Abstract
In bacteria, transcription, translation and gene regulation are highly coupled processes. The achievement of a certain functional structure at a distinct temporal and spatial position is therefore essential for RNA molecules. Proteins that facilitate this proper folding of RNA molecules are called RNA chaperones. Here a prominent example from E. coli is reviewed: the nucleoid associated protein StpA. Based on its various RNA remodeling functions, we propose a mechanistic model that explains how StpA promotes RNA folding. Through transient interactions via the RNA backbone, thereby shielding repelling charges in RNA, it pre-positions the RNA molecules for the successful formation of transition states from encounter complexes.
Introduction
Soon after catalytic RNAs were discovered, it was realized that, very much like proteins, RNAs fold into complex three-dimensional structures that are essential for their activities. Several catalytic RNAs like self-splicing introns are active in vitro without the help of proteins; however, many of these RNAs require non-physiological ionic conditions or are clearly dependent on proteins for optimal activity. Therefore the search for proteinaceous splicing factors accompanied the studies on self-splicing intron catalysis from the very beginning. Genetic screens were the most common approach to explore proteins that promote the activity of catalytic RNAs. Two main strategies were employed: screens for trans-acting mutants that result in splicing deficiency or screens for suppressors of splicing deficient intron mutants.Citation1–Citation6 A screen using splicing-deficient mutants of the T4 phage derived thymidylate synthase (td) group I intron was performed to search for E. coli proteins that could restore splicing. It resulted in the discovery of StpA, “suppressor of the td- phenotype”.Citation7 StpA is not a specific splicing factor like the maturases or Cyt-18 and CBP2. In contrast, it interacts with RNA non-specifically and promotes splicing by acting as an RNA chaperone. In this review, we will discuss the properties of this interesting protein, which reveal many of the most important characteristics of this class of proteins.
Functions of StpA in gene-regulation.
StpA-like genes have so far been found in several Gram negative bacteria while Gram positive bacteria do not seem to express homologs of this protein.Citation8 StpA's paralogy and overlapping function of the well-studied histone-like nucleoid structuring protein (H-NS) were recognized immediately.Citation9,Citation10 In the form of a homo- or hetero-dimer with StpA or other proteins H-NS shapes the structure and organization of the E. coli genome by bending and bridging DNACitation11 and both H-NS and StpA restrain DNA supercoils in vitro. Thus, H-NS and StpA exert the role of pleiotropic regulators and influence transcription of certain genes in an either positive or negative way.Citation10
In full medium, StpA is not abundant and its expression is only induced during a very concise period in mid-exponential growth phase. Expression of StpA can be enhanced through osmolytic stress or elevated temperatures and a more pronounced induction of StpA expression can be achieved in minimal growth medium.Citation12 Apart from negative auto-regulation of both H-NS and StpA genes, both proteins cross-regulate their homologue's expression in a negative way indicating that they have partially overlapping functions.Citation10 Some H-NS regulated genes have indeed been found to be regulated by StpA, while others remain unaffected. So far, no genes have been identified which are regulated by StpA only.Citation8 Consistently, while mutation of the hns gene has noticeable effects on growth and cell structure, single stpA-knockouts do not show a specific phenotype. However, growth of double hns/stpA mutants is strongly impaired under normal growth temperatures as well as under cold or heat shock. For those reasons, StpA is sometimes considered a ‘molecular backup’ of H-NS. However, the ability of H-NS to dimerize with StpA and other H-NS like proteins hints at the establishment of fine-tuning mechanisms in response to changing growth conditions as opposed to simple H-NS substitution by StpA or other H-NS like proteins.Citation11
Like H-NS, StpA is a histone-like protein that binds and bends DNA,Citation10 and therefore, it was surprising when StpA was isolated as a suppressor of an RNA-based phenotype. The ability of StpA to refold RNA molecules has been studied extensively. StpA has been shown to stimulate group I intron splicing by accelerating folding of the intron RNA both in vitro and in vivo.Citation7,Citation13 While specific splicing factors stabilize the structure of RNAs by recognizing and binding a particular RNA sequence, StpA, being an RNA chaperone loosens the structure by non-specific interactions with RNA.Citation13,Citation14 Most important for distinguishing specific splicing factors from RNA chaperones is the observation that RNA chaperones are only needed during folding and as soon as the RNA is in its native conformation, the chaperones are no longer required.Citation15 StpA can interact with many different RNA molecules and has no sequence preference.Citation16 It accelerates annealing and strand displacement of many different RNAs. In an RNA recombination assay, StpA was able to enhance copy-choice recombination during reverse transcription of HIV leading to higher reshuffling of genetic markers.Citation17 Additionally, StpA's simultaneous action on DNA and RNA could be explained by the spatial-temporal coupling of transcription and translation in bacteria. This has also been shown for the E. coli cold shock protein, CspA which also has RNA chaperone activity and regulates transcription and translation.Citation18 Recently, it was noticed that mRNAs in E. coli stay at their site of transcription attached to the chromosome during their lifetime. Thus StpA and other nucleic acid chaperones could easily act on both DNA and RNA substrates to regulate gene expression.Citation19
Structure of StpA.
