are a preferred substrate. The highly conserved R63 can also reach and recognize the ribose on the RNA backbone, whereas D65 on the L(β4-β5) loop seems to limit the space of the backbone phosphates inside the pore of the ring. This nucleotide binding pocket is not present in all LSm proteins,Citation6,Citation7,Citation10 but it is conserved in ring-forming Sm and LSm proteins from eukaryotes, and therefore, amino acids will frequently be referred to in the following by their position in the AfSm1 protein.
Figure 1. Alignment and structure of crystallized ring-forming (L)Sm and Hfq proteins. (A) Structure-based sequence alignment. Secondary structure elements and numbering above and below the alignment are from the AfSm1 and StHfq proteins, respectively. Only the crystallized (L)Sm-core of each protein is aligned. The numbers preceding each sequence refer to the aligned residues and to the total length of each protein. They also indicate the presence and size of C-terminal extensions. The crystallized sequences of StHfq and EcHfq are identical. Positions involved in RNA or nucleotide binding are boxed. Blue: Stacking partners for the nucleobase in the proximal site. Hydrogen bonds to the RNA backbone in the case of R63 and its (L)Sm homologs. Red: Base recognition by hydrogen bonds in the proximal site. Green: Constriction of the RNA backbone in the proximal site and RNA 3′-end recognition in the case of Hfq. Yellow: Conserved “R”-site on the distal surface of Hfq; Q33 in EcHfq: “A”-site on the distal surface of EcHfq, not present in many gram-positive bacteria. Gray: Conserved uridine-site on the outer rim of the proximal surface of Hfq and of archaeal LSm proteins, plus members of the basic patch on the lateral surface of StHfq and EcHfq (R16, R17, R19, K47) that is less well conserved in gram-positive bacteria. Sequence information is from the respective PDB files listed by PDB-ID, chain-ID, resolution, and species: Sm1, 1i4k_A,Citation9 2.50 Å, Archaeoglobus fulgidus, Af; LSm1–7, 4c92_ABCDEFG,Citation84 2.30 Å, and 4m75_A,Citation85 2.95 Å, Saccharomyces cerevisiae, Sc; LSm8, 4m77_A,Citation86 3.11 Å, Saccharomyces cerevisiae, Sc; Sm proteins, 2y9a_ABCDEFG,Citation87 3.60 Å, Homo sapiens, Hs; EcHfq, 2ylc_A,Citation29 1.30 Å, Salmonella typhimurium, St and Escherichia coli, Ec; PaHfq, 4j6y_A,Citation28 2.14 Å, Pseudomonas aeruginosa, Pa; SaHfq, 1kq2_A,Citation26 2.71 Å, Staphylococcus aureus, Sa; BsHfq, 3ahu_A,Citation39 2.20 Å, Bacillus subtilis, Bs; SsHfq, 3hfo_A,Citation79 1.30 Å, Synechocystis ssp., Ss. (B) LSm monomer and homoheptamer. The monomer of the archaeal AfSm1 proteinCitation9 is shown with α-helices in red and β-strands in green. N39 (sticks) marks the nucleotide binding pocket shown in (C). The loop L(β4-β5) forms a short 3–10 helix that is highly conserved. The protomers in the heptameric ring are colored with in white, gray and yellow, and helices in red for orientation. The diameter of the pore is 16.5 Å (proximal view), using the Cβ atom of D65 as a reference. (C) Nucleotide binding in the proximal site of an LSm protein. Uridine recognition is shown for AfSm1,Citation9 with optimal geometry for hydrogen bonds (dotted red). Important side chains are shown as sticks with nitrogens in blue and oxygens in red. The asterisk and double asterisk mark the preceding and following protomer in the 5′-3′ direction of the bound RNA, respectively. (D) Hfq monomer and homohexamer. Compared with the AfSm1 protein, the monomer of the bacterial StHfq proteinCitation29 reveals an extended α-helix α1, a loop L(β2-β3) that has a distinct structure, and a variable loop L(β3-β4) that is very short. Q8 (sticks) marks the modified nucleotide binding pocket shown in (E). The diameter of the pore is 12.2 Å (proximal view), using the Cβ atom of H57 as a reference. (E) Nucleotide binding in the proximal site of Hfq. Uridine recognition by hexameric StHfqCitation29 is structurally distinct from (L)Sm proteins and involves residues from two neighboring protomers. Base recognition geometry is rather poor, a potential consequence of the constricted RNA backbone conformation. A conserved water molecule is shown as a blue sphere (see also ). H57 is positioned close to the 3′-oxygen of the ribose. The alignment was done with the help of ESPRIPT,Citation110 structural analysis and figures were done with the help of COOTCitation111 and PYMOL (http://www.pymol.org).
The Structure of Hexameric Hfq Reveals Modified Nucleotide Binding Pockets at the Subunit Interfaces
The Hfq protein was originally identified as a bacterial host factor for the Qβ phage replication.Citation11 Since then, it has been implicated in numerous processes in bacterial RNA metabolism and by now it is probably one of the best studied RNA binding protein in bacteria, which has recently been reviewed extensively.Citation12-Citation21 Its most prominent role is in riboregulation by bacterial small RNAs (sRNA). The realization that Hfq shares the Sm-fold came relatively late,Citation22,Citation23 because Hfq sequences lack an important sequence signature that commonly identifies eukaryotic and archaeal (L)Sm proteinsCitation6-Citation8,Citation24 and that results from their conserved nucleotide binding pocket (). Moreover, the Hfq protein assembles into homohexameric ringsCitation22,Citation23 rather than into heptamers, as confirmed by the first crystal structures.Citation25,Citation26 These structures also revealed a modified nucleotide binding pocket as compared with the heptameric (L)Sm proteins ().Citation25,Citation27 Whereas F42 from the prototypic Escherichia coli (Ec) Hfq (the structural equivalent of H37 in AfSm1) still provides a stacking platform for the base, and whereas H57 (the equivalent of D65 in AfSm1) still is in a position to contact the RNA backbone, the equivalent of N39 is missing. Instead, the Watson-Crick edge of a bound uridine is read out by Q8* on helix α1 from the neighboring Hfq monomer (indicated by the asterisk). Q8* is highly conserved among Hfq proteins and assisted in base recognition by K56* and Q41*. The pocket shows a preference for uridine, but can also accommodate cytidine and adenine.Citation26,Citation28-Citation30 It is rather clear by now however that it strongly discriminates against guanine.Citation26,Citation28,Citation29 In the hexamer, the six-nucleotide binding pockets are located in close proximity on the inner rim of the so-called proximal surface of the ring. Collectively, they are also referred to as the proximal RNA binding site ().
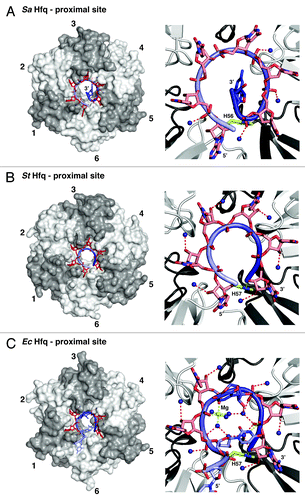
Figure 2. RNA binding and 3′-end recognition in the proximal site of Hfq. Left panels: Overview, with the protein as transparent surfaces (ring orientation as in ). Right panels: Zoom, with the protein as cartoon. (A) Dilated RNA conformation and an expelled 3′-end on SaHfq.Citation26 Hfq protomers are colored in white and gray and numbered clockwise for orientation. RNA is shown as sticks with a backbone cartoon that is colored with a gradient from light to dark blue to indicate the 5′-3′ direction. Nucleotides that reside in specific binding pockets are in red, the expelled terminal guanine is in dark blue. Dark blue spheres indicate conserved water molecules that, in the dilated conformation, contact the phosphates of the RNA backbone (hydrogen bonds dotted red). (B) Constricted RNA conformation on StHfq.Citation29 The constricted conformation allows the recognition of the 3′-end by H57 (residue and dotted hydrogen bond in lime). The conserved water molecules contact the ribose of the RNA backbone. (C) Mixed RNA conformation on EcHfq.Citation30 The irregular backbone alternates between the dilated and constricted conformations as illustrated by the contacts to the conserved waters. One of the uridines (dark blue) is expelled from its pocket, and another uridine (light blue) stabilizes the backbone conformation by a hydrogen bond (lime) to the last phosphate in the RNA chain. The 3′-hydroxyl group of the terminal adenine is recognized via a hydrogen bond to H57 (both in lime). The stabilizing Mg2+-ion and its coordination are also shown in lime, the remaining hydrogen bond network as dotted red lines.
