Abstract
The small RNA AbcR1 regulates the expression of ABC transporters in the plant pathogen Agrobacterium tumefaciens, the plant symbiont Sinorhizobium meliloti, and the human pathogen Brucella abortus. A combination of proteomic and bioinformatic approaches suggested dozens of AbcR1 targets in A. tumefaciens. Several of these newly discovered targets are involved in the uptake of amino acids, their derivatives, and sugars. Among the latter is the periplasmic sugar-binding protein ChvE, a component of the virulence signal transduction system. We examined 16 targets and their interaction with AbcR1 in close detail. In addition to the previously described mRNA interaction site of AbcR1 (M1), the CopraRNA program predicted a second functional module (M2) as target-binding site. Both M1 and M2 contain single-stranded anti-SD motifs. Using mutated AbcR1 variants, we systematically tested by band shift experiments, which sRNA region is responsible for mRNA binding and gene regulation. On the target site, we find that AbcR1 interacts with some mRNAs in the translation initiation region and with others far into their coding sequence. Our data show that AbcR1 is a versatile master regulator of nutrient uptake systems in A. tumefaciens and related bacteria.
Introduction
Within the last decade it has become increasingly clear that small RNAs (sRNAs) are equally efficient and versatile regulators of gene expression as protein-based transcription factors. Most trans-encoded sRNAs act at the post-transcriptional level by base-pairing to target mRNAs and can have a positive or negative effect on gene expression by affecting translation and/or RNA decay.Citation1,Citation2 Small RNAs typically offer only limited complementarity to their targets. A segment of only seven contiguous bases, the so-called seed region, can be sufficient to confer specificity.Citation3-Citation5 Therefore, sRNAs are well suited for regulation of multiple mRNAs. Another level of complexity is reached when a single mRNA is subject to regulation by several sRNAs.Citation6 Overall, this can lead to large sRNA-based regulatory networks that sense and respond to the nutritional status of the cell.Citation7,Citation8
The fundamental importance of sRNAs is reflected by their involvement in numerous cellular processes, like cell division (DicF), transcription (6S RNA), photosynthesis (PcrZ), stress adaption (OxyS), virulence, quorum sensing (Qrr), carbon storage (CsrBC), and phosphosugar metabolism.Citation9-Citation21 A class of genes frequently controlled by sRNAs codes for periplasmic substrate binding proteins of bacterial ABC transporters.Citation7,Citation22-Citation26 This transporter superfamily uses periplasmic solute-binding proteins to take up a wide range of substrates (sugars, amino acids and their derivatives, as well as proteins and drugs).Citation27-Citation29
Most of our knowledge on sRNAs derives from studies with Escherichia coli and Salmonella. However, deep sequencing-assisted approaches have revealed numerous sRNAs in any given bacterium or archaeon.Citation30 Experimental evidence for a regulatory function of these small-sized RNAs has been provided in a limited number of cases, for example, in Bacillus subtilis and other Gram-positives, in cyanobacteria, archaea, Rhodobacter, and Xanthomonas.Citation11,Citation31-Citation35
Genome-wide surveys have recently revealed hundreds of sRNAs in the plant pathogen Agrobacterium tumefaciens.Citation36,Citation37 This bacterium is able to induce tumors (crown galls) upon transfer of a DNA fragment (T-DNA) from its tumor-inducing (Ti) plasmid to the nuclear genome of the host plant.Citation38,Citation39 In the transformed plant cells, expression of T-DNA encoded growth factor genes results in cell proliferation and tumor formation. Additionally, plant metabolism is re-programmed to produce opines serving as carbon and nitrogen source for A. tumefaciens.Citation40,Citation41 Perception of plant-derived signals involves several bacterial factors, including the two-component system VirA/VirG, which mediates the activation of the virulence cascade, and ChvE, a periplasmic substrate binding protein that binds host-derived sugars and plays a role in activation of the virulence cascade (ChvE).Citation42-Citation45 Other putative substrate-binding proteins are involved in attachment (AttC), host defense (Atu2422 and Atu4243), and agrocinopine uptake.Citation46-Citation49 In addition to these specialized functions in plant-microbe interaction, ABC transport systems are required for regular nutrient acquisition in A. tumefaciens like in other free-living bacteria.Citation50
At least three ABC transporters in A. tumefaciens are under negative control of the sRNA AbcR1 (ABC transporter regulator 1).Citation26 Among the targets is atu2422 encoding the binding protein for GABA (γ-amino butyric acid), a plant-derived defense molecule that interferes with quorum sensing in Agrobacterium.Citation47,Citation51 AbcR1 is encoded in an intergenic region in tandem with the related sRNA AbcR2.Citation26 Both are maximally expressed in the late exponential phase. Currently, there is no evidence that AbcR2 plays a regulatory role in A. tumefaciens.
Like Agrobacterium, various Rhizobium species encode numerous sRNAs, including homologs of AbcR1 and AbcR2.Citation52-Citation56 In contrast to Agrobacterium, AbcR1 and AbcR2 in Sinorhizobium are divergently expressed, namely the first is present in exponential phase whereas the second accumulates in stationary phase suggesting that they operate at different conditions.Citation57 The amino acid binding protein LivK was found to be controlled by AbcR1 but not AbcR2.Citation57 Two other ABC transporter genes are negatively regulated by AbcR1 and AbcR2.Citation58 In Brucella abortus, both AbcR1 and AbcR2 seem to have at least some redundant function.Citation59 Microarray analysis revealed about 25 elevated transcripts, several coding for ABC transporters, in the double mutant. At least three of these transcripts can be controlled by AbcR1 or AbcR2 alone. Moreover, only the double mutant but neither single mutant was attenuated in macrophages and in mice. The commonalities and differences in AbcR1-mediated gene regulation in these model organisms certainly warrants further studies to understand the role of this conserved sRNA in a plant pathogen, a plant symbiont, and a human pathogen.
AbcR1 belongs to the large group of Hfq-associated sRNAs.Citation55,Citation59-Citation61 Hfq is an RNA chaperone that facilitates base-pairing between sRNAs and their targets.Citation62,Citation63 About 10 ABC transporter proteins were found to accumulate in an A. tumefaciens ∆hfq mutant and we wondered whether more than the previously identified three targets atu2422, atu1879, and frcC were controlled by AbcR1.Citation26,Citation61 We used a combination of proteomics and bioinformatics approaches to identify numerous new targets of AbcR1. RNA–RNA interactions studies revealed that AbcR1 uses two separate regions to address mRNAs either in the translation initiation region (TIR) or far downstream in the coding region. Our results support the function of AbcR1 as versatile master regulator to control Agrobacterium physiology.
Results
AbcR1 regulates periplasmic binding proteins of several ABC transporters
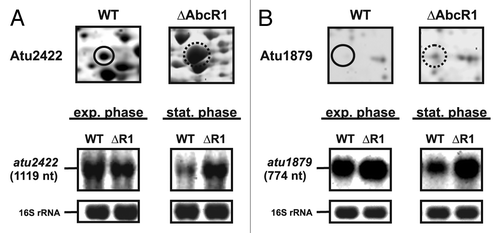
To identify new targets of AbcR1, we compared the proteomes of the marker-less AbcR1 mutant (∆AbcR1) and the wild-type (WT) strain by two-dimensional PAGE. Cultures were grown to stationary phase (OD600 of 1.5) when AbcR1 is maximally expressed.Citation26 Total protein extracts from three biological replicates were subjected to two-dimensional PAGE and the relative protein abundance was visualized by dual-channel images (Fig. S1). Proteins equally abundant in the WT and mutant appear as yellow spots, whereas proteins overrepresented in WT or ∆AbcR1 are green or red, respectively. Overall, 68 proteins were affected by the presence of AbcR1, indicating potential targets of AbcR1 (Table S1). Twenty-five were up and 43 downregulated. Twenty candidates were extracted from the gel, digested with trypsin, and subjected to mass spectrometry (). The presence of the known targets Atu2422 and Atu1879 among them validated this approach. Northern blot experiments revealed that the increased protein levels in the ∆AbcR1 mutant correlated with increased mRNA levels of atu2422 and atu1879 in stationary phase ().
Table 1. Potential AbcR1 targets in A. tumefaciens identified by 2D proteomics
Figure 1. Identification of known AbcR1 targets by 2D-PAGE. Subsections of 2D gels showing Atu2422 (A) and Atu1879 (B) from A. tumefaciens WT (closed black circle) and ∆AbcR1 deletion mutant (dotted black circle) and northern blot analyses of atu2422 (B) and atu1879 (C) transcripts in different growth phases. The WT and the ∆AbcR1 deletion mutant (∆R1) were grown to exponential (OD600: 0.5) or stationary phase (OD600: 1.5) in YEB medium. Eight µg of total RNA were separated on 1.2% denaturing agarose gels. Ethidiumbromide-stained 16S rRNAs were used as loading control.
Validation of eight new AbcR1 targets
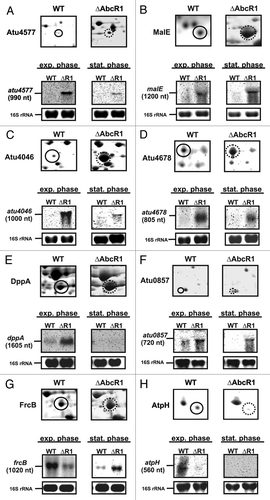
The 18 other AbcR1-dependent proteins were so-far-unknown candidates (Table S1). To recapitulate AbcR1-mediated regulation at the mRNA level, eight of the new candidates were chosen for northern blot analysis with Agrobacterium WT and ∆AbcR1 mutant grown to exponential (OD600 of 0.5) and stationary (OD600 of 1.5) phase. The mRNAs of five periplasmic binding proteins of ABC transporters (Atu4577, MalE, Atu4046, Atu4678, and DppA) showed clear AbcR1-dependent regulation consistent with elevated protein levels in the ∆AbcR1 strain (). The same was true for Atu0857, an annotated oxidoreductase (). The frcB transcript appears to be downregulated by AbcR1 in the exponential growth phase but, consistent with 2D PAGE, upregulated in stationary phase (). Reduced transcript levels of atpH in ∆AbcR1 in exponential growth supported positive regulation by AbcR1 as seen on the protein level (). Transcripts of atpH and dppA () were only detectable in exponential growth phase suggesting that they undergo a rapid turnover in later growth phases.