StpA is a modular protein with remarkable similarities to the chromatin-associated protein H-NS, sharing a 58% identity over all residues.Citation20 The protein can be subdivided into two domains (). The N-terminal domain comprising residues 1–76, has a lower identity to the respective H-NS domain (51%) and functions as the site for homo-Citation16 and heterodimerization with H-NS.Citation21 The C-terminal domain (residues 90–134) shows a higher degree of similarity to H-NS (73% identity) and forms a stable 6 kDa product after restricted proteolysis. Both domains are connected by a flexible linker (residues 76–90) that is prone to proteolysis. According to Cusick et al.Citation20 StpA's RNA chaperone activity resides in the basic C-terminal domain (CTD) of StpA, which is also responsible for RNA/DNA binding. The CTD contains 5 lysine and 3 arginine residues in a total of 46 residues. In contrast, the StpA N-terminal domain does not exhibit activity in either the annealing or the trans-splicing assays, although in the cis-splicing assay, RNA chaperone activity was also found for the N-terminal domain. Bio-informatics predictions made for several RNA chaperones indicate a high propensity of disorder within the proteins, which may be a universal property of this class of proteins and mechanistically important for their function.Citation22 Calculations using the PONDR-algorithmCitation23 to determine the overall disorder score indicate that StpA has an overall disorder of 73% and is therefore a largely unstructured protein.Citation24 A study using the meta-structure approachCitation25 resulted in the prediction of certain secondary structure elements: α-helical structures in the N-terminal domain (residues 115 to 134) and at the very C-terminal end, and a stretch of β-sheets for residue 90 to 115. Moreover, the overall compactness for StpA CTD is lower in these calculations than for folded proteins with an ARCfolded proteins (average residue compactness) around 300. Interestingly, the linker-region that is most sensitive to limited proteolysis shows no clear propensity for a certain secondary structure but a rather high measure of compactness, compared to the rest of the sequence.Citation16
Contrasting the rigorous results of the PONDR prediction is the considerably strong homology to highly structured H-NS, for which three-dimensional structures of certain domains could be determined.Citation26,Citation27 This suggests that StpA might also exhibit folded domains. Furthermore, the stabilities of the homo- and heterodimers with H-NS are remarkable.Citation21 Therefore, we assume that StpA exhibits well-defined structured domains.
It is interesting that although both proteins harbor the same amount of basic amino acids (each with 23 arginines and lysines), H-NS is less active than StpA in the applied chaperone assays, indicating a possible structure-function relationship.Citation20 The difference could be either attributed to the compensation of the positive charges by the presence of acidic residues or to the spatial distribution of the charged amino acids. The positive net-charge is lower in H-NS (H-NS-pI 5.44) than in StpA (StpA-pI 6.41; calculated using ExPASy). The spatial distribution of charged residues over the surface of the proteins shows a higher difference for the nucleic acids binding C-terminal domain harboring the RNA chaperone activity when compared to the NTD (). Moreover, the H-NS DNA binding site was mapped by NMR spectroscopic methods to a mostly positive charged surface region of the CTD of the protein. Most affected residues upon DNA binding are D101, T109, R113, T114 and A116 (see arrow marks in ).Citation28 If the homologous region of StpA is involved in nucleic acid binding, a net of two positive charges is added to the interaction surface.
Assays for RNA Chaperone Activity
The diversity of the assays that have been used is key to the understanding of the mode of action of proteins with RNA chaperone activity. Here all assays in which StpA has been tested, as well as its performance in these assays, are summarized. Besides full-length StpA, two mutant variants of the protein as well as the CTD and NTD were tested (). The NTD is responsible for dimerization and the CTD has been implicated in nucleic acid interaction. The RNA chaperone activity of StpA was shown in different in vitro and in vivo assays, comprising mono- and bimolecular reactions.
RNA Annealing and RNA Strand Displacement.
The two basic activities an RNA chaperone is expected to accomplish are the acceleration of hybridization (annealing) and the dissociation (displacement) of complementary RNA strands. Assays to monitor annealing or strand displacement are usually carried out with radioactively labeled RNAs and visualized via gel electrophoresis. In addition, complementary RNAs can be end-labeled with fluorophores and the fraction of double-stranded RNAs can be monitored via FRET. Using a pair of unstructured 21mer RNAs that self-anneal with a rate constant of kann = 106 M−1sec−1, StpA was shown to accelerate the rate constant of annealing about 4-fold.Citation16 Using longer complementary RNAs of 96 nt and 92 nt, that are capable of forming a 86 bp duplex, but do not self-anneal due to intramolecular structures, StpA accelerates the initial rate of annealing >130-fold at 37°C.Citation7 StpA shows the highest activity at about 1.8 µM. Due to the length of the substrate RNA, it is likely that the assay monitors not only the annealing reaction but also an unwinding step which is required because of secondary structures.Citation29 The use of short unstructured RNA substrates allows the discrimination between strand annealing and unfolding of pre-existing secondary structures in the complementary oligonucleotides.
In addition to annealing, StpA also stimulates strand displacement without the addition of ATP.Citation7,Citation30 In a combined FRET-based assay, a pair of complementary 21mer long RNAs anneals in the presence or absence of StpA (phase I). The addition of a non-labelled fully complementary competitor 21mer RNA initiates strand displacement (phase II). In the absence of StpA, phase II is dominated by the annealing activity, however in the presence of StpA, strand displacement is catalyzed with a rate constant of kSD = 4 × 106 M−1sec−1.
T4 td Intron Splicing.
The group I intron derived from the T4 bacteriophage thymidylate synthase (td) gene self splices in vivo and in vitro. The splicing reaction is initiated by the nucleophilic attack of guanosine at the 5′ splice site and followed by two transesterfication reactions. Splicing of the td intron is strongly dependent on the correct three-dimensional fold of the RNA. In vivo, the process is probably promoted by different trans-acting factors and is therefore fast and efficient, whereas in vitro, misfolding of the RNA renders the process slow and inefficient. Overexpression of StpA in E. coli cells expressing the td pre-mRNA leads to a 3.0-fold increase of the pre-mRNA level and a 7.4-fold increase of the mRNA level. Thus, the mRNA-to-pre-mRNA ratio was increased 2.5-fold.Citation7
Group I intron splicing in cis.