The Proximal RNA Binding Site on Hfq Hexamers Preferably Binds to the 3′-Ends of RNA
An initial crystal structure of Hfq from the gram-positive bacterium Staphylococcus aureus (Sa) revealed how the proximal RNA binding site can bind to an oligonucleotide (5′-AUUUUUG-3′) with internal uridines that was derived from the RNA binding siteCitation4 of the eukaryotic Sm ring on U snRNAs. In this structure, the six-nucleotide binding pockets of the SaHfq ring are occupied by the first six residues of the oligonucleotide, whereas the 3′-terminal guanine is expelled ().Citation26
Subsequent binding studies with Salmonella typhimurium (St) Hfq demonstrated however that the proximal site binds uridine-rich oligonucleotides much better if the 3′-terminal nucleotide is a uridine.Citation29 In this case, the 3′-terminal nucleotide occupies one of the binding pockets and the 3′-hydroxyl group of the last ribose in the chain is directly recognized by H57, as indicated by a high-resolution crystal structure of StHfq in complex with hexauridine (5′-UUUUUU-3′, ). The same recognition is possible with an adenine or cytidine in the terminal position, whereas phosphate modification of the 3′ hydroxyl group leads to a strong reduction of affinity.Citation29
The crystal structure also reveals a much more constricted backbone conformation than observed with the SaHfq complex and a different binding register for the phosphate and ribose moieties that is likely triggered by the positioning of the 3′-terminal hydroxyl group ().Citation29 These differences indicate a general difficulty to accommodate the RNA backbone in the constricted space of the pore and a trade-off between optimal nucleotide binding and optimal backbone geometry. Indeed, such a trade-off might also explain the poor base-recognition geometry observed in the crystal structuresCitation26,Citation29,Citation30 and it might be another reason for Hfq hexamers to preferably bind RNA 3′-ends, where the strain on the RNA backbone is reduced.
Regarding Hfq function, the realization that Hfq binds RNA 3′-endsCitation29,Citation31 was particularly important, because it suggested and explained how Hfq protects and stabilizes RNA transcripts with uridine-rich 3′-ends resulting from Rho-independent transcription termination.Citation32 These transcripts include the small regulatory sRNAs for the function of which Hfq plays a central role.Citation12-Citation14,Citation16,Citation18-Citation20
The trade-off between nucleotide binding and an optimal backbone geometry can also lead to more complex modes of oligonucleotide binding that might quite frequently occur in nature, but are hard to crystallize and observe. A good example is the remarkable crystal structure of a 5′-AUUUUUUA-3′ oligonucleotide bound in the proximal site of EcHfq (, the crystallized sequence of EcHfq is 100% identical to StHfq).Citation30 The structure demonstrates once more the preference of the proximal site for RNA 3′-ends and it shows how an adenine is recognized and tolerated at the 3′-end. Furthermore, the 3′-hydroxyl group is indeed recognized by H57 in the way as suggested before,Citation29 and a magnesium ion plays a crucial role in the stabilization of the irregular conformation of the RNA backbone (). Intriguingly, only four of the six nucleotide binding pockets are occupied, and the corresponding uridines are either looped out (, protomer 4) or participate in the stabilization of the RNA backbone (, protomer 1) by a hydrogen bond from the N3 position to the last phosphate five nucleotides downstream.
In summary, the preference of the proximal RNA binding site for RNA 3′-ends over internal RNA sequences likely results from the direct recognition of the 3′-hydroxyl group and from the possibility to engage the nucleotide binding pockets with less strain on the phosphoribose backbone and with more contacts to the protein surface.
The Distal Surface of Hfq Hexamers Binds Adenine-Rich RNAs
EcHfq also binds oligo-(A) tracts with high affinity.Citation16,Citation33-Citation35 This has been puzzling until it was realized that the distal surface of the Hfq hexamer provides a second RNA binding surface,Citation36 and crystal structures revealed how adenine-rich oligonucleotides are recognized ().Citation37-Citation39 Each subunit contains a conserved nucleotide binding pocket (originally termed the “R” site)Citation38 that allows the base to stack on the aromatic Y25 and that is lined by Q52, S60, and T61 to recognize the Watson-Crick edge (, sequence and numbers according to EcHfq). Experimentally, the “R”-site has been shown to be highly selective for adenine, although other nucleotides might occasionally be accommodated as well.Citation28,Citation37-Citation41
Figure 3. RNA binding on the distal surface of Hfq. (A) Recognition of an RNA with a (5′-ANA-3′)6 signature on EcHfq.Citation38 Transparent surfaces of Hfq protomers are colored in white and gray and numbered counterclockwise to relate the arrangement to . RNA is shown as sticks with a backbone cartoon that is colored with a gradient from light to dark blue to indicate the 5′-3′ direction. Nucleotides that reside in specific binding pockets are in dark red (conserved “R”-site) and light red (‘A’-site for the second linker nucleotide), the unbound linker nucleotide is in blue. (B) Recognition of an RNA with a (5′-AN-3′)6 signature on BsHfq,Citation39 a representative from a gram-positive bacterium. Nucleotides that reside in specific binding pockets are in dark red (conserved “R”-site), the unbound linker nucleotide is in blue.
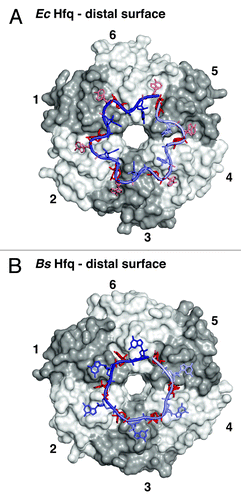
The “R”-site pockets allow EcHfq to recognize every third adenine of an oligo-(A) tract (),Citation38 whereas the “R”-site pockets on Hfq hexamers from Bacillus subtilis (Bs), Staphylococcus aureus, and probably many other gram-positive bacteria recognize every second adenine ().Citation37,Citation39 In the latter case, the nucleotide that links two recognized adenines (linker nucleotide) frequently seems to adopt the rare syn-conformation, but linker nucleotide identity does not seem to be conserved between species. In the case of EcHfq, there are two linker residues between the adenines in the “R”-site pockets. The first one of these protrudes into the bulk solvent and is not recognized specifically, whereas the second one fits into an additional shallow and adenine-specific binding pocket that is not present in BsHfq and SaHfq and that reads out the Hoogsteen-edge of the base via contacts to the peptide-backbone of Q33 on β-strand β2 ( and ).
As a consequence, the Hfq hexamers from Escherichia coli and related gram-negative bacteria preferably bind RNA sequences with an (5′-ANA-3′)n signature on their distal surface, whereas Hfq hexamers from many gram-positive bacteria seem to prefer a (5′-AN-3′)n signature.Citation37-Citation39,Citation41 Furthermore, at least four Hfq subunits need to be engaged for an optimal binding, and longer oligo-(A) tracts can be bound cooperatively by Hfq hexamers.Citation16,Citation42
Additional RNA Binding Surfaces on Hfq Hexamers are Responsible for the Specific Recognition of Regulatory sRNA
Small regulatory RNAs in bacteria (sRNAs) are characterized by (1) a usually 5′-terminal “seed” sequence that is complementary to the target mRNA, (2) a central sRNA “body,” and (3) a 3′-terminal stem-loop structure that ends in an oligo-(U) tail as a consequence of Rho-independent transcription termination.Citation19,Citation32,Citation43,Citation44 The size and shape of the RNA body varies considerably among sRNAs and they frequently contain additional uridine-rich stretches that alternate with small, often only transiently stable base-pairs and stem-loop structures.Citation42-Citation46 Nevertheless, Hfq recognizes these sRNAs specifically and distinguishes them from other RNAs in the cell.Citation22,Citation23,Citation47-Citation49 With the described RNA binding properties of the proximal and distal surfaces of the Hfq ring, the specificity of the recognition and the protection of the entire sRNA body from nuclease attackCitation12,Citation22,Citation23,Citation42,Citation43,Citation50-Citation52 were hard to explain.
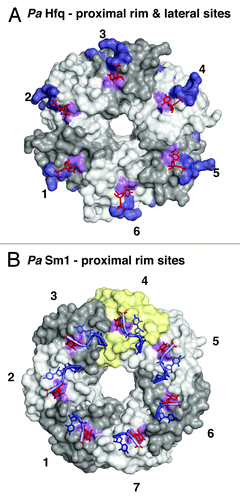
This situation changed with the realization that certain sRNAs still bind Hfq even when their 3′-end is blocked by a cyclic phosphate and even when the proximal and distal RNA binding surfaces of the Hfq hexamer are mutated or occupied by other small oligonucleotides.Citation42 Obviously, there had to be additional RNA-binding surfaces on Hfq () that were subsequently identified to include a basic patch on the lateral surface of each Hfq protomer. On StHfq and EcHfq, the basic patch consists of R16, R17, R19, and K47, and it is conserved primarily in gram-negative bacteria ( and ).Citation42 Furthermore, an additional uridine binding site was identified by crystallography on the outer rim of the proximal surface in Pseudomonas aeruginosa (Pa) Hfq, where the base stacks on the aromatic F39 in β-strand β2 (F39 in EcHfq) and where specificity is achieved by hydrogen bonds from the Watson-Crick edge of the uridine to the peptide backbone of the amino acid ( and ).Citation28 This uridine binding site is close to the basic patch and may indeed be quite conserved, because it is also found in the crystal structure of the heptameric LSm ring of the PaSm1 protein from the archaeon Pyrococcus abyssii (Y34 in AfSm1) that was co-crystallized with a heptauridine (5′-UUUUUUU-3′) oligonucleotide ( and ).Citation28,Citation53
Figure 4. Additional RNA binding surfaces on Hfq and (L)Sm proteins. (A) RNA binding on the outer rim of the proximal surface and on the lateral surface of Hfq. Transparent surfaces of Hfq protomers are colored in white and gray and numbered for orientation. A conserved uridine binding site on the proximal surface of bacterial PaHfq (Pseudomonas aeruginosa) is colored in purple (corresponding to F39 in EcHfq).Citation28 The areas corresponding to the basic patch on the lateral surface of StHfq are colored in dark blue (corresponding to R16, R17, R19, K47 in StHfq and EcHfq).Citation29 The basic patch allows the specific recognition of regulatory sRNAs and catalyzes base-pair formation and exchange. The uridines co-crystallized with PaHfq are shown as red sticks. (B) Conservation of the uridine binding site on an archaeal LSm heptamer.Citation53 The surface of the additional protomer in the homoheptameric ring of the archaeal PaSm1 (Pyrococcus abyssii) protein is shown in yellow. The conserved uridine binding site is colored in purple (corresponding to Y34 in AfSm1) and the bound uridines are shown as red sticks. Other nucleotides and the RNA backbone of the co-crystallized oligomers are drawn in blue as described before.