Figure 2. Validation of new AbcR1 targets. Subsections of 2D gels showing Atu4577 (A), MalE (B), Atu4046 (C), Atu4678 (D), DppA (E), Atu0857 (F), FrcB (G), and AtpH (H) from A. tumefaciens wild-type (closed black circles) and ∆AbcR1 mutant (dotted black circles) and corresponding northern blot analyses of target mRNAs in different growth phases. The wild-type (WT) and the ∆AbcR1 deletion mutant (∆R1) were grown and treated as in .
Overlap between AbcR1- and Hfq-dependent mRNAs
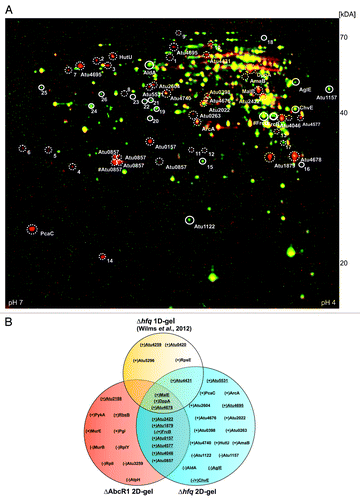
The AbcR1-dependent genes malE, atu4678, and dppA have recently been shown to be affected by Hfq.Citation61 In that study, several proteins overrepresented in the A. tumefaciens hfq mutant were isolated from 1D SDS-PAGE gels and identified as ABC transporters. This led us to assume that a more comprehensive profile of the ∆hfq proteome might reveal additional AbcR1 targets. Upon separation by 2D-SDS-PAGE, 31 putative Hfq-dependent proteins were selected and identified by mass spectrometry (). Among them were many periplasmic binding proteins of ABC transporters (Table S2) and 10 proteins identified as AbcR1 targets were also affected by the absence of Hfq () indicating that AbcR1 acts through Hfq as previously shown for the target Atu2422.Citation61
Figure 3. Altered protein synthesis in the ∆hfq mutant reveals new putative AbcR1 targets. (A) Total protein samples from stationary growth phase (OD600: 1.0) of A. tumefaciens WT and the hfq deletion mutant (∆hfq) were subjected to two-dimensional gel electrophoresis. Proteins overrepresented in WT or in ∆hfq preparations are shown in green or red, respectively. Proteins present in equal amounts in both preparations appear in yellow. Six (green spots) and 22 (red spots) proteins significantly accumulated equally in three replicates of WT (closed white circles) or in ∆hfq (dotted white circles) were analyzed via MALDI-TOF. Altered proteins that were not identified by MALDI-TOF are marked with numbers. #, proteins were identified in multiple spots. (B) Venn diagram comparing altered proteins from different proteomic approaches (∆hfq 1D-gel, ∆hfq 2D-gel, and ∆AbcR1 2D-gel) in A. tumefaciens. ABC transporter components are underlined. (+) or (-) indicate over- or underrepresentation of proteins in deletion mutants.
Validation of six more AbcR1 targets
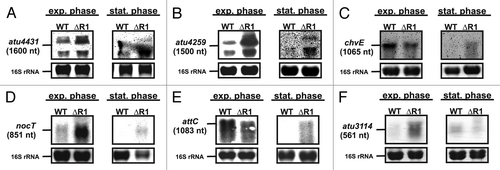
Northern blot experiments with probes against the two Hfq targets atu4431 () and atu4259 confirmed regulation by AbcR1 as their mRNAs accumulated in the sRNA mutant (; note that migration of the very abundant 16S rRNA to a similar position in the gel interferes with detection of the mRNAs and results in two bands).Citation61 One particularly interesting protein affected by Hfq was ChvE, a periplasmic sugar-binding protein involved in host sensing of A. tumefaciens.Citation43,Citation45,Citation64 Its regulatory pattern resembles that of FrcB. Both proteins were less abundant in the hfq deletion strain than in the WT (; Table S2). In contrast to most other AbcR1 targets, but like the frcB transcript (), the chvE mRNA was slightly downregulated in the absence of AbcR1 in exponential phase but clearly upregulated in stationary phase () suggesting growth phase-dependent regulation by AbcR1. Regulation of ChvE by AbcR1 raised our interest in NocT and AttC, substrate binding proteins of putative virulence-related ABC transporters required for the uptake of plant-synthesized nopaline (NocT) or for the transport spermidine and putrescine (AttC).Citation46 They were not detected by 2D PAGE analysis. However, northern blot analysis revealed that they clearly are AbcR1 targets. nocT is a typical negatively controlled AbcR1 target () whereas regulation of attC varies depending on the growth condition (). The final potential AbcR1 target was predicted by the CopraRNA algorithm (Comparative Prediction Algorithm for sRNA Targets, see below).Citation8 Atu3114 was not identified by our proteomics approaches but northern blot analysis showed AbcR1-dependent regulation ().
Figure 4. Verification of additional AbcR1 targets. Northern blot analyses of atu4413 (A), atu4259 (B), chvE (C), nocT (D), attC (E), and atu3114 (F) mRNAs in different growth phases. The wild-type and the ∆AbcR1 deletion mutant (∆R1) were grown to exponential (OD600: 0.5) or stationary phase (OD600: 1.5) in YEB medium. Eight µg of total RNA were separated on 1.2% denaturing agarose gels. Ethidiumbromide-stained 16S rRNAs were used as loading control.
CopraRNA predicts two functional AbcR1 modules and variable target-binding regions
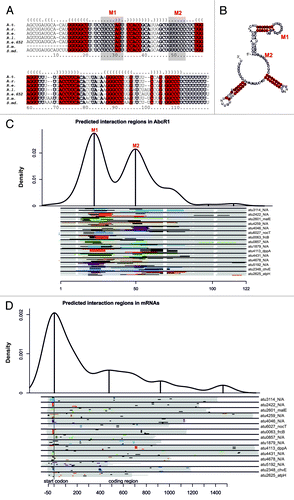
Having identified at least 16 AbcR1-dependent genes, we wondered whether they are all regulated by base pairing of the RBS with the first exposed loop of AbcR1 as documented for atu2422 and S. meliloti livK.Citation26,Citation57 To computationally predict interaction regions between AbcR1 and its target mRNAs, we made use of the recently established CopraRNA program.Citation8 It integrates phylogenetic information to predict sRNA–mRNA interactions on the genomic scale. An alignment of orthologous AbcR1 sequences from A. tumefaciens C58, Agrobacterium radiobacter K84, Rhizobium etli CFN42, Rhizobium leguminosarum bv. vicae, Rhizobium etli 652, Sinorhizobium meliloti 1021, and Sinorhizobium medicae WSM419 revealed long almost identical sequence stretches (). The secondary structures were compared using the ClustalW2 program prior to calculation of a consensus structure with the RNAalifold webserver.Citation65,Citation66 Regions highly conserved in sequence are equally conserved in structure (). Like the experimentally mapped structure of A. tumefaciens AbcR1, the sRNAs fold into three hairpins.Citation26 Apart from the atu2422 interaction site (module 1 = M1), a second conserved single-stranded region (M2) was found between the first and second hairpin. Both regions contain a UCCC motif potentially able to interact with SD-like sequences (). A domain prediction of putatively interacting sites between AbcR1 and 15 of the target mRNAs validated in this study (note that atu4577 could not be used because it is not a conserved gene) suggested that both M1 and M2 are involved in target recognition (, for visualization of detailed AbcR1 M1- and M2-target mRNA interactions in A. tumefaciens see Table S3). The predicted mRNA interaction sites preferentially lie around the SD sequence but several sites are located far into the coding region (; Table S3).
Figure 5. Comparative computational predictions suggest two functional AbcR1 modules. Sequence alignment (A) and consensus structure (B) of AbcR1 in A. tumefaciens C58 (A.t.), A. radiobacter K84 (A.r.), R. etli CFN42 (R.e.), R. leguminosarum bv. vicae (R.l.), R. etli 652 (R.e.652) S. meliloti 1021 (S.m.), and S. medicae WSM419 (S.md.). Sequence conservation is given in bold letters. Nucleotides highly conserved in structure are marked in red. The calculated structure is given in dot-bracket notation above the alignment. Grey shaded boxes (A) or gray marked nucleotides (B) represent regions M1 and M2. Visualization of the predicted binding regions in AbcR1 (C) and the experimentally verified targets of AbcR1 (D). The density plots at the top indicate the relative frequency of sRNA or mRNA nucleotide positions in the predicted AbcR1–target mRNA interactions. The density plots combine all optimal predictions for the 15 conserved verified targets in all included homologs of AbcR1 (C) or target mRNAs (D). Distinct interaction domains are indicated by local maxima marked with upright lines. Below the density plots, schematic alignments of the AbcR1 homologs (C) and the targets (D) are drawn to visualize the predicted optimal and suboptimal interactions for each organism. Each alignment contains eight lines, one for each organism included in the CopraRNA prediction. The order of the organisms in the alignment from top to bottom is: A. tumefaciens C58 (A.t.), A. radiobacter K84 (A.r.), R. etli CFN42 (R.e.), R. leguminosarum bv. vicae (R.l.), R. etli 652 (R.e.652) S. meliloti 1021 (S.m.), and S. medicae WSM419 (S.md.). The aligned regions are colored in gray and the optimal predicted interaction regions are given in different colors (for contrast only). The respective best suboptimal interaction site predictions are additionally shown by gray lines. White regions indicate gaps inside the AbcR1 alignment. Locus tag and gene name (if available) of target mRNAs are given on the right. A vertical gray line indicates the start codon. Numbering of bases in mRNA alignments is given relative to the start codon (D). For detailed visualization of optimal and suboptimal interactions for AbcR1 and its verified target mRNAs in A. tumefaciens, see Table S3.