In vitro transcribed td intron pre-mRNA is very inefficient in folding, probably due to stable misfolded intermediates. In a non-renatured sample of td intron pre-mRNA over 90% of the molecule are not able to splice efficiently. RNA chaperones can resolve the misfolded structures and thereby elevate the fraction of fast-reacting molecules. At a concentration of 1.4 µM, StpA was shown to accelerate splicing of the td intron 30–45-fold compared to splicing of the RNA alone, leaving only a small fraction of RNA molecules in the slow reacting conformation.Citation16,Citation31
Group I intron splicing in trans. The td intron RNA structure is modular and can thereby be partitioned into pieces, which can refold and assemble into an active intron ribozyme.Citation32 However, assembly of the RNA is inefficient at 37°C, whereas at 55°C or in the presence of an RNA chaperone, a splicing-competent conformation is restored and effective trans splicing can occur.Citation15 StpA strongly promotes trans-splicing in vitro, with a peak activity at about 2 µM protein.Citation7
In vivo folding trap. Splicing of the td intron in vivo is strongly dependent on translation of the pre-mRNA, probably due to a folding trap consisting of a 9 bp interaction between exon and intron sequences. The ribosome disrupts this interaction during translation enabling a splicing competent intron conformation. A stop-codon directly upstream of the folding trap prevents its resolution by the ribosome and traps the intron in a slow reacting conformation with a splicing efficiency of less than 1%. StpA rescues the stop codon mutant but not the intronic mutants, disrupting the thermal stability of the three-dimensional structure of the intron. This distinguishes it from proteins like CYT-18 which specifically recognize and stabilize the intron structure, thereby rescuing activity of both mutants.Citation33
Mechanisms
From assays towards the understanding of mechanisms.
Annealing and strand displacement refer to different events in the folding of an RNA molecule and are therefore discussed individually in this review. It has to be kept in mind, that the rate-limiting step in RNA folding is the dissociation of formed base pairs. Depending on the structural context of the RNA interaction (e.g., domain architecture, structure of the transition state, large or many extended regions etc.,), annealing becomes a second important step in the folding reaction, and both functionalities must coincide. StpA has been tested in various assays using a large variety of different substrates resulting in more or less complex reactions, making it difficult to distinguish between strand annealing, strand dissociation and the influence of StpA on tertiary interactions. Moreover, it is probable that the two processes of annealing and displacement affect each other to a certain extent. Not only might the strand displacement activity of a chaperone support annealing by the opening of mispaired nucleotides, but the displacement of one strand by a competitor might also be strongly dependent on annealing. Even further, annealing and strand displacement may be part of one and the same process (see the discussion in the last section with the generalized model).
In order to discuss possible mechanisms of how StpA chaperones RNA folding, a closer look into the mechanisms of RNA folding itself is necessary. Within milliseconds after synthesis, significant amounts of the RNA molecule fold to form various possible secondary structures. In many regions, the structure may initially consist primarily of small base-paired elements. Over time, ranging from microseconds to minutes,Citation34–Citation37 parts of the molecule will continue to fold into compact structures stabilized by tertiary interactions. Via such folding routes, RNA is prone to adopt non-native stable conformations which may act as folding traps.Citation38,Citation39 To overcome these folding traps, the RNA molecules have to partially unfold and refold to reach the final functional state. Furthermore, many ribonucleic acids are able to adopt more than one single three-dimensional structure.Citation40,Citation41 These alternative structures have again very similar thermodynamic stabilities but show substantially different dynamics and function. Structural transitions between a single or several different metastable RNA states and the final functional state often constitute the rate-limiting steps on the folding pathway towards a functional RNA fold.Citation38 The mechanisms by which RNA molecules refold consist of three distinct steps: (a) the strand displacement that disrupts stabilizing interactions in the starting state, (b) formation of a transition state and (c) strand association to form the final state.Citation30,Citation42 Depending on the RNA sequence, strand dissociation and association can occur simultaneously or sequentially.Citation35 As mentioned above these processes are rate limiting and therefore catalysis of the folding process is required, as the RNA folding problem is not only an in vitro artifact but also occurs in vivo.Citation33,Citation43 There are various mechanisms under discussion as to how proteins facilitate the remodeling of RNA conformations. The proposed models, which are not mutually exclusive, can be grouped into two main classes: (i) RNA-binding proteins act as cofactors, bind tightly to the RNA and become part of the native RNA-protein complex or (ii) proteins that interact only transiently with the RNA (most probably with the transition state) and thereby lower the activation energy in the remodeling reaction ().Citation44 In general, RNA chaperones are defined as polypeptidic modulators of RNA conformations and molecular associations (referring to Gething et al.Citation45). The mode of action of these chaperones seems to be as manifold as the interactions that shape RNAs. Here we will discuss the mechanistic properties of the RNA chaperone StpA, from its RNA binding properties to its ability to accelerate annealing and strand displacement of RNAs.
RNA binding properties of StpA.