Importantly, the basic patch is accessible by oligonucleotides both from the proximal and from the distal surface of hexameric Hfq,Citation42,Citation54 and the respective arginines with their intrinsic ability to stack and pair with nucleotide bases were shown by mutational analysis to participate actively in the nucleation of RNA helices.Citation54 Consequently, the basic patch (possibly extended by F39) may therefore not only bind and expose single-stranded uridine-rich sequences from the sRNA body, but it may also recognize and stabilize transient base-pairs and stem-loop structures as they occur within the sRNA body or between the seed and target sequences as they begin to anneal.Citation42,Citation54
Multiple Roles of Hfq in Riboregulation
Given the abundance of Hfq in the bacterial cell and its rather general and structure-specific RNA binding properties, it is not surprising that Hfq has been identified in a very large number of regulatory processes where RNA is involved.Citation12-Citation14,Citation16,Citation19,Citation20 The most important functions and selected examples are listed below with increasing complexity ().
Figure 5. Roles of Hfq in riboregulation. (A) Schematic structure of an sRNA with a 5′-terminal seed region and a 3′-terminal terminator stem-loop structure. sRNAs compete rapidly for Hfq binding (blue arrow). (B) Hfq/sRNA complex. The sRNA is shown with its 3′-end anchored in the proximal site of Hfq and its body adapted to the lateral sites (small red arrows). (C) mRNA targeting complex with an mRNA bound to the distal surface of Hfq. The lateral sites assist in the (de-) formation of base-pairs (red asterisks). The topology of the complexes may vary from case to case;Citation72 the directionality of the RNA on the proximal and distal surfaces of Hfq corresponds to the crystal structures. Figure modified from Sauer, et al.Citation42
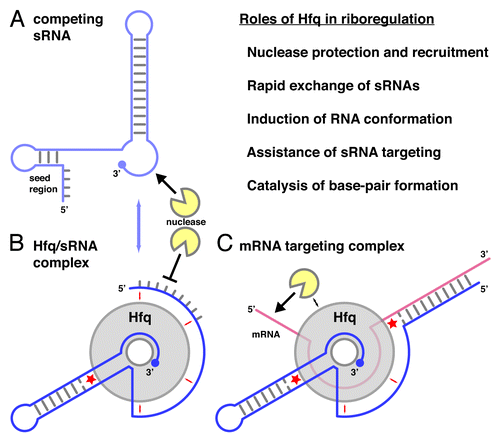
Protection against and recruitment of nucleases
One of the easiest ways for Hfq to affect RNA stability and gene expression is the attachment to an RNA molecule and its protection from nuclease attack. For example, it has long been known that Hfq stabilizes A-tracts and oligo-(A) tails, which likely happens via the distal surface and arguably in a cooperative manner.Citation16,Citation33-Citation35,Citation42,Citation55-Citation57 Furthermore, the proximal binding site can protect the 3′-oligo-(U) ends of Rho-independent transcripts from the attack of PNPase,Citation50 and uridine-rich sequences that are present in the interior of both mRNAs and sRNAs and are protected from RNaseE probably because they interact with the additional RNA-binding surfaces of Hfq.Citation16,Citation34,Citation51 In particular, many sRNAs are strongly destabilized in the absence of Hfq.Citation22,Citation23,Citation52
On the other hand, Hfq also interacts with PNPase and RNaseE,Citation12,Citation13,Citation19,Citation58,Citation59 potentially even directly.Citation60 In the case of the RyhB sRNA, this property of Hfq is exploited specifically for the targeted degradation of sodB mRNA by RNaseE. This works even in the absence of sodB mRNA translation, outlining a pathway of active, sRNA-induced mRNA degradation.Citation61
Rapid recycling of sRNAs on Hfq
In the bacterial cell, the amount of Hfq is considered to be limiting such that numerous sRNAs compete for Hfq binding.Citation20,Citation46,Citation62-Citation64 Consequently, the ability of Hfq to rapidly exchange bound sRNAs while maintaining a high binding affinity and specificity becomes an important property.Citation20,Citation62 This property allows Hfq to act like a hub, integrating different sRNA responses and it allows the cell to switch rapidly between the regulation by different sRNAs, simply because freshly induced sRNA molecules will displace previous sRNAs from Hfq, leading to their rapid degradation.Citation20,Citation42,Citation46,Citation62-Citation64 Mechanistically, the dynamic interaction with sRNAs and their rapid competitive displacement can be explained by a “peeling” mechanism, where the sRNA disengages the lateral surfaces consecutively one by one rather than breaking all of the contacts with Hfq in a single step.Citation42,Citation62
Induction of conformational changes
Another important function of Hfq is its action as an RNA chaperone,Citation65-Citation68 inducing conformational changes in the bound RNA. A recent example of a positional effect is from the 5′ leader sequence of rpoS mRNA, where the distal surface of Hfq binds to an upstream (5′-ANA-3′)4 motif and stabilizes an RNA conformation that allows sRNAs (DsrA, RprA, ArcZ) to act on a downstream target site.Citation69 Alternatively, Hfq may also interfere directly with the formation of an inhibitory secondary structure and expose the bound nucleotides in a conformation that is optimal for base-pairing with a target RNA as suggested for the lateral binding surface of Hfq.Citation42
Assistance of sRNA targeting
A single Hfq ring can stably bind two RNA molecules at the same time, such as an oligo-(A)20 RNA on the distal surface and a RybB sRNA that is 3′-anchored in the proximal site and covers several of the basic patches.Citation42 Consequently, Hfq can stimulate sRNA targeting not only by conformational activation, but also by bringing the binding partners closely together in a ternary sRNA-Hfq-mRNA complex that should exist at least transiently before further conformational changes and downstream events take place.Citation19,Citation36,Citation42,Citation70 Indeed, many mRNAs that are known sRNA targets contain adenine-rich (5′-ANA-3′)n motifs in their 5′-UTRs close to the translation start site,Citation19,Citation38,Citation47,Citation71 supporting the notion that Hfq actively assists bound sRNAs to associate with their mRNA targets.
Direct catalysis of base-pair formation
Finally, the basic patch not only recognizes transient base-pairs and stem-loops as part of the sRNA body,Citation42,Citation45 but the respective arginine residues also promote RNA-helix formation between distinct RNA oligonucleotides that are anchored on the proximal and distal surfaces of the Hfq hexamer, respectively. In this sense, the basic patch has enzyme-like active site properties, stabilizing initial base-pairs as a transition state.Citation54
Future Challenges Regarding Hfq
It becomes clear from this list that the contribution of Hfq to riboregulation is highly complex and variable for each RNA partner, and this will continue to present a challenge for future investigations. Mutational approaches in vivo reveal considerable differences on how individual sRNAs interact with Hfq,Citation72 an observation which is supported by structural studies in solution.Citation73,Citation74 However, to capture the dynamics of sRNA Hfq interactions requires novel approaches such as single molecule techniques. These would also allow it to address the roles of the presumably disordered C-terminal extensions that are present in many gram-negative bacteria (). In the simplest view, these C-terminal tails might act as a “molecular bumper” or as a protective cage around a bound sRNA, but more specific roles in riboregulation have also been suggested.Citation75,Citation76
Furthermore, the roles of Hfq in many bacterial species may differ substantially from what has been learned from the gram-negative species that have been studied most intensively. For example, in the Hfq hexamers from the gram-positive Staphylococcus aureus, the C-terminal tails are short, the basic patch on the lateral surface is only poorly conserved (),Citation42 and the distal surface reveals an altered mode of RNA recognition.Citation37 Functional assays suggest a much less prominent role of Hfq-bound sRNAs in Staphylococcus aureus riboregulation.Citation77,Citation78 Another example is cyanobacterial Hfq (Synechocystis, SsHfq) that appears not to be involved in riboregulation at allCitation79 and many bacterial pathogens such as Helicobacter pylori do not even encode a protein with an Sm fold.Citation80,Citation81 It shall be highly interesting to learn how riboregulation is achieved in these species.