AbcR1 discriminates between target mRNAs through two target-binding regions
A series of band shift experiments was used to experimentally validate the algorithmically predicted RNA–RNA interactions. Four different in vitro synthesized AbcR1 RNAs were used: the WT RNA, Mut1 with a UCC-AAA exchange in M1, Mut2 with a UCCC-AAAA exchange in M2, and the combined Mut1+2 exchange (). Band shift experiments were performed to verify interactions between AbcR1 variants and their target mRNAs. In the first round, the 32P-labeled AbcR1 variants were incubated with increasing concentrations of four different targets predicted to be addressed around the TIR. The target RNAs consisted of 100 to 150 nucleotides containing the predicted interaction region. As expected, band shifts with AbcR1 and AbcR1 Mut2 but not with the Mut1 and Mut1+2 RNAs confirmed complex formation between the TIRs of atu2422 and frcB with AbcR1 region M1 (). Conversely, the atu4678 and chvE TIRs were shown to interact with AbcR1 region M2 ().
Figure 6. Binding of target mRNAs at the translation initiation region by two distinct functional modules. (A) Secondary structures of WT AbcR1 and the variants Mut1, Mut2, and Mut1+2. Band shift experiments with AbcR1 variants and atu2422 (B), frcB (B), atu4678 (C), chvE (C), and atpH (D) mRNA fragments (~-50/+100 nt relative to the AUG start codon). Predicted IntaRNA duplexes formed by AbcR1 and target mRNAs are shown to the left. Numbering of mRNA nucleotides is given relative to the AUG/GUG start codon. 32P-labeled AbcR1 variants (< 0.05 pmol) were incubated with increasing concentrations of unlabeled target RNAs at 30 °C for 20 min. Final concentrations of unlabeled RNA were added in 100 (lanes 2), 200 (lanes 3), and 400 (lanes 4) fold excess. Samples shown in lanes 1 were incubated with water (control).
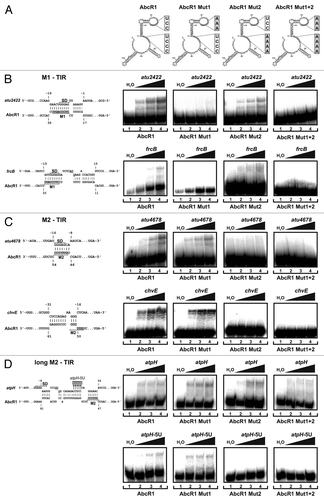
Underrepresentation of AtpH protein and atpH mRNA in the absence of AbcR1 () suggested positive regulation by the sRNA. CopraRNA predicted an interaction between the TIR of atpH and AbcR1 region M2 including the adjacent hairpin 2 (). In agreement with this prediction, mutations in M1 or M2 alone and even in both M1 and M2 were not sufficient to fully abrogate AbcR1–atpH interaction suggesting an extended interaction (). The sRNA–mRNA interaction was lost in the simultaneous presence of a mutation in M2 of AbcR1 and in the atpH TIR (atpH-5U; ).
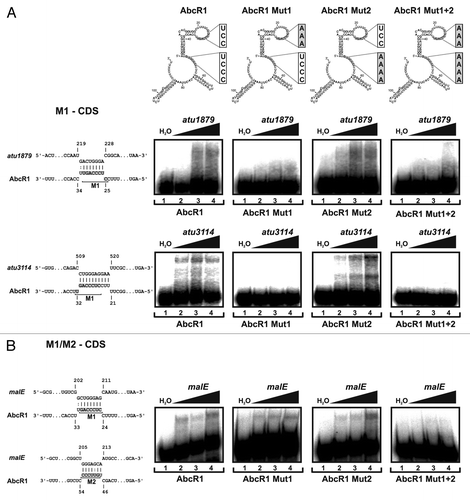
In a second round of experiments, three mRNAs predicted to interact with AbcR1 in their CDS were tested. One hundred and fifty nt-long RNAs containing the predicted interaction region were incubated with the radiolabeled AbcR1 variants. Interaction of M1-containing AbcR1 in the CDS of atu1879 () explains why the TIR of atu1879 could not be shifted in our previous study.Citation26 Region M1 also interacts with the CDS of atu3114. Extending the CopraRNA prediction, the CDS region of malE was not only able to interact with module M1 but also retarded the M2 RNA () suggesting that both modules are able to initiate seed pairing. Consistent with this assumption, the Mut1+2 RNA was unable to shift the malE fragment.
Figure 7. Binding of target mRNAs in the coding sequence by AbcR1. (Top) Secondary structures of AbcR1 wild-type, the variants Mut1, Mut2, and Mut1+2. Band shift experiments with AbcR1 variants and atu1879 (A), atu3114 (A), and malE (B) mRNA fragments (~150 nt). Predicted IntaRNA duplexes formed by AbcR1 and target mRNAs are shown to the left. Numbering of mRNA nucleotides is given relative to the start codon. 32P-labeled AbcR1 variants (< 0.05 pmol) were incubated with increasing concentrations of unlabeled target RNAs at 30 °C for 20 min. Final concentrations of unlabeled RNA were added in 100 (lanes 2), 200 (lanes 3) and 400 (lanes 4) fold excess. Samples from lanes 1 were incubated with water (control).
In vivo verification of target binding by AbcR1 modules M1 and M2
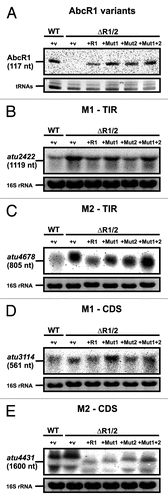
To validate the in vitro results on the interaction of AbcR1 with its target mRNAs in vivo, we used an A. tumefaciens ΔAbcR1/2 double mutant complemented with the empty vector (+v in ) or a plasmid constitutively expressing one of the four AbcR1 variants (+AbcR1, +Mut1, +Mut2, or +Mut1+2). Production of the AbcR1 transcripts was confirmed by northern blot analysis (). The mRNA levels of four different AbcR1 targets were determined by northern blot analysis. Consistent with the band shift experiments ( and ), region M1 was responsible for regulation of atu2422 and atu3114 (). In accordance with the band shift results (), atu4678 was predominantly controlled via M2 (). The same was true for atu4431, which was predicted to bind the M2 region AbcR1 in in its coding sequence ().
Figure 8. In vivo validation of AbcR1 modules 1 and 2. Northern blot analyses of AbcR1 (A), atu2422 (B), atu4678 (C), atu3114 (D), and atu4431 (E) transcripts from cultures of the A. tumefaciens wild-type (WT) or the ΔAbcR1/2 deletion mutant (ΔR1/2) complemented with a plasmid expressing different AbcR1 variants (+R1, +Mut1, +Mut2, +Mut1+2). The strains were grown in YEB medium. +v: control strains harboring the empty vector. Eight μg of total RNA were separated on 1.2% denaturing agarose gels. Ethidiumbromide-stained tRNAs or 16S rRNAs were used as loading control.
Binding sites of AbcR1 in the CDS contain SD-like sequences
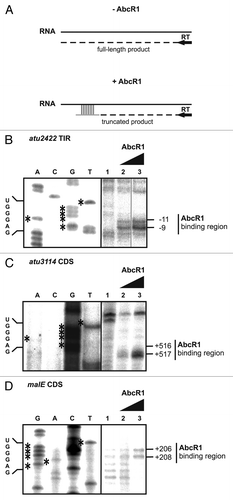
To precisely map the AbcR1-binding positions in the CDS of selected target mRNAs, we used an in vitro reverse transcription approach. The principle is illustrated in . Target mRNA fragments of 100 or 150 nt length were annealed to end-labeled primers complementary to regions upstream of the predicted interaction region followed by cDNA synthesis. Truncated products upon addition of two different concentrations of AbcR1 prior to reverse transcription were mapped in reference to a sequence reaction run on the same gel. In a control experiment (), the mapped atu2422–AbcR1 interaction site corresponded to the previously reported site overlapping the SD sequence of the mRNA.Citation26 As an example for an mRNA targeted far within the CDS, atu3114 was used. The CopraRNA-predicted region around 516 nt in the open reading frame was found to interact with AbcR1, thus resulting in prematurely terminated cDNA fragments (). With the malE RNA, the presence of AbcR1 led to truncated cDNA products corresponding to a CDS region around +207 (). The mapped interactions sites show that AbcR1 addresses SD-like UGGGAG motifs (see sequence to the left of ) regardless of their position in the mRNA.