No specific RNA recognition sequence is known for StpA. In a Genomic SELEX screenCitation16 no binding motifs could be significantly enriched suggesting that StpA does not exhibit any sequence-dependent RNA-binding. StpA rather has a broad specificity to RNA molecules of any sequence. The dissociation constants measured for different target molecules are in the low µM range. Interestingly, for unstructured RNAs (intronless mRNA) or small ssRNA oligonucleotides the dissociation constant is lower than that for more structured RNAs (short exons carrying a folded intron), KD = 0.58 µM vs. KD = 0.73 µM, respectively. Furthermore, in a filter-binding assay the retention is dramatically reduced by a factor of 8, if a highly structured and compact RNA is tested in comparison to the more unstructured mRNA. The same low binding affinity is monitored for structured short RNAs such as small stable duplexes and hairpins.Citation16 In all assays the optimal concentration for the action of StpA is close to but above the determined dissociation constants (between 1.4 µM and 8 µM).Citation7,Citation10,Citation20 Similar results were also reported for other proteins with RNA chaperone activity (e.g., Ncp7), in which an inhibitory effect is shown for concentrations beyond this optimum.Citation14
Dissociation constants for the interaction between StpA and DNA are in the same range as between StpA and RNA (KD = 0.7 µM, for the H-NS/DNA interaction the dissociation constant is determined to be KD = 2.8 µM). Other proteins with RNA chaperone activity (e.g., Nucleolin) have been shown to be both RNACitation46 and DNACitation47 chaperones. Since StpA interacts with both RNA and DNA with comparable dissociation constants the most probable interaction occurs at the phosphate-backbone interface of the nucleic acid. In line with the proposed interaction via the phosphate-backbone, the potency of interaction between StpA and RNA is highly dependent on the ionic strength of the solution. Mono- as well as divalent-ions can compete for the RNA's interaction with the protein and can also induce similar changes on the protein structure as RNA (Fürtig B, unpublished).Citation16 The increase of magnesium ion concentration reduces the efficient binding of RNA to StpA by a factor of ∼4. When increasing the concentration of magnesium from 0.1 mM to 0.5 mM, the same behavior is observed as during an increase in concentration of monovalent ions from 25 mM to 250 mM.Citation16 Assuming that the interaction with the phosphate backbone is the dominant recognition mode, then shielding the charge of the phosphate backbone could contribute to the RNA annealing and displacement activities by reducing either inter- or/and intramolecular repulsions.Citation48
From a dual binding assay, which probes the interaction of StpA with two non-complementary 21mer RNAsCitation16,Citation30 it can be deduced that the protein is able to interact with two different strands of RNA at the same time. The dual binding assay further suggests an additional interesting fact: although the amplitudes of the annealing reaction and the dual binding reaction differ strongly, the rate constants for both reactions are identical (kobs,ann = kobs,bind = 4 × 106 M−1s−1). This suggests that the rate-limiting step for StpA catalyzed RNA annealing is the binding step. In other words, binding of the RNA to protein induces a structural change in the RNA that makes it favorable for annealing (for further discussion see the generalized model section).
Aside from the low dissociation constant and the broad specificity of StpA, the concept that the interactions between protein and RNA are only of transient nature is founded on the observation that the protein does not need to stay bound, when the folding reaction of the RNA is completed. It can be digested without changing the reaction dynamics or ‘un-folding’ the chaperoned RNA.Citation7 This also implies that in ribozyme-based assays the physical folding steps are affected but the functional steps are not. Furthermore, a mutant of StpA (G126V), which has a dramatically reduced binding affinity to RNA (KD >10 µM), harbors an increased RNA chaperone activity.Citation16 This implies that the tighter the binding strength of a protein is to RNA, the more reduced is its ability to promote RNA folding as an RNA chaperone. This correlation between RNA chaperone activity and weak RNA bindingCitation16,Citation49–Citation51 gives strong evidence that the transient nature of RNA-protein interaction is a necessity for RNA structure remodelling. In fact, the nature of the StpA-RNA interaction fits well with the notion that ‘transient complexes’ are dominated by long-range electrostatic interactions.Citation52
RNA annealing.
So far, three different, but not necessarily mutually exclusive, mechanisms for the acceleration of complementary RNA annealing have been proposed: (a) the active attraction of RNA molecules to increase the encounter frequency between them, (b) stabilization of the annealing transition state by shielding the negative RNA backbone charges (often referred to as ‘matchmaker activity’) and (c) ‘conversion’ of the RNA into an annealing-prone conformation.Citation53–Citation55 Studies of annealing acceleration that used different proteins suggest not a general applicability of one of the above mentioned scenarios, but instead the co-existence of several (mixed) mechanisms.
In all the applied annealing assays the protein was used in a 25- to 200- fold molar excess over RNA single strands. Furthermore, a threshold StpA concentration of 0.6 µM was found in a mixed annealing and strand displacement assay below which no reaction acceleration could be detected.Citation7 It is generally assumed for RNA annealers (as well as for chaperones) that several molecules of protein bind one RNA molecule. This ‘RNA coating’ is presumed to be a necessity for acceleration of annealing (and strand displacement).
Several authors described the importance of positively charged amino acids for nucleic acid annealing activity.Citation55–Citation59 In fact, the CTD of StpA is rich in basic residues and carries a net charge of +3. We analyzed RNA annealing acceleration by the short HIV-1 Tat protein-derived peptide Tat(44–61) (Doetsch et al. paper in preparation). Our results stress not only the importance of the overall charge of the protein, but also suggest a distinct spatial arrangement of basic residues within the peptide. The same relation between positive charges and annealing activity may also apply to StpA: Interestingly, H-NS is less active than StpA in annealing assays,Citation20 which could be explained by the different distributions of positive amino acids within their sequence (). RNA annealing activity is sometimes connected to the ability of proteins to actively increase the encounter frequency between complementary RNA molecules.Citation29 One potential mechanism for the StpA-catalyzed annealing reaction is an increase of local concentration of the RNA molecules, as proposed by Mayer et al.Citation16 and Rajkowitsch et al.Citation30 It was suggested that for simultaneous RNA binding and thus RNA annealing acceleration, dimerization of the protein is indispensable. This hypothesis is supported by the dimerization-deficient StpA L30P mutant that is inactive in annealing assays. These results oppose those from the Belfort lab, reporting that both CTD and NTD individually are inactive in annealing assays.Citation30 The CTD's lack of annealing activity was explained by its inability to bind two RNAs simultaneously. Since StpA dimerization is attributed to the NTD, the CTD alone may have only one RNA binding platform. However, more recent experiments show that the CTD is active in annealing (Fürtig B, et al. unpublished results). The contradicting performance of the CTD might be explained with different protein preparations resulting in different concentrations of active protein, as well as with the use of different salt concentrations in the applied annealing assays. StpA's activity, like that of other RNA chaperones, is inhibited by low amounts of MgCl2 and NaCl. Therefore, the hypothesis that annealing activity is conferred by simultaneous RNA binding needs further confirmation.
RNA displacement.