Finally, in species where Hfq does exist, its impact on the molecular biology of the entire system deserves particular attention.Citation82 Indeed, the entire transcriptome is affected by the presence of Hfq, because RNA molecules have not only evolved to positively engage the protein and exploit it for riboregulation, but apparently also to prevent the inadvertent association with Hfq by chance. Examples can be seen by the underrepresentation of (5′-ANA-3′)n stretches in protein coding regions of mRNAsCitation47,Citation71 or in the sequestration of the uridine-rich 3′-tail into a stable stem-loop structure in GlmY sRNA to prevent Hfq association in the context of amino sugar pathway regulation.Citation83
The Recognition of RNA 3′-Ends by Eukaryotic (L)Sm Proteins
Recently, crystal structures became available of the LSm1–7Citation84,Citation85 and the LSm2–8 ring,Citation86 the latter also in complex with a short RNA oligonucleotide (5′-GCUUUU-3′). Together with the previously published structures of the U1 and U4 snRNPs that contain the Sm ring,Citation87-Citation89 these data allow a first comparison of the hetero-heptameric rings from eukaryotic species (), especially with respect to the mechanism of RNA 3′-end recognition that has been demonstrated for Hfq.Citation29,Citation31
Figure 6. RNA binding in the proximal sites of eukaryotic Sm and LSm heteroheptamers. (A) The Sm ring.Citation87 All seven nucleotide binding pockets are specifically occupied by the residues from the Sm-site in U snRNAs. The human Sm ring was co-crystallized with engineered U1 or U4 snRNA, which continues through the pore to the distal side. Right panel: Superposition of the seven nucleotide binding pockets including their nucleotide ligands, illustrating the strong variation and frequently suboptimal base-recognition geometry. The upper and lower stacking partner as well as the central asparagine are labeled and numbered in brackets according to the sequence in AfSm1. (B) The LSm2–8 ring.Citation86 Only four of the nucleotide binding pockets are specifically occupied by uridines from the bound oligonucleotide (red sticks). The 3′-terminal nucleotide contacts both LSm3 and LSm6 with good base-recognition geometry in the LSm3 pocket. Right panel as in (A). The LSm2–8 ring is highly similar to the LSm1–7 ring, which has LSm8 (yellow) exchanged against LSm1 and which has been crystallized in the absence of RNACitation84,Citation85 (sequences for both rings are from Saccharomyces cerevisiae). (C) Left panel: Superposition of the LSm2–8 ring (RNA from light to dark blue) onto the Sm ring (RNA from light to dark red), emphasizing the similarity of the protein backbone (shown as cartoon) and illustrating the differences in RNA binding. Right panel: Superposition of the LSm2–8 ring (RNA from light to dark blue) onto the LSm1–7 ring (no RNA bound), illustrating the LSm1 C-terminal helix that obstructs the central pore on the distal surface of the ring. The position of Pat1-binding on LSm2 and LSm3 of the LSm1–7 ring is indicated.
The Sm Ring in U snRNPs
The Sm ring () is specialized for the recognition of the Sm site on U snRNAs,Citation4 which is a single-stranded and defined RNA sequence (5′-AUUUUUG-3′ in the crystallized structuresCitation87-Citation89), located internally, in between base-paired parts of U4 RNA that are fixed above and below the Sm ring. As a consequence, the Sm ring needs to assemble around the Sm site on U snRNAs, a process for which several assembly factors have been identified in mammalian cells.Citation90,Citation91 The Sm ring consists of the protomers SmE, SmG, SmD3, SmBB’, SmD1, SmD2, and SmF, listed in the order of engagement by the RNA in the 5′ to 3′ direction. The RNA enters via the protomer SmE from the proximal surface and exits via the protomer SmF before threading through the pore to the distal side (). Each protomer binds a nucleotide in its binding pocket. The pockets are in the same structural positions as in the archaeal AfSm1 prototype () and the amino acid corresponding to asparagine N39 that contacts the nucleotide at the Watson-Crick edge is strictly conserved. Similarly conserved is R63, the arginine that serves as the lower π-stacking partner and that contacts the phosphoribose backbone. An exception is SmE that binds the adenine at the RNA entrance and which contains a lysine instead. The upper π-stacking partner corresponding to H37 in AfSm1 is less well conserved, but it provides π-electrons for base-stacking in all cases except SmD1, which contains a serine (). Importantly, the pockets reveal considerable variation in the orientation of their binding partner, even for the five central uridines where not every Watson-Crick edge is read out with optimal geometry ().Citation87 Similar to the situation in bacterial Hfq (), there appears to be a trade-off between optimal base recognition by the conserved asparagine and an ideal backbone conformation in the constricted space of the pore. For the Sm ring, the placement of the phosphoribose backbone inside the narrow pore might pose a particular challenge because it assembles around an internal RNA binding site with the need to accommodate the continuing 3′-sequence. As a consequence, the more limited conformational freedom of the RNA molecule in the absence of a 3′-end might be the reason why none of the bases is in an optimal position with respect to the interacting asparagine (N39 in AfSm1).
The LSm2–8 Ring
In contrast to the Sm ring, the LSm2–8 ring () assembles in the absence of RNACitation92 and it specifically recognizes the processed 3′-end of the spliceosomal U6 snRNA, which terminates with five uridines and a 2’-3′-cyclic phosphate instead of the usual 3′-hydroxyl group.Citation93 This substrate specificity is reminiscent of the bacterial Hfq rings, only that a terminal phosphate decreases the affinity in the case of Hfq, whereas it increases the affinity in the case of the LSm2–8 ring.Citation29,Citation93 Based on sequence similarity, the LSm protomers (LSm5, LSm7, LSm4, LSm8, LSm2, LSm3, and LSm6) can be aligned to the corresponding Sm protomers (SmE, SmG, SmD3, SmBB’, SmD1, SmD2, and SmF), and they assemble in the same order ().Citation86 The preference for uridines is reflected by conserved nucleotide binding pockets, with the exceptions of LSm7 and LSm8, which lack the aromatic π-stacking partner in the upper position, and especially LSm5, where the lower π-stacking partner, the highly conserved arginine (R63 in AfSm1), is replaced by a serine (). The LSm2–8 ring was co-crystallized with 5′-GCUUUU-3′ and 5′-GCUUU-3′ RNA oligonucleotides that terminate with a 3′-hydroxyl group.Citation86 The structures reveal that the 3′-terminal uridine of these substrates is preferentially bound by the LSm3 protomer (), which shows the best base-recognition geometry and permits a contact between the 2’ hydroxyl group of the terminal ribose and the conserved arginine of LSm6. LSm3 is the last protomer in the 5′-3′ direction of the RNA where this contact is possible. If LSm6 accommodated the terminal uridine, LSm5 could not provide the corresponding contact because it lacks the arginine.Citation86 Uracil base recognition geometry deteriorates with increasing distance from the 3′-end,Citation86 consistent again with the notion that increasing conformational constraints on the phosphoribose backbone lead to suboptimal use of the uracil binding pockets (). As a consequence, substrates with terminal uridines are preferred over substrates with internal uridines, and uridine substitutions in the terminal position have the strongest effect on affinity,Citation86 as previously observed for Hfq.Citation29
However, the present structures do not show how the physiological RNA substrate with its five terminal uridines and the 2’-3′-cyclic phosphateCitation93 would fit into the ring. Zhou, et al.Citation86 argue that LSm3 could also accommodate the cyclic phosphate if the conserved arginine from LSm6 adjusts its position. However, there is an attractive alternative where the additional uridine would enter the empty binding pocket on LSm6, placing the terminal phosphate for specific recognition into the space that is left by the missing arginine from LSm5. In this case, all five uridines of the substrate would be recognized without the need to change the path for RNA entry into the LSm ring between the LSm7 and LSm4 protomers. It should be easy to experimentally distinguish between these two alternatives.
The LSm1–7 Ring
Whereas the Sm ring and the LSm2–8 ring form specific and stable complexes with spliceosomal U snRNAs in the nucleus, the LSm1–7 ring is not involved in mRNA splicing.Citation6-Citation8 Instead, it plays an important role in cytoplasmic mRNA decay, linking mRNA deadenylation to decapping.Citation94,Citation95 LSm1–7 binds and protects the 3′-ends of deadenylated mRNA substrates and it initiates mRNA decapping most likely through its interaction with the structured, C-terminal domain of the decapping activator Pat1,Citation96,Citation97 with which it forms a stable complex that has recently been crystallized ().Citation84
LSm1–7 appears to recognize a variety of RNA 3′-ends.Citation21 It participates in the 3′-oligouridinylation-dependent decay of mRNAsCitation98,Citation99 that has been described for the special case of mammalian histone mRNAs,Citation100 for the mRNAs of certain proteins in the yeast Saccharomyces pombe,Citation101 and for many oligoadenylated mRNAs in the plant Arabidopsis thaliana.Citation102 Most importantly, however, LSm1–7 is thought to trigger the decapping of 3′-oligoadenylated mRNA decay intermediates, which result from mRNA deadenylation by the PAN2-PAN3 and CCR4-NOT complexes in the mRNA 5′-3′ decay pathway.Citation103-Citation105 Such decay intermediates can be recognized directly by the LSm1–7/Pat1 complex, as suggested by in vitro analysis.Citation106 Furthermore, very recent data suggest that oligoadenylated mRNA decay intermediates may get oligouridinylated quite frequently also in mammalian cells,Citation107 providing an additional explanation for the association of the LSm1–7 ring with these RNA substrates.