Figure 9. Precise mapping of AbcR1 binding sites in target mRNAs. (A) Principle of AbcR1 binding-site mapping by toeprinting analysis. -AbcR1, without AbcR1; reverse transcription (RT) starting from a primer complementary to the target mRNA sequence transcribes a full-length product. +AbcR1, pairing of AbcR1 with the target sequence terminates reverse transcription (truncated product). AbcR1 binding-site mapping on atu2422 (B), atu3114 (C), and malE (D) RNA fragments was performed as described in Experimental procedures. The position of truncated products is indicated to the right. mRNA nucleotides involved in M1 binding are shown to the left. Concentrations of AbcR1 RNAs were 1.5 pmol µl−1 (lane 2) and 2.5 pmol µl−1 (lane 3).
AbcR1 promotes degradation of target mRNAs when bound to the TIR or CDS
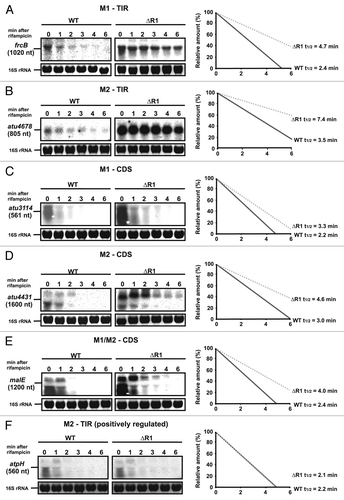
The previously identified target mRNAs of atu2422 and atu1879 were significantly stabilized in the absence of AbcR1 in vivo.Citation26 This provided evidence that interaction of M1 with the TIR (atu2422) or CDS (atu1879) accelerates mRNA turnover and led us to study the effect of AbcR1 on the stability of various target mRNAs. We selected one example each for M1-TIR, M2-TIR, M1-CDS, M2-TIR, and M1/M2-CDS interactions and determined mRNA degradation in the presence or absence of AbcR1 after transcription was stopped by addition of rifampicin.
Interaction of AbcR1 with the TIR via M1 (frcB, ) or M2 (atu4678, ) destabilizes the target mRNAs as shown by their elevated stability in the absence of the sRNA. The same is true when the CDS is bound by AbcR1 either by M1 (atu3114, ), M2 (atu4431, ), or M1 or M2 (malE, ) suggesting that negative regulation by AbcR1 involves RNA degradation regardless of whether the TIR or CDS is targeted. Contrary to these negatively regulated transcripts, stability of the positively regulated atpH transcripts was not influenced by AbcR1 ().
Figure 10. AbcR1 M1 and M2 target mRNAs in the TIR and the CDS for degradation. Northern blot analyses of frcB (A), atu4678 (B), atu3114 (C), atu4431 (D), malE (E), and atpH (F) transcripts from cultures treated with rifampicin. Cultures of the A. tumefaciens wild-type (WT) or the ΔAbcR1 deletion mutant (ΔR1) were grown to exponential or stationary (in case of frcB) growth phase in YEB medium and treated with rifampicin (250 mg ml-1). Total RNA fractions were collected at the indicated time points. Eight μg of total RNA were separated on 1.2% denaturing agarose gels. Ethidiumbromide-stained 16S rRNAs were used as loading control. Quantification of transcript stabilities and their calculated half-lives are given to the right.
Discussion
Global approaches like proteomics or microarrays and bioinformatic predictions are commonly used for sRNA target identification.Citation2,Citation8,Citation67 In this study, we employed a combination of global proteomics and comparative biocomputational predictions for identifying targets of AbcR1 in the plant-pathogen A. tumefaciens. Validation of 14 targets via northern blot hybridization enlarged the set of currently known AbcR1 targets to 16 mRNAs. Although several target mRNAs of AbcR1 have been reported in Brucella and Sinorhizobium, the mode of action of this conserved sRNA has not yet been studied.Citation57,Citation59 Our study uncovered two distinct target-binding sites in AbcR1 and variable interacting loci in the controlled transcripts.
AbcR1 targets different sites of mRNAs through two functional modules
Many Hfq-associated sRNAs contain one single-stranded domain able to interact with multiple target mRNAs.Citation5,Citation14,Citation68-Citation71 Other sRNAs have several functional domains that base pair with different sets of target mRNAs in E. coli, Salmonella, and Vibrio harveyi.Citation7,Citation16,Citation72,Citation73
Previously, only one conserved target-binding region strategically positioned in the first exposed hairpin loop of AbcR1 has been reported.Citation26,Citation59 Here, we exploited the recently established comparative target prediction tool CopraRNA, which has been previously used to predict the two and three interaction regions of GcvB and Spot42, respectively.Citation7,Citation8,Citation73 Strikingly, the two computationally predicted and experimentally verified modules M1 and M2 are highly conserved among Rhizobiaceae suggesting that the two functional modules are not limited to AbcR1 in A. tumefaciens. The more distantly related AbcR1 sequence from Brucella was not included in the CopraRNA predictions because it exhibits less sequence identity to A. tumefaciens AbcR1 than the homologs from Sinorhizobium and Rhizobium species. The existence of two single-stranded M1- and M2-like regions in the predicted secondary structure of B. abortus AbcR1, however, suggests that two functional AbcR1 modules are not restricted to plant-associated bacteria.Citation59
On the target site, our study revealed that AbcR1 binding regions are scattered throughout the TIR and CDS. Although interference with translation by mRNA binding around the SD sequence is considered the most common control mechanism of sRNAs, targeting of coding sequences has been described in enterobacteria, for example, MicC and ompD, ArcZ-tpx, RybB-fadL, SgrS-manX, and SdsR-ompD.Citation14,Citation71,Citation74-Citation76 Two exposed UC-rich interaction regions in AbcR1 and the potential to interact with SD-like regions in the TIR or CDS allows pervasive gene regulation by this sRNA in A. tumefaciens.
AbcR1: A conserved master regulator of ABC transporters
There is increasing evidence that sRNAs are more than single target regulators, but rather act on multiple trans-encoded targets and rewire entire transcriptional networks.Citation2,Citation77,Citation78 Many well-studied sRNAs in enterobacteria control large sets of functionally related target mRNAs; for example, RyhB regulates mRNAs encoding iron-binding proteins involved in iron homeostasis, OmrA/OmrB regulate mRNAs encoding proteins for outer membrane protein synthesis, and GcvB controls genes for amino acid biosynthesis and transport.Citation7,Citation22-Citation24,Citation69,Citation70,Citation79-Citation83
Homologs of AbcR1 from S. meliloti, R. etli, and B. abortus are similar in sequence and structure.Citation52-Citation54,Citation56,Citation59,Citation84 A functional classification of target mRNAs (ABC transport system) was initially described for the AbcR sRNAs in A. tumefaciens and B. abortus 2308.Citation26,Citation59 The experimental verification of 14 AbcR1 targets encoding periplasmic transport proteins carrying sugars, amino acids, and opines supports the function of AbcR1 as a key regulator for these transport systems ().
Figure 11. The AbcR1 regulon of A. tumefaciens. AbcR1 controls mRNAs of periplasmic substrate-binding proteins of 14 ABC transporters (sugars and amino acids to the left and right, respectively), an annotated oxidoreductase (atu0857) and AtpH. Module 1 (red) and module 2 (blue) dependent genes are sorted toward the top and bottom of the schematic cell, respectively. The interaction region (TIR or CDS) and the mode of action (repression or activation) are indicated. Dashed lines refer to computationally predicted interactions.
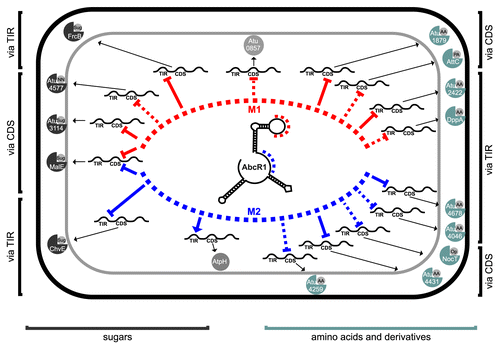
Our previous study described a potential role of AbcR1 in plant defense, quorum sensing, and virulence of A. tumefaciens because the AbcR1 target atu2422 codes for binding protein of an importer of GABA, a plant defense molecule.Citation26,Citation85-Citation88 We now find that AbcR1 also silences synthesis of ChvE (), a regulator in sugar-dependent activation of the virulence cascade as well as other virulence-related ABC transporters (NocT and AttC).Citation43,Citation45 This strengthens the hypothesis that AbcR1 is involved in plant–microbe interactions and post-infection nutrient acquisition.
Although most currently known sRNAs block translation of target mRNAs by interfering with ribosome binding, several sRNAs can activate gene expression.Citation89 They can, for example, bind upstream of the TIR and remodel an intrinsic inhibitory mRNA structure such that the sequestered ribosome binding site is liberated (DsrA and RprA).Citation90-Citation92 Recently, new translation-independent pathways of mRNA activations have been reported for cfa through RydC and for yigL through SgrS in Salmonella.Citation19,Citation93 In enterobacteria, well-characterized sRNAs like RyhB and ArcZ repress some target mRNAs, but activate translation of shiA (RyhB) and rpoS (ArcZ).Citation69,Citation74,Citation79,Citation80,Citation94 In addition to the many negatively controlled AbcR1 targets in A. tumefaciens we found atpH as positively regulated gene. It is predicted to encode the delta subunit of the ATP synthase. In E. coli, this subunit plays a key role in the assembly of the H+-translocating F0F1 ATP synthase.Citation95 Although it remains unknown how AbcR1 controls atpH expression, for instance, AbcR1 does not alter atpH mRNA stability directly, the extensive interaction region between AbcR1 M2 and the TIR of atpH is indicative of a direct mechanism. Control of multiple ABC transporters and the ATP synthase suggests that AbcR1 coordinates nutrient acquisition and energy conversion in A. tumefaciens.