As indicated above, the opening of stretches of base-paired nucleotides and subsequent exchange of one of the pairing partners are fundamental steps in RNA folding. Most often, the opening process is energetically disfavored and characterized with a low rate constant, that decreases exponentially with the length of the stretch that has to be opened.Citation60 Catalysis of this process is beneficial in order to increase the overall folding rate of RNAs and is therefore an important feature of RNA chaperones.Citation61 Using a FRET assay, an orders-of-magnitude increase in strand displacement activity could be monitored for StpA.Citation30
Mechanistically, strand exchange has to involve open or at least partially open RNA conformations as intermediate or transition states. This reduction in the extent of structure is necessary to speed up the zippering process.Citation49 In the open states, nucleotides are exposed and subsequently available for new interactions. This effect was seen for StpA. Upon interaction with StpA the td group I intron showed a higher accessibility to modifying agents, such as DMS, for tertiary structure elements and a reduced compactness of the overall fold ().Citation13,Citation31 Interestingly, the sensitivity of an RNA towards StpA correlates with the three dimensional structural stability of the RNA.Citation62 Intron mutants with lower thermal stability showed a decreased splicing efficiency in the presence of StpA. This effect could be reverted by dropping the temperature from 37°C to 25°C, indicating that the opening of RNA structures by StpA is only beneficial for folding up to a certain degree.Citation62 This result is consistent with the notion that there is an optimal concentration for StpA beyond which folding efficiency is again decreased. Even so, it is also notable that StpA can not unfold stable RNAs completely.Citation62 These results show a clear difference between StpA and specific binding proteins such as tRNA synthetase Cyt-18, which acts in an opposite way, by binding to the RNA and stabilizing tertiary elements in the RNA ().Citation13
A Generalized Model for StpA Activities
All these findings can be used to define a generalized mechanism for StpA promoting RNA annealing and strand displacement.
The astounding identity of rate constants.
The rate constants of the StpA catalyzed annealing, strand displacement and dual binding reactions hint at which step StpA influences RNA folding.Citation16,Citation30 Strikingly, all three rate constants are identical within error, namely about 4 × 106 M−1sec−1, suggesting that all three activities are subject to the same rate-limiting step in the presence of StpA.
The RNA-only folding scenario.
To explain this phenomenon, we first consider the RNA folding reaction simplified as the annealing of two complementary RNA molecules (). On their way to the formation of the thermodynamically most stable complex, the RNA molecules form a first encounter complex, which might either proceed into a transition state or fall apart. While the encounter complex is characterized by long-range (mainly electrostatic) interactions, the transition state contains RNAs that have already formed initial base pairs.Citation52 The formation of the final duplex evolving from the transition state is assumed to be very fast.Citation63–Citation65 The annealing rate constants of complementary RNAs are very small and in the conformationally-controlled regime. This means that the reaction velocity is restricted by necessary RNA refolding events which have to take place before the transition state formation.Citation52 For example, long 69mer RNAs with some degree of internal secondary structures anneal with a rate of 3.4 × 103 M−1sec−1 and a 21mer unstructured RNA anneals with a rate of 106 M−1sec−1 in the absence of RNA chaperones.Citation30,Citation54 We therefore assume that the rate-limiting step of the folding reaction is the conversion from encounter complex to transition state. We suggest that the restricting processes are conformational changes in both RNA molecules that are necessary for base pair formation. Apart from folding into the most stable conformation, the RNAs have the propensity to form other more or less stable duplexes. Thus, RNA molecules might either get trapped in alternative folds or they might form duplexes that fall apart due to low stability ().
The effect of StpA on RNA annealing.
In the presence of StpA () colliding RNA molecules are most probably not ‘naked’ but instead coated with one or more StpA molecules. Due to electrostatic attraction between RNA and StpA and the strong molar excess of protein over RNA, it is very likely that most RNAs are in an StpA-bound state. Thus, the encounter complex contains, besides the two RNAs, additional StpA molecules. The rate constants for annealing and strand displacement lie within the diffusion-controlled regime, meaning that StpA changes the rate limiting step of RNA folding. Thus, we assume that StpA acts on the conversion between encounter and transition state. Considering the role of basic amino acid residues in StpA's chaperone activity, we suggest that the protein alters RNA conformation in such way that the probability of progression into the transition state is increased and less encounter complexes fall apart. In addition to its conformational influence on the RNA, StpA could also act through shielding of negative RNA backbone charges and thus stabilize the first encounter complex. In summary, StpA induces conformational changes in the RNA that overcome structural barriers that prohibit base pair formation and thus render the RNA strands prone for annealing.
The catalysis of strand displacement by StpA and its connection with RNA annealing.
Another difference from the ‘RNA only’ scenario is the propensity of StpA to open up stable duplexes (). This results in the refolding of the alternative duplexes so that the most prominent product of annealing will be the thermodynamically most stable structure. Since StpA works in a sequence unspecific way, one has to keep in mind that all base paired regions are susceptible to being opened up, including the thermodynamically most stable duplex. This is the basis for strand displacement: when a competing RNA strand is present (especially when it is in molar excess) it will be able to invade a just partly opened duplex and thus replace one of the strands. Thus, the strand displacement reaction is strongly coupled to an annealing process.
The connection of annealing, strand displacement and dual binding.
In the presence of StpA both reactions, annealing and strand displacement, become diffusion-controlled while steps subsequent to encounter complex/transition state formation are very fast and thus not rate-limiting anymore. This means, the reactions are only dependent on the velocity with which RNA molecules collide. The process of StpA-coated RNA molecules meeting each other is what is measured with the dual binding assay. That annealing, strand displacement and dual binding share the same rate-limiting step is the reason for their identical rate constants. In summary, we propose that StpA does not accelerate diffusion of RNA molecules towards each other but instead pre-positions RNAs and stabilizes the encounter complex and thus increases the probability of successful RNA remodeling.