Structurally, the LSm1–7 ring is highly similar to the LSm2–8 ring, with an rmsd of 0.89 Å for the entire ring (461 Cα atoms) and an rmsd of 0.97 Å for the alternative protomers LSm1 and LSm8 (54 Cα atoms).Citation85 Similar to the C-terminal extension of LSm8, the LSm1 C-terminal extension is positioned to obstruct the pore on the distal side of the ring, probably preventing an RNA 3′-end from exiting on this side.Citation84 In the case of LSm1, the C-terminal extension forms a long helix that lies across the pore, reaching out for the distal surface of the LSm6 protomer ().Citation84,Citation85 The nucleotide binding pocket of LSm1 is intact and, in contrast to LSm8, even contains an aromatic π-stacking partner in the upper position again (). Consequently, both the LSm1–7 and the LSm2–8 ring present an arc of six putative nucleotide binding pockets that starts with LSm7 and ends with LSm6, with the notable exception of LSm5 that lacks the otherwise highly conserved arginine in the lower stacking position (R63 in AfSm1, see above and ). It is therefore surprising that LSm1–7 binds octauridine (5′-UUUUUUUU-3′) with an affinity of only ~2 μM, i.e., ~100-fold less well than the LSm2–8 ring.Citation85
Although at present one can only speculate about the cause of the reduced affinity, an increased difficulty to accommodate the phosphoribose backbone in the constricted space of the pore might be a possible explanation. It is possible that even minor differences in the relative orientation of the nucleotide binding pockets have important effects. More likely, however, residues inside the pore, like the bulky E107 of LSm1 (S59 in LSm8, D65 in AfSm1, ) might obstruct the path of the RNA backbone, promoting it to loop out of the LSm ring and preventing the consecutive interaction of more than two to three nucleotides with their pockets. Such a scenario is feasible as illustrated by the crystal structure of the EcHfq ring with 5′-GUUUUUA-3′ RNA ().Citation30 This structure also provides an example of how an adenine can be accommodated in the 3′-terminal position, and it demonstrates how an upstream uridine can help to stabilize the RNA backbone of the terminal nucleotides by a hydrogen bond from its N3 position.
The described scenario therefore would not only account for the lower affinity of the LSm1–7 ring for uridine-rich RNA 3′-ends,Citation85 but it also suggests how the LSm1–7 ring could recognize 3′-ends on oligoadenylated mRNA.Citation106 In this case, the ring would simultaneously bind two or three upstream uridines, as are frequently found near the end of mRNA 3′-UTRs,Citation108 and one to three 3′-terminal adenines that could fit the nucleotide binding pockets reasonably well. The intermediate nucleotides would be looped out like the single uridine in the EcHfq structure (), increasing the difficulty to bind both elements simultaneously with an increasing number of adenines in the loop. Like this, the LSm1–7 ring could sense and protect the 3′-end of the RNA molecule and distinguish oligoadenylated from polyadenylated substrates, which in vitro are known to bind less well.Citation106 Additional contributions that increase substrate specificity and compensate for the generally lower RNA affinity of the LSm1–7 ring probably come from Pat1 (). Pat1 associates with the protomers LSm2 and LSm3Citation84,Citation109 and also binds RNA independently.Citation96 Again, it should be fairly easy to test whether the reconstituted LSm1–7 ring still recognizes the 3′-terminal nucleotide on its suggested RNA substrates and what are the contributions of the Pat1 protein.
Final Remarks
The present overview demonstrates that Hfq and the ring-forming (L)Sm proteins share many common principles. These include the common protein fold and the assembly into hexameric and heptameric rings with similar RNA recognition principles on the proximal surface. Hfq and LSm rings are suited particularly well to recognize and bind RNA 3′-ends, combining direct sequence readout with the recognition of the 3′-terminal ribose and RNA backbone distorsion.
However, many rings also acquired highly specialized functions. In eukaryotes, this includes the sequence-specific Sm ring, where the RNA continues through the central pore, but also rare LSm proteins that assemble with the described ones into modified rings of novel function.Citation8 In archaea, even the most basic RNA binding functions of the LSm rings remain to be explored, whereas bacterial Hfq rings are probably the functionally most diverse members of the family.Citation12-Citation21 In many species, Hfq has become a central player in riboregulation and has evolved additional RNA binding surfaces on the distal and lateral surfaces of the ring. It remains an important challenge to determine precisely how Hfq recognizes its sRNA partners in these species. Furthermore, the interactions of Hfq and of the other (L)Sm rings with additional, modulating protein partners like RNaseE and Pat1 is just at the beginning to be understood in molecular terms.
Clearly, there still is much to be learned on how these tiny rings express their power to rule the RNA world.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
I am very grateful to Evelyn Sauer and I thank Regina Büttner for their scientific contributions to our work on Hfq over the past 6 y. I also thank Jörg Vogel for getting us started on the topic in the context of the DFG Priority Program SPP1258 (Sensory and Regulatory RNAs in Prokaryotes). Scientific exchange and funding via the DFG SPP1258 Priority Program is gratefully acknowledged. I also thank the members of the group and of the department for discussions as well as Elisa Izaurralde and the Max Planck Society for continued support.
References
- Notman DD, Kurata N, Tan EM. Profiles of antinuclear antibodies in systemic rheumatic diseases. Ann Intern Med 1975; 83:464 - 9; http://dx.doi.org/10.7326/0003-4819-83-4-464; PMID: 1080976
- Lerner MR, Steitz JA. Antibodies to small nuclear RNAs complexed with proteins are produced by patients with systemic lupus erythematosus. Proc Natl Acad Sci U S A 1979; 76:5495 - 9; http://dx.doi.org/10.1073/pnas.76.11.5495; PMID: 316537
- Lührmann R, Kastner B, Bach M. Structure of spliceosomal snRNPs and their role in pre-mRNA splicing. Biochim Biophys Acta 1990; 1087:265 - 92; http://dx.doi.org/10.1016/0167-4781(90)90001-I; PMID: 2147394
- Liautard JP, Sri-Widada J, Brunel C, Jeanteur P. Structural organization of ribonucleoproteins containing small nuclear RNAs from HeLa cells. Proteins interact closely with a similar structural domain of U1, U2, U4 and U5 small nuclear RNAs. J Mol Biol 1982; 162:623 - 43; http://dx.doi.org/10.1016/0022-2836(82)90392-8; PMID: 6187926
- Kambach C, Walke S, Young R, Avis JM, de la Fortelle E, Raker VA, Lührmann R, Li J, Nagai K. Crystal structures of two Sm protein complexes and their implications for the assembly of the spliceosomal snRNPs. Cell 1999; 96:375 - 87; http://dx.doi.org/10.1016/S0092-8674(00)80550-4; PMID: 10025403
- Albrecht M, Lengauer T. Novel Sm-like proteins with long C-terminal tails and associated methyltransferases. FEBS Lett 2004; 569:18 - 26; http://dx.doi.org/10.1016/j.febslet.2004.03.126; PMID: 15225602
- Anantharaman V, Aravind L. Novel conserved domains in proteins with predicted roles in eukaryotic cell-cycle regulation, decapping and RNA stability. BMC Genomics 2004; 5:45; http://dx.doi.org/10.1186/1471-2164-5-45; PMID: 15257761
- Wilusz CJ, Wilusz J. Eukaryotic Lsm proteins: lessons from bacteria. Nat Struct Mol Biol 2005; 12:1031 - 6; http://dx.doi.org/10.1038/nsmb1037; PMID: 16327775