Experimental Procedures
Bacterial growth conditions
Bacterial strains and antibiotics used in this study are listed in Table S5. E. coli was grown in LB medium at 37 °C. A. tumefaciens strains were cultivated in YEB medium at 30 °C.
Strain and vector constructions
The ∆AbcR1 and ∆hfq mutant strains were constructed in previous studies.Citation26,Citation61 Runoff plasmids as templates for in vitro transcription of AbcR1 or target mRNA fragments were flanked by the T7-promoter sequence (GAAATTAATA CGACTCACTA TAGGG) and an EcoRV site PCR-amplified with primers listed in Table S4 and subcloned into pUC18.Citation96 AbcR1 variants were constructed via site-directed mutagenesis using the primers listed in Table S4.
Protein preparation
Cells of A. tumefaciens wild-type, ∆AbcR1, and ∆hfq were grown in 30 ml YEB medium at 30 °C to an OD600 of 1.5. Culture volumes of 30 ml were harvested, washed three times in 30 ml of TE-buffer (100 mM Tris and 1 mM EDTA), and finally resuspended in 4 ml of TE-buffer with 1.39 mM PMSF and 0.2 mM DTT. Cells were disrupted by three passes through a chilled French press. The lysates were centrifuged at 10 000 × g for 30 min to remove the cell debris. Protein concentrations were determined by Bradford assays.Citation97
Two-dimensional PAGE and Mass Spectrometry
Total proteins extracts of A. tumefaciens wild-type, ∆AbcR1, and ∆hfq cells were concentrated by chloroform/methanol precipitation up to 600 μg μl−1.Citation98 Isoelectric focusing and SDS-PAGE were performed as described previously.Citation99 Protein solutions were loaded on Immobiline DryStrip pH 4–7, 24 cm (GE Healthcare). After isoelectric focusing, proteins were subjected to 12.5% SDS-PAGE, and the spots were visualized using RuBPS (C72H42N6Na4O18RuS6) staining. Protein spots were scanned using a Typhoon TRIO (GE Healthcare) and were quantified with the Delta two-dimensional software (version 4.0, Decodon). Selected protein spots were excised from the gel, and protein identification using mass spectrometry was performed by MALDI-TOF mass spectrometry as described previously.Citation100
RNA preparation and northern analysis
Cells were harvested, washed, and frozen in liquid nitrogen as described previously.Citation26 Isolation of total RNA was done by using the hot acid phenol method.Citation101 Northern analyses were performed as previously described.Citation26 To measure mRNA stability, rifampicin was added to the cell cultures in a final concentration of 250 mg ml−1 and samples for RNA isolation were collected before (0 min) and 1, 2, 3, 4, and 6 min after addition of the transcriptional inhibitor rifampicin. In order to determine the half-life of the specific mRNAs, the amount of transcripts present at each time point was quantified using the Image software Alpha Ease FC (Alpha Innotech). The primers used for RNA probe generation are listed in Table S4 in the supplemental material.
Gel shift experiments
The sRNAs AbcR1 WT, QC1, QC2, and QC1+2 and the target mRNA fragments (comprising ~150 nucleotides in the TIR or in the CDS) were synthesized in vitro by runoff transcription with T7 RNA polymerase from the linearized plasmids listed in Table S4. 5‘ end labeling of AbcR1 WT or AbcR1 variants (QC1, QC2, and QC1+2) with 32P was performed as described.Citation102 RNA band shift experiments were performed in 1x structure buffer (Ambion) in a total reaction mixture volume of 15 µl as follows. 5′ end labeled AbcR1 (corresponding to 5000 c.p.m.) and 1 µg of tRNA were incubated in the presence of unlabeled target mRNA fragments (~150 nt) at 30 °C for 20 min. The final concentrations of added unlabeled RNA fragments are given in the figure legends. Prior to gel loading, the binding reactions were mixed 4.5 µl of native loading dye (50% glycerol, 0.5× TBE, 0.1% bromophenol blue and 0.1% xylene cyanol) and run on native 6% polyacrylamide gels in 0.5× TBE buffer at 300 V for 1.5–3 h.
Mapping of sRNA-binding sites
Mapping of AbcR1-binding sites were performed like previously described “toeprint analysis” with some modifications.Citation103 Annealing mixtures contained 0.5 pmol unlabeled atu2422 (50 nt +/− from the AUG start codon), malE (+100/+250 relative to AUG start codon), or atu3114 (+437/+588 relative to AUG start codon) mRNA fragments and 1 pmol of 5′ end labeled primer runoff_atu2422_rv, runoff_malE2_rv, and runoff_atu3114_rv in VD buffer without magnesium. Annealing mixtures were heated for 3 min at 80 °C and snap frozen in a frozen plastic box. After incubation on ice for 20 min, different concentrations (listed in figure legends) of AbcR1, WT, or water (as negative control) were added and incubated at 37 °C for 20 min. After addition of 2 µl MMLV-Mix (VD + Mg2+, BSA, dNTPs and MMLV reverse transcriptase [USB]), cDNA synthesis were performed at 37 °C for 10 min. Reactions were stopped by adding formamide loading dye and reaction aliquots were separated on a denaturing 8% polyacrylamide gel. Reverse transcription cDNA products were identified by comparison with sequences generated with the same 5′ end labeled primer.
Bioinformatic tools
Alignments of sequences were generated by the ClustalW software obtained from http://www.ebi.ac.uk/Tools/msa/clustalw2/. sRNA–mRNA duplexes were predicted by the IntaRNA webserver from http://rna.informatik.uni-freiburg.de:8080/v1/IntaRNA.jsp.Citation104 Secondary structures and consensus structures were computed with mfold http://mfold.rna.albany.edu/?q=mfold/RNA-Folding-Form and RNAalifold http://rna.tbi.univie.ac.at/cgi-bin/RNAalifold.cgi.Citation65,Citation105 Comparative predictions for AbcR1 binding sites and targets were done with CopraRNA http://rna.informatik.uni-freiburg.de/CopraRNA/Input.jsp
Additional material
Download Zip (369.1 KB)Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Rosemarie Gurski for excellent technical assistance, Nicole Frankenberg-Dinkel and Julia Bandow for sharing proteomic infrastructure and expertise; Knut Büttner (Greifswald, Germany) for MALDI-MS analysis and Philip Möller for critical reading of the manuscript. Parts of this study were conducted in the Central Unit for Ion beams and Radionucleotides (RUBION). This work was supported by a grant from the German Research Foundation (DFG priority program SPP 1258: Sensory and regulatory RNAs in Prokaryotes) to F.N. and W.R.H. and by the Bundesministerium für Bildung und Forschung (BMBF) Grant 0316165 to W.R.H.
References
- Lalaouna D, Simoneau-Roy M, Lafontaine D, Massé E. Regulatory RNAs and target mRNA decay in prokaryotes. Biochim Biophys Acta 2013; 1829:742 - 7; http://dx.doi.org/10.1016/j.bbagrm.2013.02.013; PMID: 23500183
- Storz G, Vogel J, Wassarman KM. Regulation by small RNAs in bacteria: expanding frontiers. Mol Cell 2011; 43:880 - 91; http://dx.doi.org/10.1016/j.molcel.2011.08.022; PMID: 21925377
- Kawamoto H, Koide Y, Morita T, Aiba H. Base-pairing requirement for RNA silencing by a bacterial small RNA and acceleration of duplex formation by Hfq. Mol Microbiol 2006; 61:1013 - 22; http://dx.doi.org/10.1111/j.1365-2958.2006.05288.x; PMID: 16859494
- Bouvier M, Sharma CM, Mika F, Nierhaus KH, Vogel J. Small RNA binding to 5′ mRNA coding region inhibits translational initiation. Mol Cell 2008; 32:827 - 37; http://dx.doi.org/10.1016/j.molcel.2008.10.027; PMID: 19111662
- Balbontín R, Fiorini F, Figueroa-Bossi N, Casadesús J, Bossi L. Recognition of heptameric seed sequence underlies multi-target regulation by RybB small RNA in Salmonella enterica.. Mol Microbiol 2010; 78:380 - 94; http://dx.doi.org/10.1111/j.1365-2958.2010.07342.x; PMID: 20979336
- Boehm A, Vogel J. The csgD mRNA as a hub for signal integration via multiple small RNAs. Mol Microbiol 2012; 84:1 - 5; http://dx.doi.org/10.1111/j.1365-2958.2012.08033.x; PMID: 22414234