Figures and Tables
Figure 1 Domain architecture of StpA in comparison to H-NS: (A) displays the primary structure of both proteins; in a color-coded scheme the distribution of charged amino-acids is displayed, blue and red bars indicate positively (R and K) and negatively (D and E) charged amino-acids, respectively; the green letters separate every ten amino-acids; the over-bars indicate the domain structure of StpA as discussed in the text and the grey under-bars show the domains of H-NS for which high-resolution structures (pdb-codes: 1ni8 and 1 hns) are available; residues involved in H-NS DNA binding are highlighted by arrow heads; in (B) the three-dimensional structures are displayed in cartoon representation; the color-coding on the surface representation equates the electrostatic potential; residues involved in H-NS DNA binding are highlighted by stick representation.
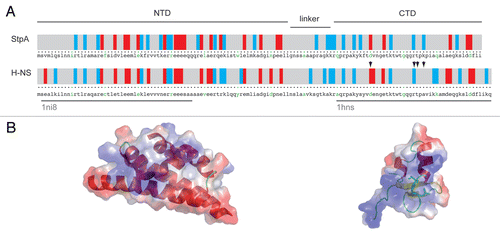
Figure 2 Two simplistic models of how proteins facilitate the remodeling of RNA conformations. (A) Energy landscape for a tight binding protein that acts as a co-factor for RNA and through binding induces a conformational change that is more stable (red) then the respective RNA conformation in the absence of the protein (blue). (B) As an experimental example for such a scenario the changes in the td intron structure upon binding of Cyt-18 are shown. Without the protein residues, A46 and A47 are moderately accessible to DMS modifications (blue line). In the presence of Cyt-18 these residues become involved in a stable tertiary interaction and are protected (red line). No apparent changes are monitored for A48 which is part of a secondary structure element. (C) Energy landscape for proteins that interact only transiently with the transition state of an RNA and thereby lowers the activation energy in the remodeling reaction (blue versus red reaction path, for RNA alone and RNA/protein, respectively). (D) As an experimental example for this scenario the changes in the td intron structure upon interaction with StpA are shown. Without the protein residues A46 and A47 are moderately accessible to DMS modifications. In the presence of StpA these residues sample more open conformations and are easier to access by DMS (red lines). No apparent changes are monitored for A48 as it is involved in a secondary structure interaction; (B and C) are adapted from reference (Waldsich et al.).Citation13
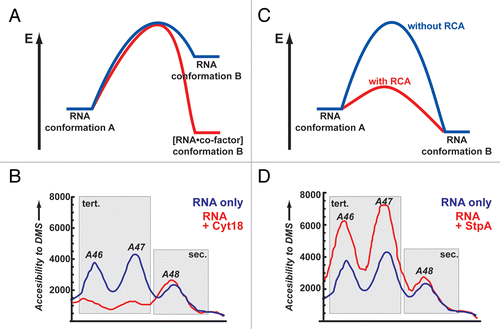
Figure 3 A Generalized Model for StpA Activities. (A) Two annealing RNAs (R1 and R2 with complementary sequences) run through different states before they form a duplex. In addition to the most stable double-strand, alternative duplexes (indicated by the subfix ‘alt’) can form and, depending on their thermodynamic stability, eventually fall apart again. (B) Proposed mechanisms for StpA-facilitated RNA-RNA annealing and strand displacement. Partial opening of the R1R2 duplex (indicated through parentheses) allows the R1-complementary R3 RNA to invade the double-strand.
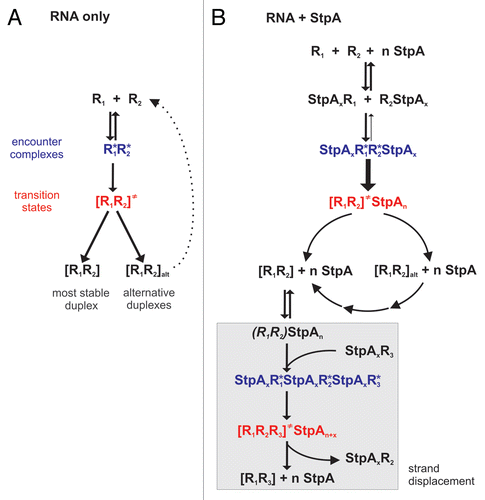
Table 1 The table summarizes the results of various in vitro chaperone assays for WT StpA, the N terminal domain (NTD) and the C terminal domain (CTD) of StpA as well as two StpA mutants G126V and L30P
Acknowledgements
We would like to thank all members of the Schroeder Lab for helpful discussions on the topic of RNA chaperones. We are indebted to Jennifer L. Boots and Bob Zimmermann for critical reading of the manuscript and helpful comments. This work is supported by F.W.F. through a Lise Meitner-Position (M1157-B12) to B.F. and grant F1703 to R.S. and by the European Community (EU-NMR, Contract # RII3-026145). M.D. is funded by the University of Vienna.