- Törö I, Thore S, Mayer C, Basquin J, Séraphin B, Suck D. RNA binding in an Sm core domain: X-ray structure and functional analysis of an archaeal Sm protein complex. EMBO J 2001; 20:2293 - 303; http://dx.doi.org/10.1093/emboj/20.9.2293; PMID: 11331594
- Tritschler F, Eulalio A, Truffault V, Hartmann MD, Helms S, Schmidt S, Coles M, Izaurralde E, Weichenrieder O. A divergent Sm fold in EDC3 proteins mediates DCP1 binding and P-body targeting. Mol Cell Biol 2007; 27:8600 - 11; http://dx.doi.org/10.1128/MCB.01506-07; PMID: 17923697
- Franze de Fernandez MT, Eoyang L, August JT. Factor fraction required for the synthesis of bacteriophage Qbeta-RNA. Nature 1968; 219:588 - 90; http://dx.doi.org/10.1038/219588a0; PMID: 4874917
- Bandyra KJ, Luisi BF. Licensing and due process in the turnover of bacterial RNA. RNA Biol 2013; 10:627 - 35; http://dx.doi.org/10.4161/rna.24393; PMID: 23580162
- De Lay N, Schu DJ, Gottesman S. Bacterial small RNA-based negative regulation: Hfq and its accomplices. J Biol Chem 2013; 288:7996 - 8003; http://dx.doi.org/10.1074/jbc.R112.441386; PMID: 23362267
- Gottesman S, Storz G. Bacterial small RNA regulators: versatile roles and rapidly evolving variations. Cold Spring Harb Perspect Biol 2011; 3:3; http://dx.doi.org/10.1101/cshperspect.a003798; PMID: 20980440
- Mura C, Randolph PS, Patterson J, Cozen AE. Archaeal and eukaryotic homologs of Hfq: A structural and evolutionary perspective on Sm function. RNA Biol 2013; 10:636 - 51; http://dx.doi.org/10.4161/rna.24538; PMID: 23579284
- Régnier P, Hajnsdorf E. The interplay of Hfq, poly(A) polymerase I and exoribonucleases at the 3′ ends of RNAs resulting from Rho-independent termination: A tentative model. RNA Biol 2013; 10:602 - 9; http://dx.doi.org/10.4161/rna.23664; PMID: 23392248
- Sauer E. Special focus Hfq. RNA Biol 2013; 10:590 - 1; http://dx.doi.org/10.4161/rna.24617; PMID: 23774896
- Sauer E. Structure and RNA-binding properties of the bacterial LSm protein Hfq. RNA Biol 2013; 10:610 - 8; http://dx.doi.org/10.4161/rna.24201; PMID: 23535768
- Vogel J, Luisi BF. Hfq and its constellation of RNA. Nat Rev Microbiol 2011; 9:578 - 89; http://dx.doi.org/10.1038/nrmicro2615; PMID: 21760622
- Wagner EG. Cycling of RNAs on Hfq. RNA Biol 2013; 10:619 - 26; http://dx.doi.org/10.4161/rna.24044; PMID: 23466677
- Wilusz CJ, Wilusz J. Lsm proteins and Hfq: Life at the 3′ end. RNA Biol 2013; 10:592 - 601; http://dx.doi.org/10.4161/rna.23695; PMID: 23392247
- Møller T, Franch T, Højrup P, Keene DR, Bächinger HP, Brennan RG, Valentin-Hansen P. Hfq: a bacterial Sm-like protein that mediates RNA-RNA interaction. Mol Cell 2002; 9:23 - 30; http://dx.doi.org/10.1016/S1097-2765(01)00436-1; PMID: 11804583
- Zhang A, Wassarman KM, Ortega J, Steven AC, Storz G. The Sm-like Hfq protein increases OxyS RNA interaction with target mRNAs. Mol Cell 2002; 9:11 - 22; http://dx.doi.org/10.1016/S1097-2765(01)00437-3; PMID: 11804582
- Hermann H, Fabrizio P, Raker VA, Foulaki K, Hornig H, Brahms H, Lührmann R. snRNP Sm proteins share two evolutionarily conserved sequence motifs which are involved in Sm protein-protein interactions. EMBO J 1995; 14:2076 - 88; PMID: 7744013
- Sauter C, Basquin J, Suck D. Sm-like proteins in Eubacteria: the crystal structure of the Hfq protein from Escherichia coli. Nucleic Acids Res 2003; 31:4091 - 8; http://dx.doi.org/10.1093/nar/gkg480; PMID: 12853626
- Schumacher MA, Pearson RF, Møller T, Valentin-Hansen P, Brennan RG. Structures of the pleiotropic translational regulator Hfq and an Hfq-RNA complex: a bacterial Sm-like protein. EMBO J 2002; 21:3546 - 56; http://dx.doi.org/10.1093/emboj/cdf322; PMID: 12093755
- Murina VN, Nikulin AD. RNA-binding Sm-like proteins of bacteria and archaea. similarity and difference in structure and function. Biochemistry (Mosc) 2011; 76:1434 - 49; http://dx.doi.org/10.1134/S0006297911130050; PMID: 22339597
- Murina V, Lekontseva N, Nikulin A. Hfq binds ribonucleotides in three different RNA-binding sites. Acta Crystallogr D Biol Crystallogr 2013; 69:1504 - 13; http://dx.doi.org/10.1107/S090744491301010X; PMID: 23897473
- Sauer E, Weichenrieder O. Structural basis for RNA 3′-end recognition by Hfq. Proc Natl Acad Sci U S A 2011; 108:13065 - 70; http://dx.doi.org/10.1073/pnas.1103420108; PMID: 21737752
- Wang W, Wang L, Zou Y, Zhang J, Gong Q, Wu J, Shi Y. Cooperation of Escherichia coli Hfq hexamers in DsrA binding. Genes Dev 2011; 25:2106 - 17; http://dx.doi.org/10.1101/gad.16746011; PMID: 21979921
- Otaka H, Ishikawa H, Morita T, Aiba H. PolyU tail of rho-independent terminator of bacterial small RNAs is essential for Hfq action. Proc Natl Acad Sci U S A 2011; 108:13059 - 64; http://dx.doi.org/10.1073/pnas.1107050108; PMID: 21788484
- Wilson KS, von Hippel PH. Transcription termination at intrinsic terminators: the role of the RNA hairpin. Proc Natl Acad Sci U S A 1995; 92:8793 - 7; http://dx.doi.org/10.1073/pnas.92.19.8793; PMID: 7568019
- de Haseth PL, Uhlenbeck OC. Interaction of Escherichia coli host factor protein with oligoriboadenylates. Biochemistry 1980; 19:6138 - 46; http://dx.doi.org/10.1021/bi00567a029; PMID: 6162476
- Folichon M, Arluison V, Pellegrini O, Huntzinger E, Régnier P, Hajnsdorf E. The poly(A) binding protein Hfq protects RNA from RNase E and exoribonucleolytic degradation. Nucleic Acids Res 2003; 31:7302 - 10; http://dx.doi.org/10.1093/nar/gkg915; PMID: 14654705
- Le Derout J, Folichon M, Briani F, Dehò G, Régnier P, Hajnsdorf E. Hfq affects the length and the frequency of short oligo(A) tails at the 3′ end of Escherichia coli rpsO mRNAs. Nucleic Acids Res 2003; 31:4017 - 23; http://dx.doi.org/10.1093/nar/gkg456; PMID: 12853618
- Mikulecky PJ, Kaw MK, Brescia CC, Takach JC, Sledjeski DD, Feig AL. Escherichia coli Hfq has distinct interaction surfaces for DsrA, rpoS and poly(A) RNAs. Nat Struct Mol Biol 2004; 11:1206 - 14; http://dx.doi.org/10.1038/nsmb858; PMID: 15531892
- Horstmann N, Orans J, Valentin-Hansen P, Shelburne SA 3rd, Brennan RG. Structural mechanism of Staphylococcus aureus Hfq binding to an RNA A-tract. Nucleic Acids Res 2012; 40:11023 - 35; http://dx.doi.org/10.1093/nar/gks809; PMID: 22965117
- Link TM, Valentin-Hansen P, Brennan RG. Structure of Escherichia coli Hfq bound to polyriboadenylate RNA. Proc Natl Acad Sci U S A 2009; 106:19292 - 7; http://dx.doi.org/10.1073/pnas.0908744106; PMID: 19889981
- Someya T, Baba S, Fujimoto M, Kawai G, Kumasaka T, Nakamura K. Crystal structure of Hfq from Bacillus subtilis in complex with SELEX-derived RNA aptamer: insight into RNA-binding properties of bacterial Hfq. Nucleic Acids Res 2012; 40:1856 - 67; http://dx.doi.org/10.1093/nar/gkr892; PMID: 22053080
- Hämmerle H, Beich-Frandsen M, Večerek B, Rajkowitsch L, Carugo O, Djinović-Carugo K, Bläsi U. Structural and biochemical studies on ATP binding and hydrolysis by the Escherichia coli RNA chaperone Hfq. PLoS One 2012; 7:e50892; http://dx.doi.org/10.1371/journal.pone.0050892; PMID: 23226421
- Robinson KE, Orans J, Kovach AR, Link TM, Brennan RG. Mapping Hfq-RNA interaction surfaces using tryptophan fluorescence quenching. Nucleic Acids Res 2014; 42:2736 - 49; PMID: 24288369
- Sauer E, Schmidt S, Weichenrieder O. Small RNA binding to the lateral surface of Hfq hexamers and structural rearrangements upon mRNA target recognition. Proc Natl Acad Sci U S A 2012; 109:9396 - 401; http://dx.doi.org/10.1073/pnas.1202521109; PMID: 22645344
- Balbontín R, Fiorini F, Figueroa-Bossi N, Casadesús J, Bossi L. Recognition of heptameric seed sequence underlies multi-target regulation by RybB small RNA in Salmonella enterica. Mol Microbiol 2010; 78:380 - 94; http://dx.doi.org/10.1111/j.1365-2958.2010.07342.x; PMID: 20979336