- Sharma CM, Papenfort K, Pernitzsch SR, Mollenkopf HJ, Hinton JCD, Vogel J. Pervasive post-transcriptional control of genes involved in amino acid metabolism by the Hfq-dependent GcvB small RNA. Mol Microbiol 2011; 81:1144 - 65; http://dx.doi.org/10.1111/j.1365-2958.2011.07751.x; PMID: 21696468
- Wright PR, Richter AS, Papenfort K, Mann M, Vogel J, Hess WR, Backofen R, Georg J. Comparative genomics boosts target prediction for bacterial small RNAs. Proc Natl Acad Sci U S A 2013; 110:E3487 - 96; http://dx.doi.org/10.1073/pnas.1303248110; PMID: 23980183
- Bouché F, Bouché JP. Genetic evidence that DicF, a second division inhibitor encoded by the Escherichia coli dicB operon, is probably RNA. Mol Microbiol 1989; 3:991 - 4; http://dx.doi.org/10.1111/j.1365-2958.1989.tb00249.x; PMID: 2477663
- Wassarman KM, Storz G. 6S RNA regulates E. coli RNA polymerase activity. Cell 2000; 101:613 - 23; http://dx.doi.org/10.1016/S0092-8674(00)80873-9; PMID: 10892648
- Mank NN, Berghoff BA, Hermanns YN, Klug G. Regulation of bacterial photosynthesis genes by the small noncoding RNA PcrZ. Proc Natl Acad Sci U S A 2012; 109:16306 - 11; http://dx.doi.org/10.1073/pnas.1207067109; PMID: 22988125
- Altuvia S, Zhang A, Argaman L, Tiwari A, Storz G. The Escherichia coli OxyS regulatory RNA represses fhlA translation by blocking ribosome binding. EMBO J 1998; 17:6069 - 75; http://dx.doi.org/10.1093/emboj/17.20.6069; PMID: 9774350
- Harris JF, Micheva-Viteva S, Li N, Hong-Geller E. Small RNA-mediated regulation of host-pathogen interactions. Virulence 2013; 4:785 - 95; http://dx.doi.org/10.4161/viru.26119; PMID: 23958954
- Papenfort K, Bouvier M, Mika F, Sharma CM, Vogel J. Evidence for an autonomous 5′ target recognition domain in an Hfq-associated small RNA. Proc Natl Acad Sci U S A 2010; 107:20435 - 40; http://dx.doi.org/10.1073/pnas.1009784107; PMID: 21059903
- Lenz DH, Mok KC, Lilley BN, Kulkarni RV, Wingreen NS, Bassler BL. The small RNA chaperone Hfq and multiple small RNAs control quorum sensing in Vibrio harveyi and Vibrio cholerae.. Cell 2004; 118:69 - 82; http://dx.doi.org/10.1016/j.cell.2004.06.009; PMID: 15242645
- Shao Y, Feng L, Rutherford ST, Papenfort K, Bassler BL. Functional determinants of the quorum-sensing non-coding RNAs and their roles in target regulation. EMBO J 2013; 32:2158 - 71; http://dx.doi.org/10.1038/emboj.2013.155; PMID: 23838640
- Weilbacher T, Suzuki K, Dubey AK, Wang X, Gudapaty S, Morozov I, Baker CS, Georgellis D, Babitzke P, Romeo T. A novel sRNA component of the carbon storage regulatory system of Escherichia coli.. Mol Microbiol 2003; 48:657 - 70; http://dx.doi.org/10.1046/j.1365-2958.2003.03459.x; PMID: 12694612
- Edwards AN, Patterson-Fortin LM, Vakulskas CA, Mercante JW, Potrykus K, Vinella D, Camacho MI, Fields JA, Thompson SA, Georgellis D, et al. Circuitry linking the Csr and stringent response global regulatory systems. Mol Microbiol 2011; 80:1561 - 80; http://dx.doi.org/10.1111/j.1365-2958.2011.07663.x; PMID: 21488981
- Papenfort K, Sun Y, Miyakoshi M, Vanderpool CK, Vogel J. Small RNA-mediated activation of sugar phosphatase mRNA regulates glucose homeostasis. Cell 2013; 153:426 - 37; http://dx.doi.org/10.1016/j.cell.2013.03.003; PMID: 23582330
- Göpel Y, Papenfort K, Reichenbach B, Vogel J, Görke B. Targeted decay of a regulatory small RNA by an adaptor protein for RNase E and counteraction by an anti-adaptor RNA. Genes Dev 2013; 27:552 - 64; http://dx.doi.org/10.1101/gad.210112.112; PMID: 23475961
- Sun Y, Vanderpool CK. Physiological consequences of multiple-target regulation by the small RNA SgrS in Escherichia coli.. J Bacteriol 2013; 195:4804 - 15; http://dx.doi.org/10.1128/JB.00722-13; PMID: 23873911
- Urbanowski ML, Stauffer LT, Stauffer GV. The gcvB gene encodes a small untranslated RNA involved in expression of the dipeptide and oligopeptide transport systems in Escherichia coli.. Mol Microbiol 2000; 37:856 - 68; http://dx.doi.org/10.1046/j.1365-2958.2000.02051.x; PMID: 10972807
- Sharma CM, Darfeuille F, Plantinga TH, Vogel J. A small RNA regulates multiple ABC transporter mRNAs by targeting C/A-rich elements inside and upstream of ribosome-binding sites. Genes Dev 2007; 21:2804 - 17; http://dx.doi.org/10.1101/gad.447207; PMID: 17974919
- Pulvermacher SC, Stauffer LT, Stauffer GV. Role of the sRNA GcvB in regulation of cycA in Escherichia coli.. Microbiology 2009; 155:106 - 14; http://dx.doi.org/10.1099/mic.0.023598-0; PMID: 19118351
- Antal M, Bordeau V, Douchin V, Felden B. A small bacterial RNA regulates a putative ABC transporter. J Biol Chem 2005; 280:7901 - 8; http://dx.doi.org/10.1074/jbc.M413071200; PMID: 15618228
- Wilms I, Voss B, Hess WR, Leichert LI, Narberhaus F. Small RNA-mediated control of the Agrobacterium tumefaciens GABA binding protein. Mol Microbiol 2011; 80:492 - 506; http://dx.doi.org/10.1111/j.1365-2958.2011.07589.x; PMID: 21320185
- Saier MH Jr.. Families of transmembrane sugar transport proteins. Mol Microbiol 2000; 35:699 - 710; http://dx.doi.org/10.1046/j.1365-2958.2000.01759.x; PMID: 10692148
- Davidson AL, Chen J. ATP-binding cassette transporters in bacteria. Annu Rev Biochem 2004; 73:241 - 68; http://dx.doi.org/10.1146/annurev.biochem.73.011303.073626; PMID: 15189142
- Hosie AH, Poole PS. Bacterial ABC transporters of amino acids. Res Microbiol 2001; 152:259 - 70; http://dx.doi.org/10.1016/S0923-2508(01)01197-4; PMID: 11421273
- Narberhaus F, Vogel J. Regulatory RNAs in prokaryotes: here, there and everywhere. Mol Microbiol 2009; 74:261 - 9; http://dx.doi.org/10.1111/j.1365-2958.2009.06869.x; PMID: 19732342
- Gimpel M, Heidrich N, Mäder U, Krügel H, Brantl S. A dual-function sRNA from B. subtilis: SR1 acts as a peptide encoding mRNA on the gapA operon. Mol Microbiol 2010; 76:990 - 1009; http://dx.doi.org/10.1111/j.1365-2958.2010.07158.x; PMID: 20444087
- Romby P, Charpentier E. An overview of RNAs with regulatory functions in gram-positive bacteria. Cell Mol Life Sci 2010; 67:217 - 37; http://dx.doi.org/10.1007/s00018-009-0162-8; PMID: 19859665
- Dühring U, Axmann IM, Hess WR, Wilde A. An internal antisense RNA regulates expression of the photosynthesis gene isiA.. Proc Natl Acad Sci U S A 2006; 103:7054 - 8; http://dx.doi.org/10.1073/pnas.0600927103; PMID: 16636284
- Jäger D, Pernitzsch SR, Richter AS, Backofen R, Sharma CM, Schmitz RA. An archaeal sRNA targeting cis- and trans-encoded mRNAs via two distinct domains. Nucleic Acids Res 2012; 40:10964 - 79; http://dx.doi.org/10.1093/nar/gks847; PMID: 22965121
- Schmidtke C, Abendroth U, Brock J, Serrania J, Becker A, Bonas U. Small RNA sX13: a multifaceted regulator of virulence in the plant pathogen Xanthomonas.. PLoS Pathog 2013; 9:e1003626; http://dx.doi.org/10.1371/journal.ppat.1003626; PMID: 24068933
- Lee K, Huang X, Yang C, Lee D, Ho V, Nobuta K, Fan JB, Wang K. A genome-wide survey of highly expressed non-coding RNAs and biological validation of selected candidates in Agrobacterium tumefaciens.. PLoS One 2013; 8:e70720; http://dx.doi.org/10.1371/journal.pone.0070720; PMID: 23950988
- Wilms I, Overlöper A, Nowrousian M, Sharma CM, Narberhaus F. Deep sequencing uncovers numerous small RNAs on all four replicons of the plant pathogen Agrobacterium tumefaciens.. RNA Biol 2012; 9:446 - 57; http://dx.doi.org/10.4161/rna.17212; PMID: 22336765
- Pitzschke A, Hirt H. New insights into an old story: Agrobacterium-induced tumour formation in plants by plant transformation. EMBO J 2010; 29:1021 - 32; http://dx.doi.org/10.1038/emboj.2010.8; PMID: 20150897
- Lacroix B, Citovsky V. The roles of bacterial and host plant factors in Agrobacterium-mediated genetic transformation. Int J Dev Biol 2013; 57:467 - 81; http://dx.doi.org/10.1387/ijdb.130199bl; PMID: 24166430
- Zupan J, Muth TR, Draper O, Zambryski P. The transfer of DNA from agrobacterium tumefaciens into plants: a feast of fundamental insights. Plant J 2000; 23:11 - 28; http://dx.doi.org/10.1046/j.1365-313x.2000.00808.x; PMID: 10929098