References
- Akins RA, Lambowitz AM. A protein required for splicing group I introns in Neurospora mitochondria is mitochondrial tyrosyl-tRNA synthetase or a derivative thereof. Cell 1987; 50:331 - 345
- Dujardin G, Jacq C, Slonimski PP. Single base substitution in an intron of oxidase gene compensates splicing defects of the cytochrome b gene. Nature 1982; 298:628 - 632
- Labouesse M, Netter P, Schroeder R. Molecular basis of the ‘box effect’, A maturase deficiency leading to the absence of splicing of two introns located in two split genes of yeast mitochondrial DNA. European journal of biochemistry/FEBS 1984; 144:85 - 93
- Lazowska J, Jacq C, Slonimski PP. Sequence of introns and flanking exons in wild-type and box3 mutants of cytochrome b reveals an interlaced splicing protein coded by an intron. Cell 1980; 22:333 - 348
- Seraphin B, Simon M, Faye G. MSS18, a yeast nuclear gene involved in the splicing of intron aI5 beta of the mitochondrial cox1 transcript. EMBO J 1988; 7:1455 - 1464
- De la Salle H, Jacq C, Slonimski PP. Critical sequences within mitochontrial introns: pleiotropic mRNA maturase and cis-dominant signals of the box intron controlling reductase and oxidase. Cell 1982; 721 - 732
- Zhang A, Derbyshire V, Salvo JL, Belfort M. Escherichia coli protein StpA stimulates self-splicing by promoting RNA assembly in vitro. RNA 1995; 1:783 - 793
- Dorman CJ, Hinton JC, Free A. Domain organization and oligomerization among H-NS-like nucleoid-associated proteins in bacteria. Trends Microbiol 1999; 7:124 - 128
- Zhang A, Belfort M. Nucleotide sequence of a newly-identified Escherichia coli gene, stpA, encoding an H-NS-like protein. Nucleic Acids Res 1992; 20:6735
- Zhang A, Rimsky S, Reaban ME, Buc H, Belfort M. Escherichia coli protein analogs StpA and H-NS: regulatory loops, similar and disparate effects on nucleic acid dynamics. EMBO J 1996; 15:1340 - 1349
- Muller CM, Schneider G, Dobrindt U, Emody L, Hacker J, Uhlin BE. Differential effects and interactions of endogenous and horizontally acquired H-NS-like proteins in pathogenic Escherichia coli. Mol Microbiol 75:280 - 293
- Free A, Dorman CJ. The Escherichia coli stpA gene is transiently expressed during growth in rich medium and is induced in minimal medium and by stress conditions. Journal of bacteriology 1997; 179:909 - 918
- Waldsich C, Grossberger R, Schroeder R. RNA chaperone StpA loosens interactions of the tertiary structure in the td group I intron in vivo. Genes and Dev 2002; 16:2300 - 2312
- Herschlag D, Khosla M, Tsuchihashi Z, Karpel RL. An RNA chaperone activity of non-specific RNA binding proteins in hammerhead ribozyme catalysis. EMBO J 1994; 13:2913 - 2924
- Coetzee T, Herschlag D, Belfort M. Escherichia coli proteins, including ribosomal protein S12, facilitate in vitro splicing of phage T4 introns by acting as RNA chaperones. Genes Dev 1994; 8:1575 - 1588
- Mayer O, Rajkowitsch L, Lorenz C, Konrat R, Schroeder R. RNA chaperone activity and RNA-binding properties of the E. coli protein StpA. Nucleic Acids Res 2007; 35:1257 - 1269
- Negroni M, Buc H. Copy-choice recombination by reverse transcriptases: reshuffling of genetic markers mediated by RNA chaperones. Proc Natl Acad Sci USA 2000; 97:6385 - 6390
- Jiang W, Hou Y, Inouye M. CspA, the major cold-shock protein of Escherichia coli, is an RNA chaperone. J Biol Chem 1997; 272:196 - 202
- Llopis PM, Jackson AF, Sliusarenko O, Surovtsev I, Heinritz J, Emonet T, et al. Spatial organization of the flow of genetic information in bacteria. Nature 2010; 466:77 - 81
- Cusick ME, Belfort M. Domain structure and RNA annealing activity of the Escherichia coli regulatory protein StpA. Mol Microbiol 1998; 28:847 - 857
- Leonard PG, Ono S, Gor J, Perkins SJ, Ladbury JE. Investigation of the self-association and hetero-association interactions of H-NS and StpA from Enterobacteria. Mol Microbiol 2009; 73:165 - 179
- Dyson HJ, Wright PE. Intrinsically unstructured proteins and their functions. Nat Rev Mol Cell Biol 2005; 6:197 - 208
- Garner E, Romero P, Dunker AK, Brown C, Obradovic Z. Predicting Binding Regions within Disordered Proteins. Genome informatics 1999; 10:41 - 50
- Tompa P, Csermely P. The role of structural disorder in the function of RNA and protein chaperones. FASEB J 2004; 18:1169 - 1175
- Konrat R. The protein meta-structure: a novel concept for chemical and molecular biology. Cell Mol Life Sci 2009; 66:3625 - 3639
- Bloch V, Yang Y, Margeat E, Chavanieu A, Auge MT, Robert B, et al. The H-NS dimerization domain defines a new fold contributing to DNA recognition. Nat Struct Biol 2003; 10:212 - 218
- Shindo H, Iwaki T, Ieda R, Kurumizaka H, Ueguchi C, Mizuno T, et al. Solution structure of the DNA binding domain of a nucleoid-associated protein, H-NS, from Escherichia coli. FEBS letters 1995; 360:125 - 131
- Sette M, Spurio R, Trotta E, Brandizi C, Brandi A, Pon CL, et al. Sequence-specific recognition of DNA by the C-terminal domain of nucleoid-associated protein H-NS. J Biol Chem 2009; 284:30453 - 30462
- Cristofari G, Darlix JL. The ubiquitous nature of RNA chaperone proteins. Prog Nucleic Acid Res Mol Biol 2002; 72:223 - 268
- Rajkowitsch L, Schroeder R. Dissecting RNA chaperone activity. RNA 2007; 13:2053 - 2060
- Mayer O, Waldsich C, Grossberger R, Schroeder R. Folding of the td pre-RNA with the help of the RNA chaperone StpA. Biochem Soc Trans 2002; 30:1175 - 1180