- Papenfort K, Bouvier M, Mika F, Sharma CM, Vogel J. Evidence for an autonomous 5′ target recognition domain in an Hfq-associated small RNA. Proc Natl Acad Sci U S A 2010; 107:20435 - 40; http://dx.doi.org/10.1073/pnas.1009784107; PMID: 21059903
- Ishikawa H, Otaka H, Maki K, Morita T, Aiba H. The functional Hfq-binding module of bacterial sRNAs consists of a double or single hairpin preceded by a U-rich sequence and followed by a 3′ poly(U) tail. RNA 2012; 18:1062 - 74; http://dx.doi.org/10.1261/rna.031575.111; PMID: 22454537
- Olejniczak M. Despite similar binding to the Hfq protein regulatory RNAs widely differ in their competition performance. Biochemistry 2011; 50:4427 - 40; http://dx.doi.org/10.1021/bi102043f; PMID: 21510661
- Sittka A, Lucchini S, Papenfort K, Sharma CM, Rolle K, Binnewies TT, Hinton JC, Vogel J. Deep sequencing analysis of small noncoding RNA and mRNA targets of the global post-transcriptional regulator, Hfq. PLoS Genet 2008; 4:e1000163; http://dx.doi.org/10.1371/journal.pgen.1000163; PMID: 18725932
- Sittka A, Sharma CM, Rolle K, Vogel J. Deep sequencing of Salmonella RNA associated with heterologous Hfq proteins in vivo reveals small RNAs as a major target class and identifies RNA processing phenotypes. RNA Biol 2009; 6:266 - 75; http://dx.doi.org/10.4161/rna.6.3.8332; PMID: 19333007
- Zhang A, Wassarman KM, Rosenow C, Tjaden BC, Storz G, Gottesman S. Global analysis of small RNA and mRNA targets of Hfq. Mol Microbiol 2003; 50:1111 - 24; http://dx.doi.org/10.1046/j.1365-2958.2003.03734.x; PMID: 14622403
- Andrade JM, Pobre V, Matos AM, Arraiano CM. The crucial role of PNPase in the degradation of small RNAs that are not associated with Hfq. RNA 2012; 18:844 - 55; http://dx.doi.org/10.1261/rna.029413.111; PMID: 22355164
- Moll I, Afonyushkin T, Vytvytska O, Kaberdin VR, Bläsi U. Coincident Hfq binding and RNase E cleavage sites on mRNA and small regulatory RNAs. RNA 2003; 9:1308 - 14; http://dx.doi.org/10.1261/rna.5850703; PMID: 14561880
- Urban JH, Vogel J. Translational control and target recognition by Escherichia coli small RNAs in vivo. Nucleic Acids Res 2007; 35:1018 - 37; http://dx.doi.org/10.1093/nar/gkl1040; PMID: 17264113
- Thore S, Mayer C, Sauter C, Weeks S, Suck D. Crystal structures of the Pyrococcus abyssi Sm core and its complex with RNA. Common features of RNA binding in archaea and eukarya. J Biol Chem 2003; 278:1239 - 47; http://dx.doi.org/10.1074/jbc.M207685200; PMID: 12409299
- Panja S, Schu DJ, Woodson SA. Conserved arginines on the rim of Hfq catalyze base pair formation and exchange. Nucleic Acids Res 2013; 41:7536 - 46; http://dx.doi.org/10.1093/nar/gkt521; PMID: 23771143
- de Haseth PL, Uhlenbeck OC. Interaction of Escherichia coli host factor protein with Q β ribonucleic acid. Biochemistry 1980; 19:6146 - 51; http://dx.doi.org/10.1021/bi00567a030; PMID: 6162477
- Folichon M, Allemand F, Régnier P, Hajnsdorf E. Stimulation of poly(A) synthesis by Escherichia coli poly(A)polymerase I is correlated with Hfq binding to poly(A) tails. FEBS J 2005; 272:454 - 63; http://dx.doi.org/10.1111/j.1742-4658.2004.04485.x; PMID: 15654883
- Mohanty BK, Maples VF, Kushner SR. The Sm-like protein Hfq regulates polyadenylation dependent mRNA decay in Escherichia coli. Mol Microbiol 2004; 54:905 - 20; http://dx.doi.org/10.1111/j.1365-2958.2004.04337.x; PMID: 15522076
- De Lay N, Gottesman S. Role of polynucleotide phosphorylase in sRNA function in Escherichia coli. RNA 2011; 17:1172 - 89; http://dx.doi.org/10.1261/rna.2531211; PMID: 21527671
- Ikeda Y, Yagi M, Morita T, Aiba H. Hfq binding at RhlB-recognition region of RNase E is crucial for the rapid degradation of target mRNAs mediated by sRNAs in Escherichia coli. Mol Microbiol 2011; 79:419 - 32; http://dx.doi.org/10.1111/j.1365-2958.2010.07454.x; PMID: 21219461
- Morita T, Maki K, Aiba H. RNase E-based ribonucleoprotein complexes: mechanical basis of mRNA destabilization mediated by bacterial noncoding RNAs. Genes Dev 2005; 19:2176 - 86; http://dx.doi.org/10.1101/gad.1330405; PMID: 16166379
- Prévost K, Desnoyers G, Jacques JF, Lavoie F, Massé E. Small RNA-induced mRNA degradation achieved through both translation block and activated cleavage. Genes Dev 2011; 25:385 - 96; http://dx.doi.org/10.1101/gad.2001711; PMID: 21289064
- Fender A, Elf J, Hampel K, Zimmermann B, Wagner EG. RNAs actively cycle on the Sm-like protein Hfq. Genes Dev 2010; 24:2621 - 6; http://dx.doi.org/10.1101/gad.591310; PMID: 21123649
- Hussein R, Lim HN. Disruption of small RNA signaling caused by competition for Hfq. Proc Natl Acad Sci U S A 2011; 108:1110 - 5; http://dx.doi.org/10.1073/pnas.1010082108; PMID: 21189298
- Moon K, Gottesman S. Competition among Hfq-binding small RNAs in Escherichia coli. Mol Microbiol 2011; 82:1545 - 62; http://dx.doi.org/10.1111/j.1365-2958.2011.07907.x; PMID: 22040174
- Geissmann TA, Touati D. Hfq, a new chaperoning role: binding to messenger RNA determines access for small RNA regulator. EMBO J 2004; 23:396 - 405; http://dx.doi.org/10.1038/sj.emboj.7600058; PMID: 14739933
- Moll I, Leitsch D, Steinhauser T, Bläsi U. RNA chaperone activity of the Sm-like Hfq protein. EMBO Rep 2003; 4:284 - 9; http://dx.doi.org/10.1038/sj.embor.embor772; PMID: 12634847
- Rajkowitsch L, Chen D, Stampfl S, Semrad K, Waldsich C, Mayer O, Jantsch MF, Konrat R, Bläsi U, Schroeder R. RNA chaperones, RNA annealers and RNA helicases. RNA Biol 2007; 4:118 - 30; http://dx.doi.org/10.4161/rna.4.3.5445; PMID: 18347437
- Rajkowitsch L, Schroeder R. Dissecting RNA chaperone activity. RNA 2007; 13:2053 - 60; http://dx.doi.org/10.1261/rna.671807; PMID: 17901153
- Peng Y, Soper TJ, Woodson SA. Positional effects of AAN motifs in rpoS regulation by sRNAs and Hfq. J Mol Biol 2014; 426:275 - 85; http://dx.doi.org/10.1016/j.jmb.2013.08.026; PMID: 24051417
- Bandyra KJ, Said N, Pfeiffer V, Górna MW, Vogel J, Luisi BF. The seed region of a small RNA drives the controlled destruction of the target mRNA by the endoribonuclease RNase E. Mol Cell 2012; 47:943 - 53; http://dx.doi.org/10.1016/j.molcel.2012.07.015; PMID: 22902561
- Lorenz C, Gesell T, Zimmermann B, Schoeberl U, Bilusic I, Rajkowitsch L, Waldsich C, von Haeseler A, Schroeder R. Genomic SELEX for Hfq-binding RNAs identifies genomic aptamers predominantly in antisense transcripts. Nucleic Acids Res 2010; 38:3794 - 808; http://dx.doi.org/10.1093/nar/gkq032; PMID: 20348540
- Zhang A, Schu DJ, Tjaden BC, Storz G, Gottesman S. Mutations in interaction surfaces differentially impact E. coli Hfq association with small RNAs and their mRNA targets. J Mol Biol 2013; 425:3678 - 97; http://dx.doi.org/10.1016/j.jmb.2013.01.006; PMID: 23318956
- Henderson CA, Vincent HA, Casamento A, Stone CM, Phillips JO, Cary PD, Sobott F, Gowers DM, Taylor JE, Callaghan AJ. Hfq binding changes the structure of Escherichia coli small noncoding RNAs OxyS and RprA, which are involved in the riboregulation of rpoS. RNA 2013; 19:1089 - 104; http://dx.doi.org/10.1261/rna.034595.112; PMID: 23804244
- Vincent HA, Henderson CA, Stone CM, Cary PD, Gowers DM, Sobott F, Taylor JE, Callaghan AJ. The low-resolution solution structure of Vibrio cholerae Hfq in complex with Qrr1 sRNA. Nucleic Acids Res 2012; 40:8698 - 710; http://dx.doi.org/10.1093/nar/gks582; PMID: 22730296
- Beich-Frandsen M, Večerek B, Konarev PV, Sjöblom B, Kloiber K, Hämmerle H, Rajkowitsch L, Miles AJ, Kontaxis G, Wallace BA, et al. Structural insights into the dynamics and function of the C-terminus of the E. coli RNA chaperone Hfq. Nucleic Acids Res 2011; 39:4900 - 15; http://dx.doi.org/10.1093/nar/gkq1346; PMID: 21330354