- McCullen CA, Binns AN. Agrobacterium tumefaciens and plant cell interactions and activities required for interkingdom macromolecular transfer. Annu Rev Cell Dev Biol 2006; 22:101 - 27; http://dx.doi.org/10.1146/annurev.cellbio.22.011105.102022; PMID: 16709150
- Doty SL, Yu MC, Lundin JI, Heath JD, Nester EW. Mutational analysis of the input domain of the VirA protein of Agrobacterium tumefaciens.. J Bacteriol 1996; 178:961 - 70; PMID: 8576069
- Hu X, Zhao J, DeGrado WF, Binns AN. Agrobacterium tumefaciens recognizes its host environment using ChvE to bind diverse plant sugars as virulence signals. Proc Natl Acad Sci U S A 2013; 110:678 - 83; http://dx.doi.org/10.1073/pnas.1215033110; PMID: 23267119
- Citovsky V, Kozlovsky SV, Lacroix B, Zaltsman A, Dafny-Yelin M, Vyas S, Tovkach A, Tzfira T. Biological systems of the host cell involved in Agrobacterium infection. Cell Microbiol 2007; 9:9 - 20; http://dx.doi.org/10.1111/j.1462-5822.2006.00830.x; PMID: 17222189
- He F, Nair GR, Soto CS, Chang Y, Hsu L, Ronzone E, DeGrado WF, Binns AN. Molecular basis of ChvE function in sugar binding, sugar utilization, and virulence in Agrobacterium tumefaciens.. J Bacteriol 2009; 191:5802 - 13; http://dx.doi.org/10.1128/JB.00451-09; PMID: 19633083
- Matthysse AG, Yarnall HA, Young N. Requirement for genes with homology to ABC transport systems for attachment and virulence of Agrobacterium tumefaciens.. J Bacteriol 1996; 178:5302 - 8; PMID: 8752352
- Planamente S, Moréra S, Faure D. In planta fitness-cost of the Atu4232-regulon encoding for a selective GABA-binding sensor in Agrobacterium.. Commun Integr Biol 2013; 6:e23692; http://dx.doi.org/10.4161/cib.23692; PMID: 23710277
- Kim H, Farrand SK. Characterization of the acc operon from the nopaline-type Ti plasmid pTiC58, which encodes utilization of agrocinopines A and B and susceptibility to agrocin 84. J Bacteriol 1997; 179:7559 - 72; PMID: 9393724
- Hayman GT, Beck von Bodman S, Kim H, Jiang P, Farrand SK. Genetic analysis of the agrocinopine catabolic region of Agrobacterium tumefaciens Ti plasmid pTiC58, which encodes genes required for opine and agrocin 84 transport. J Bacteriol 1993; 175:5575 - 84; PMID: 8366042
- Schneider E, Eckey V, Weidlich D, Wiesemann N, Vahedi-Faridi A, Thaben P, Saenger W. Receptor-transporter interactions of canonical ATP-binding cassette import systems in prokaryotes. Eur J Cell Biol 2012; 91:311 - 7; http://dx.doi.org/10.1016/j.ejcb.2011.02.008; PMID: 21561685
- Chevrot R, Rosen R, Haudecoeur E, Cirou A, Shelp BJ, Ron E, Faure D. GABA controls the level of quorum-sensing signal in Agrobacterium tumefaciens.. Proc Natl Acad Sci U S A 2006; 103:7460 - 4; http://dx.doi.org/10.1073/pnas.0600313103; PMID: 16645034
- del Val C, Rivas E, Torres-Quesada O, Toro N, Jiménez-Zurdo JI. Identification of differentially expressed small non-coding RNAs in the legume endosymbiont Sinorhizobium meliloti by comparative genomics. Mol Microbiol 2007; 66:1080 - 91; http://dx.doi.org/10.1111/j.1365-2958.2007.05978.x; PMID: 17971083
- Ulvé VM, Sevin EW, Chéron A, Barloy-Hubler F. Identification of chromosomal alpha-proteobacterial small RNAs by comparative genome analysis and detection in Sinorhizobium meliloti strain 1021. BMC Genomics 2007; 8:467; http://dx.doi.org/10.1186/1471-2164-8-467; PMID: 18093320
- Valverde C, Livny J, Schlüter JP, Reinkensmeier J, Becker A, Parisi G. Prediction of Sinorhizobium meliloti sRNA genes and experimental detection in strain 2011. BMC Genomics 2008; 9:416; http://dx.doi.org/10.1186/1471-2164-9-416; PMID: 18793445
- Voss B, Hölscher M, Baumgarth B, Kalbfleisch A, Kaya C, Hess WR, Becker A, Evguenieva-Hackenberg E. Expression of small RNAs in Rhizobiales and protection of a small RNA and its degradation products by Hfq in Sinorhizobium meliloti.. Biochem Biophys Res Commun 2009; 390:331 - 6; http://dx.doi.org/10.1016/j.bbrc.2009.09.125; PMID: 19800865
- Vercruysse M, Fauvart M, Cloots L, Engelen K, Thijs IM, Marchal K, Michiels J. Genome-wide detection of predicted non-coding RNAs in Rhizobium etli expressed during free-living and host-associated growth using a high-resolution tiling array. BMC Genomics 2010; 11:53; http://dx.doi.org/10.1186/1471-2164-11-53; PMID: 20089193
- Torres-Quesada O, Millán V, Nisa-Martínez R, Bardou F, Crespi M, Toro N, Jiménez-Zurdo JI. Independent activity of the homologous small regulatory RNAs AbcR1 and AbcR2 in the legume symbiont Sinorhizobium meliloti.. PLoS One 2013; 8:e68147; http://dx.doi.org/10.1371/journal.pone.0068147; PMID: 23869210
- Torres-Quesada O, Reinkensmeier J, Schlüter JP, Robledo M, Peregrina A, Giegerich R, Toro N, Becker A, Jiménez-Zurdo JI. Genome-wide profiling of Hfq-binding RNAs uncovers extensive post-transcriptional rewiring of major stress response and symbiotic regulons in Sinorhizobium meliloti. RNA Biol 2014; 11; http://dx.doi.org/10.4161/rna.28239; PMID: 24786641
- Caswell CC, Gaines JM, Ciborowski P, Smith D, Borchers CH, Roux CM, Sayood K, Dunman PM, Roop Ii RM. Identification of two small regulatory RNAs linked to virulence in Brucella abortus 2308. Mol Microbiol 2012; 85:345 - 60; http://dx.doi.org/10.1111/j.1365-2958.2012.08117.x; PMID: 22690807
- Torres-Quesada O, Oruezabal RI, Peregrina A, Jofré E, Lloret J, Rivilla R, Toro N, Jiménez-Zurdo JI. The Sinorhizobium meliloti RNA chaperone Hfq influences central carbon metabolism and the symbiotic interaction with alfalfa. BMC Microbiol 2010; 10:71; http://dx.doi.org/10.1186/1471-2180-10-71; PMID: 20205931
- Wilms I, Möller P, Stock AM, Gurski R, Lai EM, Narberhaus F. Hfq influences multiple transport systems and virulence in the plant pathogen Agrobacterium tumefaciens. J Bacteriol 2012; 194:5209 - 17; http://dx.doi.org/10.1128/JB.00510-12; PMID: 22821981
- Vogel J, Luisi BF. Hfq and its constellation of RNA. Nat Rev Microbiol 2011; 9:578 - 89; http://dx.doi.org/10.1038/nrmicro2615; PMID: 21760622
- Møller T, Franch T, Højrup P, Keene DR, Bächinger HP, Brennan RG, Valentin-Hansen P. Hfq: a bacterial Sm-like protein that mediates RNA-RNA interaction. Mol Cell 2002; 9:23 - 30; http://dx.doi.org/10.1016/S1097-2765(01)00436-1; PMID: 11804583
- Garfinkel DJ, Nester EW. Agrobacterium tumefaciens mutants affected in crown gall tumorigenesis and octopine catabolism. J Bacteriol 1980; 144:732 - 43; PMID: 6253441
- Bernhart SH, Hofacker IL, Will S, Gruber AR, Stadler PF. RNAalifold: improved consensus structure prediction for RNA alignments. BMC Bioinformatics 2008; 9:474; http://dx.doi.org/10.1186/1471-2105-9-474; PMID: 19014431
- Goujon M, McWilliam H, Li W, Valentin F, Squizzato S, Paern J, Lopez R. A new bioinformatics analysis tools framework at EMBL-EBI. Nucleic Acids Res 2010; 38:W695-9; http://dx.doi.org/10.1093/nar/gkq313; PMID: 20439314
- Vogel J, Wagner EG. Target identification of small noncoding RNAs in bacteria. Curr Opin Microbiol 2007; 10:262 - 70; http://dx.doi.org/10.1016/j.mib.2007.06.001; PMID: 17574901
- Lease RA, Cusick ME, Belfort M. Riboregulation in Escherichia coli: DsrA RNA acts by RNA:RNA interactions at multiple loci. Proc Natl Acad Sci U S A 1998; 95:12456 - 61; http://dx.doi.org/10.1073/pnas.95.21.12456; PMID: 9770507
- Massé E, Gottesman S. A small RNA regulates the expression of genes involved in iron metabolism in Escherichia coli.. Proc Natl Acad Sci U S A 2002; 99:4620 - 5; http://dx.doi.org/10.1073/pnas.032066599; PMID: 11917098
- Guillier M, Gottesman S. The 5′ end of two redundant sRNAs is involved in the regulation of multiple targets, including their own regulator. Nucleic Acids Res 2008; 36:6781 - 94; http://dx.doi.org/10.1093/nar/gkn742; PMID: 18953042
- Pfeiffer V, Papenfort K, Lucchini S, Hinton JC, Vogel J. Coding sequence targeting by MicC RNA reveals bacterial mRNA silencing downstream of translational initiation. Nat Struct Mol Biol 2009; 16:840 - 6; http://dx.doi.org/10.1038/nsmb.1631; PMID: 19620966