- Salvo JL, Coetzee T, Belfort M. Deletion-tolerance and trans-splicing of the bacteriophage T4 td intron. Analysis of the P6-L6a region. J Mol Biol 1990; 211:537 - 549
- Clodi E, Semrad K, Schroeder R. Assaying RNA chaperone activity in vivo using a novel RNA folding trap. EMBO J 1999; 18:3776 - 3782
- Webb AE, Weeks KM. A collapsed state functions to self-chaperone RNA folding into a native ribonucleoprotein complex. Nat Struct Biol 2001; 8:135 - 140
- Furtig B, Buck J, Manoharan V, Bermel W, Jaschke A, Wenter P, et al. Time-resolved NMR studies of RNA folding. Biopolymers 2007; 86:360 - 383
- Woodson SA. RNA folding and ribosome assembly. Current opinion in chemical biology 2008; 12:667 - 673
- Williamson JR. Biophysical studies of bacterial ribosome assembly. Current opinion in structural biology 2008; 18:299 - 304
- Zarrinkar PP, Williamson JR. Kinetic intermediates in RNA folding. Science 1994; 265:918 - 924
- Treiber DK, Rook MS, Zarrinkar PP, Williamson JR. Kinetic intermediates trapped by native interactions in RNA folding. Science 1998; 279:1943 - 1946
- Mayer O, Windbichler N, Wank H, Schroeder R. Silvermann SK. Protein-induced RNA Switches in Nature. Nucleic Acid Switches and Sensors: Landes Bioscience 2005;
- Schwalbe H, Buck J, Furtig B, Noeske J, Wohnert J. Structures of RNA switches: insight into molecular recognition and tertiary structure. Angewandte Chemie (International ed) 2007; 46:1212 - 1219
- Wong CH, Hendrix M, Priestley ES, Greenberg WA. Specificity of aminoglycoside antibiotics for the A-site of the decoding region of ribosomal RNA. Chem Biol 1998; 5:397 - 406
- Semrad K, Schroeder R. A ribosomal function is necessary for efficient splicing of the T4 phage thymidylate synthase intron in vivo. Genes Dev 1998; 12:1327 - 1337
- Duncan CD, Weeks KM. Nonhierarchical ribonucleoprotein assembly suggests a strain-propagation model for protein-facilitated RNA folding. Biochemistry 49:5418 - 5425
- Gething MJ, Sambrook J. Protein folding in the cell. Nature 1992; 355:33 - 45
- Ghisolfi L, Joseph G, Amalric F, Erard M. The glycine-rich domain of nucleolin has an unusual supersecondary structure responsible for its RNA-helix-destabilizing properties. J Biol Chem 1992; 267:2955 - 2959
- Sapp M, Knippers R, Richter A. DNA binding properties of a 110 kDa nucleolar protein. Nucleic Acids Res 1986; 14:6803 - 6820
- Portman DS, Dreyfuss G. RNA annealing activities in HeLa nuclei. EMBO J 1994; 13:213 - 221
- Munroe SH, Dong XF. Heterogeneous nuclear ribonucleoprotein A1 catalyzes RNA.RNA annealing. Proc Natl Acad Sci USA 1992; 89:895 - 899
- Cobianchi F, Calvio C, Stoppini M, Buvoli M, Riva S. Phosphorylation of human hnRNP protein A1 abrogates in vitro strand annealing activity. Nucleic Acids Res 1993; 21:949 - 955
- Ammerman ML, Fisk JC, Read LK. gRNA/pre-mRNA annealing and RNA chaperone activities of RBP16. RNA 2008; 14:1069 - 1080
- Schreiber G, Haran G, Zhou HX. Fundamental aspects of protein-protein association kinetics. Chem Rev 2009; 109:839 - 860
- Nedbal W, Frey M, Willemann B, Zentgraf H, Sczakiel G. Mechanistic insights into p53-promoted RNA-RNA annealing. J Mol Biol 1997; 266:677 - 687
- Nedbal W, Homann M, Sczakiel G. The association of complementary ribonucleic acids can be strongly increased without lowering Arrhenius activation energies or significantly altering structures. Biochemistry 1997; 36:13552 - 13557
- Muller UF, Goringer HU. Mechanism of the gBP21-mediated RNA/RNA annealing reaction: matchmaking and charge reduction. Nucleic Acids Res 2002; 30:447 - 455
- Hitti E, Neunteufel A, Jantsch MF. The double-stranded RNA-binding protein X1rbpa promotes RNA strand annealing. Nucleic Acids Res 1998; 26:4382 - 4388
- Lee CG, Zamore PD, Green MR, Hurwitz J. RNA annealing activity is intrinsically associated with U2AF. J Biol Chem 1993; 268:13472 - 13478
- Muller UF, Lambert L, Goringer HU. Annealing of RNA editing substrates facilitated by guide RNA-binding protein gBP21. EMBO J 2001; 20:1394 - 1404
- Schumacher MA, Karamooz E, Zikova A, Trantirek L, Lukes J. Crystal structures of T. brucei MRP1/MRP2 guide-RNA binding complex reveal RNA matchmaking mechanism. Cell 2006; 126:701 - 711
- Furtig B, Wenter P, Reymond L, Richter C, Pitsch S, Schwalbe H. Conformational dynamics of bistable RNAs studied by time-resolved NMR spectroscopy. Journal of the American Chemical Society 2007; 129:16222 - 16229
- Rajkowitsch L, Chen D, Stampfl S, Semrad K, Waldsich C, Mayer O, et al. RNA chaperones, RNA annealers and RNA helicases. RNA Biol 2007; 4:118 - 130
- Grossberger R, Mayer O, Waldsich C, Semrad K, Schroeder R. Influence of RNA structural stability on the RNA chaperone activity of the E. coli portein StpA. Nuc Acids Res 2005; 33:2280 - 2289
- Mohan S, Hsiao C, VanDeusen H, Gallagher R, Krohn E, Kalahar B, et al. Mechanism of RNA double helix-propagation at atomic resolution. The journal of physical chemistry 2009; 113:2614 - 2623
- Porschke D. A direct measurement of the unzippering rate of a nucleic acid double helix. Biophysical chemistry 1974; 2:97 - 101
- Porschke D. Model calculations on the kinetics of oligonucleotide double helix coil transitions. Evidence for a fast chain sliding reaction. Biophysical chemistry 1974; 2:83 - 96