- Olsen AS, Møller-Jensen J, Brennan RG, Valentin-Hansen P. C-terminally truncated derivatives of Escherichia coli Hfq are proficient in riboregulation. J Mol Biol 2010; 404:173 - 82; http://dx.doi.org/10.1016/j.jmb.2010.09.038; PMID: 20888338
- Jousselin A, Metzinger L, Felden B. On the facultative requirement of the bacterial RNA chaperone, Hfq. Trends Microbiol 2009; 17:399 - 405; http://dx.doi.org/10.1016/j.tim.2009.06.003; PMID: 19733080
- Tomasini A, François P, Howden BP, Fechter P, Romby P, Caldelari I. The importance of regulatory RNAs in Staphylococcus aureus. Infect Genet Evol 2014; 21:616 - 26; http://dx.doi.org/10.1016/j.meegid.2013.11.016; PMID: 24291227
- Bøggild A, Overgaard M, Valentin-Hansen P, Brodersen DE. Cyanobacteria contain a structural homologue of the Hfq protein with altered RNA-binding properties. FEBS J 2009; 276:3904 - 15; http://dx.doi.org/10.1111/j.1742-4658.2009.07104.x; PMID: 19777643
- Chao Y, Vogel J. The role of Hfq in bacterial pathogens. Curr Opin Microbiol 2010; 13:24 - 33; http://dx.doi.org/10.1016/j.mib.2010.01.001; PMID: 20080057
- Sharma CM, Hoffmann S, Darfeuille F, Reignier J, Findeiss S, Sittka A, Chabas S, Reiche K, Hackermüller J, Reinhardt R, et al. The primary transcriptome of the major human pathogen Helicobacter pylori. Nature 2010; 464:250 - 5; http://dx.doi.org/10.1038/nature08756; PMID: 20164839
- Tsui HC, Leung HC, Winkler ME. Characterization of broadly pleiotropic phenotypes caused by an hfq insertion mutation in Escherichia coli K-12. Mol Microbiol 1994; 13:35 - 49; http://dx.doi.org/10.1111/j.1365-2958.1994.tb00400.x; PMID: 7984093
- Göpel Y, Papenfort K, Reichenbach B, Vogel J, Görke B. Targeted decay of a regulatory small RNA by an adaptor protein for RNase E and counteraction by an anti-adaptor RNA. Genes Dev 2013; 27:552 - 64; http://dx.doi.org/10.1101/gad.210112.112; PMID: 23475961
- Sharif H, Conti E. Architecture of the Lsm1-7-Pat1 complex: a conserved assembly in eukaryotic mRNA turnover. Cell Rep 2013; 5:283 - 91; http://dx.doi.org/10.1016/j.celrep.2013.10.004; PMID: 24139796
- Zhou L, Zhou Y, Hang J, Wan R, Lu G, Yan C, Shi Y. Crystal structure and biochemical analysis of the heptameric Lsm1-7 complex. Cell Res 2014; 24:497 - 500; http://dx.doi.org/10.1038/cr.2014.18; PMID: 24513854
- Zhou L, Hang J, Zhou Y, Wan R, Lu G, Yin P, Yan C, Shi Y. Crystal structures of the Lsm complex bound to the 3′ end sequence of U6 small nuclear RNA. Nature 2014; 506:116 - 20; http://dx.doi.org/10.1038/nature12803; PMID: 24240276
- Leung AK, Nagai K, Li J. Structure of the spliceosomal U4 snRNP core domain and its implication for snRNP biogenesis. Nature 2011; 473:536 - 9; http://dx.doi.org/10.1038/nature09956; PMID: 21516107
- Pomeranz Krummel DA, Oubridge C, Leung AK, Li J, Nagai K. Crystal structure of human spliceosomal U1 snRNP at 5.5 A resolution. Nature 2009; 458:475 - 80; http://dx.doi.org/10.1038/nature07851; PMID: 19325628
- Weber G, Trowitzsch S, Kastner B, Lührmann R, Wahl MC. Functional organization of the Sm core in the crystal structure of human U1 snRNP. EMBO J 2010; 29:4172 - 84; http://dx.doi.org/10.1038/emboj.2010.295; PMID: 21113136
- Meister G, Eggert C, Fischer U. SMN-mediated assembly of RNPs: a complex story. Trends Cell Biol 2002; 12:472 - 8; http://dx.doi.org/10.1016/S0962-8924(02)02371-1; PMID: 12441251
- Pellizzoni L, Yong J, Dreyfuss G. Essential role for the SMN complex in the specificity of snRNP assembly. Science 2002; 298:1775 - 9; http://dx.doi.org/10.1126/science.1074962; PMID: 12459587
- Zaric B, Chami M, Rémigy H, Engel A, Ballmer-Hofer K, Winkler FK, Kambach C. Reconstitution of two recombinant LSm protein complexes reveals aspects of their architecture, assembly, and function. J Biol Chem 2005; 280:16066 - 75; http://dx.doi.org/10.1074/jbc.M414481200; PMID: 15711010
- Licht K, Medenbach J, Lührmann R, Kambach C, Bindereif A. 3′-cyclic phosphorylation of U6 snRNA leads to recruitment of recycling factor p110 through LSm proteins. RNA 2008; 14:1532 - 8; http://dx.doi.org/10.1261/rna.1129608; PMID: 18567812
- Tharun S, He W, Mayes AE, Lennertz P, Beggs JD, Parker R. Yeast Sm-like proteins function in mRNA decapping and decay. Nature 2000; 404:515 - 8; http://dx.doi.org/10.1038/35006676; PMID: 10761922
- Tharun S, Parker R. Targeting an mRNA for decapping: displacement of translation factors and association of the Lsm1p-7p complex on deadenylated yeast mRNAs. Mol Cell 2001; 8:1075 - 83; http://dx.doi.org/10.1016/S1097-2765(01)00395-1; PMID: 11741542
- Braun JE, Tritschler F, Haas G, Igreja C, Truffault V, Weichenrieder O, Izaurralde E. The C-terminal α-α superhelix of Pat is required for mRNA decapping in metazoa. EMBO J 2010; 29:2368 - 80; http://dx.doi.org/10.1038/emboj.2010.124; PMID: 20543818
- He W, Parker R. The yeast cytoplasmic LsmI/Pat1p complex protects mRNA 3′ termini from partial degradation. Genetics 2001; 158:1445 - 55; PMID: 11514438
- Song MG, Kiledjian M. 3′ Terminal oligo U-tract-mediated stimulation of decapping. RNA 2007; 13:2356 - 65; http://dx.doi.org/10.1261/rna.765807; PMID: 17942740
- Wilusz CJ, Wilusz J. New ways to meet your (3′) end oligouridylation as a step on the path to destruction. Genes Dev 2008; 22:1 - 7; http://dx.doi.org/10.1101/gad.1634508; PMID: 18172159
- Mullen TE, Marzluff WF. Degradation of histone mRNA requires oligouridylation followed by decapping and simultaneous degradation of the mRNA both 5′ to 3′ and 3′ to 5′. Genes Dev 2008; 22:50 - 65; http://dx.doi.org/10.1101/gad.1622708; PMID: 18172165
- Rissland OS, Norbury CJ. Decapping is preceded by 3′ uridylation in a novel pathway of bulk mRNA turnover. Nat Struct Mol Biol 2009; 16:616 - 23; http://dx.doi.org/10.1038/nsmb.1601; PMID: 19430462
- Sement FM, Ferrier E, Zuber H, Merret R, Alioua M, Deragon JM, Bousquet-Antonelli C, Lange H, Gagliardi D. Uridylation prevents 3′ trimming of oligoadenylated mRNAs. Nucleic Acids Res 2013; 41:7115 - 27; http://dx.doi.org/10.1093/nar/gkt465; PMID: 23748567
- Jonas S, Izaurralde E. The role of disordered protein regions in the assembly of decapping complexes and RNP granules. Genes Dev 2013; 27:2628 - 41; http://dx.doi.org/10.1101/gad.227843.113; PMID: 24352420
- Tharun S. Lsm1-7-Pat1 complex: a link between 3′ and 5′-ends in mRNA decay?. RNA Biol 2009; 6:228 - 32; http://dx.doi.org/10.4161/rna.6.3.8282; PMID: 19279404
- Wahle E, Winkler GS. RNA decay machines: deadenylation by the Ccr4-not and Pan2-Pan3 complexes. Biochim Biophys Acta 2013; 1829:561 - 70; http://dx.doi.org/10.1016/j.bbagrm.2013.01.003; PMID: 23337855
- Chowdhury A, Mukhopadhyay J, Tharun S. The decapping activator Lsm1p-7p-Pat1p complex has the intrinsic ability to distinguish between oligoadenylated and polyadenylated RNAs. RNA 2007; 13:998 - 1016; http://dx.doi.org/10.1261/rna.502507; PMID: 17513695
- Chang H, Lim J, Ha M, Kim VN. TAIL-seq: genome-wide determination of poly(A) tail length and 3′ end modifications. Mol Cell 2014; 53:1044 - 52; http://dx.doi.org/10.1016/j.molcel.2014.02.007; PMID: 24582499
- Tian B, Graber JH. Signals for pre-mRNA cleavage and polyadenylation. Wiley Interdiscip Rev RNA 2012; 3:385 - 96; http://dx.doi.org/10.1002/wrna.116; PMID: 22012871
- Wu D, Muhlrad D, Bowler MW, Jiang S, Liu Z, Parker R, Song H. Lsm2 and Lsm3 bridge the interaction of the Lsm1-7 complex with Pat1 for decapping activation. Cell Res 2014; 24:233 - 46; http://dx.doi.org/10.1038/cr.2013.152; PMID: 24247251
- Gouet P, Robert X, Courcelle E. ESPript/ENDscript: Extracting and rendering sequence and 3D information from atomic structures of proteins. Nucleic Acids Res 2003; 31:3320 - 3; http://dx.doi.org/10.1093/nar/gkg556; PMID: 12824317
- Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr 2010; 66:486 - 501; http://dx.doi.org/10.1107/S0907444910007493; PMID: 20383002