- Durand S, Storz G. Reprogramming of anaerobic metabolism by the FnrS small RNA. Mol Microbiol 2010; 75:1215 - 31; http://dx.doi.org/10.1111/j.1365-2958.2010.07044.x; PMID: 20070527
- Beisel CL, Storz G. The base-pairing RNA spot 42 participates in a multioutput feedforward loop to help enact catabolite repression in Escherichia coli.. Mol Cell 2011; 41:286 - 97; http://dx.doi.org/10.1016/j.molcel.2010.12.027; PMID: 21292161
- Papenfort K, Said N, Welsink T, Lucchini S, Hinton JC, Vogel J. Specific and pleiotropic patterns of mRNA regulation by ArcZ, a conserved, Hfq-dependent small RNA. Mol Microbiol 2009; 74:139 - 58; http://dx.doi.org/10.1111/j.1365-2958.2009.06857.x; PMID: 19732340
- Rice JB, Vanderpool CK. The small RNA SgrS controls sugar-phosphate accumulation by regulating multiple PTS genes. Nucleic Acids Res 2011; 39:3806 - 19; http://dx.doi.org/10.1093/nar/gkq1219; PMID: 21245045
- Fröhlich KS, Papenfort K, Berger AA, Vogel J. A conserved RpoS-dependent small RNA controls the synthesis of major porin OmpD. Nucleic Acids Res 2012; 40:3623 - 40; http://dx.doi.org/10.1093/nar/gkr1156; PMID: 22180532
- Gottesman S, Storz G. Bacterial small RNA regulators: versatile roles and rapidly evolving variations. Cold Spring Harb Perspect Biol 2011; 3:3; http://dx.doi.org/10.1101/cshperspect.a003798; PMID: 20980440
- Papenfort K, Vogel J. Multiple target regulation by small noncoding RNAs rewires gene expression at the post-transcriptional level. Res Microbiol 2009; 160:278 - 87; http://dx.doi.org/10.1016/j.resmic.2009.03.004; PMID: 19366629
- Massé E, Vanderpool CK, Gottesman S. Effect of RyhB small RNA on global iron use in Escherichia coli.. J Bacteriol 2005; 187:6962 - 71; http://dx.doi.org/10.1128/JB.187.20.6962-6971.2005; PMID: 16199566
- Prévost K, Salvail H, Desnoyers G, Jacques JF, Phaneuf E, Massé E. The small RNA RyhB activates the translation of shiA mRNA encoding a permease of shikimate, a compound involved in siderophore synthesis. Mol Microbiol 2007; 64:1260 - 73; http://dx.doi.org/10.1111/j.1365-2958.2007.05733.x; PMID: 17542919
- Desnoyers G, Morissette A, Prévost K, Massé E. Small RNA-induced differential degradation of the polycistronic mRNA iscRSUA. EMBO J 2009; 28:1551 - 61; http://dx.doi.org/10.1038/emboj.2009.116; PMID: 19407815
- Salvail H, Lanthier-Bourbonnais P, Sobota JM, Caza M, Benjamin JA, Mendieta ME, Lépine F, Dozois CM, Imlay J, Massé E. A small RNA promotes siderophore production through transcriptional and metabolic remodeling. Proc Natl Acad Sci U S A 2010; 107:15223 - 8; http://dx.doi.org/10.1073/pnas.1007805107; PMID: 20696910
- Guillier M, Gottesman S, Storz G. Modulating the outer membrane with small RNAs. Genes Dev 2006; 20:2338 - 48; http://dx.doi.org/10.1101/gad.1457506; PMID: 16951250
- Schlüter JP, Reinkensmeier J, Daschkey S, Evguenieva-Hackenberg E, Janssen S, Jänicke S, Becker JD, Giegerich R, Becker A. A genome-wide survey of sRNAs in the symbiotic nitrogen-fixing alpha-proteobacterium Sinorhizobium meliloti.. BMC Genomics 2010; 11:245; http://dx.doi.org/10.1186/1471-2164-11-245; PMID: 20398411
- Shelp BJ, Bown AW, Faure D. Extracellular gamma-aminobutyrate mediates communication between plants and other organisms. Plant Physiol 2006; 142:1350 - 2; http://dx.doi.org/10.1104/pp.106.088955; PMID: 17151138
- Moréra S, Gueguen-Chaignon V, Raffoux A, Faure D. Cloning, purification, crystallization and preliminary X-ray analysis of a bacterial GABA receptor with a Venus flytrap fold. Acta Crystallogr Sect F Struct Biol Cryst Commun 2008; 64:1153 - 5; http://dx.doi.org/10.1107/S1744309108036555; PMID: 19052373
- Yuan ZC, Haudecoeur E, Faure D, Kerr KF, Nester EW. Comparative transcriptome analysis of Agrobacterium tumefaciens in response to plant signal salicylic acid, indole-3-acetic acid and gamma-amino butyric acid reveals signalling cross-talk and Agrobacterium--plant co-evolution. Cell Microbiol 2008; 10:2339 - 54; http://dx.doi.org/10.1111/j.1462-5822.2008.01215.x; PMID: 18671824
- Planamente S, Vigouroux A, Mondy S, Nicaise M, Faure D, Moréra S. A conserved mechanism of GABA binding and antagonism is revealed by structure-function analysis of the periplasmic binding protein Atu2422 in Agrobacterium tumefaciens.. J Biol Chem 2010; 285:30294 - 303; http://dx.doi.org/10.1074/jbc.M110.140715; PMID: 20630861
- Fröhlich KS, Vogel J. Activation of gene expression by small RNA. Curr Opin Microbiol 2009; 12:674 - 82; http://dx.doi.org/10.1016/j.mib.2009.09.009; PMID: 19880344
- Majdalani N, Hernandez D, Gottesman S. Regulation and mode of action of the second small RNA activator of RpoS translation, RprA. Mol Microbiol 2002; 46:813 - 26; http://dx.doi.org/10.1046/j.1365-2958.2002.03203.x; PMID: 12410838
- Sledjeski D, Gottesman S. A small RNA acts as an antisilencer of the H-NS-silenced rcsA gene of Escherichia coli.. Proc Natl Acad Sci U S A 1995; 92:2003 - 7; http://dx.doi.org/10.1073/pnas.92.6.2003; PMID: 7534408
- Sledjeski DD, Gupta A, Gottesman S. The small RNA, DsrA, is essential for the low temperature expression of RpoS during exponential growth in Escherichia coli.. EMBO J 1996; 15:3993 - 4000; PMID: 8670904
- Fröhlich KS, Papenfort K, Fekete A, Vogel J. A small RNA activates CFA synthase by isoform-specific mRNA stabilization. EMBO J 2013; 32:2963 - 79; http://dx.doi.org/10.1038/emboj.2013.222; PMID: 24141880
- Mandin P, Gottesman S. Integrating anaerobic/aerobic sensing and the general stress response through the ArcZ small RNA. EMBO J 2010; 29:3094 - 107; http://dx.doi.org/10.1038/emboj.2010.179; PMID: 20683441
- Hilbers F, Eggers R, Pradela K, Friedrich K, Herkenhoff-Hesselmann B, Becker E, Deckers-Hebestreit G. Subunit δ is the key player for assembly of the H(+)-translocating unit of Escherichia coli F(O)F1 ATP synthase. J Biol Chem 2013; 288:25880 - 94; http://dx.doi.org/10.1074/jbc.M113.484675; PMID: 23864656
- Norrander J, Kempe T, Messing J. Construction of improved M13 vectors using oligodeoxynucleotide-directed mutagenesis. Gene 1983; 26:101 - 6; http://dx.doi.org/10.1016/0378-1119(83)90040-9; PMID: 6323249
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 1976; 72:248 - 54; http://dx.doi.org/10.1016/0003-2697(76)90527-3; PMID: 942051
- Wessel D, Flügge UI. A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal Biochem 1984; 138:141 - 3; http://dx.doi.org/10.1016/0003-2697(84)90782-6; PMID: 6731838
- Bandow JE, Baker JD, Berth M, Painter C, Sepulveda OJ, Clark KA, Kilty I, VanBogelen RA. Improved image analysis workflow for 2-D gels enables large-scale 2-D gel-based proteomics studies--COPD biomarker discovery study. Proteomics 2008; 8:3030 - 41; http://dx.doi.org/10.1002/pmic.200701184; PMID: 18618493
- Klüsener S, Hacker S, Tsai YL, Bandow JE, Gust R, Lai EM, Narberhaus F. Proteomic and transcriptomic characterization of a virulence-deficient phosphatidylcholine-negative Agrobacterium tumefaciens mutant. Mol Genet Genomics 2010; 283:575 - 89; http://dx.doi.org/10.1007/s00438-010-0542-7; PMID: 20437057
- Aiba H, Adhya S, de Crombrugghe B. Evidence for two functional gal promoters in intact Escherichia coli cells. J Biol Chem 1981; 256:11905 - 10; PMID: 6271763
- Brantl S, Wagner EG. Antisense RNA-mediated transcriptional attenuation occurs faster than stable antisense/target RNA pairing: an in vitro study of plasmid pIP501. EMBO J 1994; 13:3599 - 607; PMID: 7520390
- Hartz D, McPheeters DS, Traut R, Gold L. Extension inhibition analysis of translation initiation complexes. Methods Enzymol 1988; 164:419 - 25; http://dx.doi.org/10.1016/S0076-6879(88)64058-4; PMID: 2468068
- Wright PR, Georg J, Mann M, Sorescu DA, Richter AS, Lott S, et al. CopraRNA and IntaRNA: predicting small RNA targets, networks and interaction domains. Nucleic Acids Res 2014; http://dx.doi.org/10.1093/nar/gku359; PMID: 24838564
- Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 2003; 31:3406 - 15; http://dx.doi.org/10.1093/nar/gkg595; PMID: 12824337
