Abstract
Efficient gene expression requires that, during their lifetime, mRNAs associate with different sets of RNA binding proteins to form messenger ribonucleoprotein particles (mRNPs). The protein components of mRNPs are essential for the correct post-transcriptional function and regulation of mRNAs. mRNPs are constitutively remodeled during the maturation of the mRNA in the nucleus and downstream steps in the cytoplasm, and can also change depending on the cellular environment. Here we review the current understanding of the biochemical and structural aspects of a central mRNP component and regulator, the exon junction complex (EJC).
Introduction
Eukaryotic cells regulate the expression of protein encoding genes, tuning the amount and lifetime of any given gene product in response to intracellular and extracellular signals. This regulation is of utmost importance for differentiation, proliferation and morphogenesis, and allows organisms to adapt to new conditions. Besides transcriptional regulation, regulation at the post-transcriptional level is of key importance for the cell. It encompasses mRNA processing and export, surveillance, turnover, silencing and localization. It is now apparent that all of these steps are mechanistically linked. The link is physically provided by interactions between the set of proteins that are bound to the mRNAs to form large multimolecular assemblies, known as mRNPs (reviewed in ref. Citation1 and Citation2).
In metazoa, a central component of mRNPs on all mRNAs that are produced from intron-containing genes is the exon junction complex (E°C) (reviewed in ref. Citation3 and Citation4). The EJC is deposited onto the mRNA upon splicing and travels with the mRNP to the cytoplasm where it acts as an effector for downstream events in mRNA metabolism. EJCs are then disassembled and recycled into the nucleus. The EJC is formed by the assembly of four conserved core proteins, MAGO, Y14, eIF4AIII and Barentsz (BTZ).Citation5–Citation13 In humans, all EJC core components are involved in nonsense-mediated mRNA decay (NMD), a pathway that detects and rapidly degrades mRNAs containing premature stop codons (reviewed in ref. Citation14 and Citation15). Conversely, in Drosophila the core components are essential for the localized translation of oskar mRNA during embryonic developmentCitation10,Citation16–Citation21 but not for NMD.Citation22 The EJC can also increase translation efficiency and mRNA export.Citation23–Citation29
In recent years, biochemical and structural studies have provided mechanistic insights into the assembly and function of the EJC and its recycling into the nucleus. Here we review the aspects related to the assembly and disassembly of the EJC and to the recycling of its components as derived from recent biochemical and structural data.
Assembly of the EJC
In the nucleus, the EJC core is placed 20–24 nucleotides upstream of the splice junction site and thus marks the positions of former introns for the gene expression machinery.Citation3 It has been suggested that the EJC is initially recruited to intronic sequences by the interaction with a spliceosomal component, the intron binding protein IBP160.Citation30 eIF4AIII and the MAGO-Y14 heterodimer are recruited to the RNA before the exon ligation step, i.e., the second step of splicing.Citation31–Citation34 eIF4AIII RNA association involves a conformational change (see EJC disassembly) likely induced by the spliceosome and by ATP binding.Citation33 These initial steps position the EJC at the correct binding site,Citation32,Citation33 which is essential for downstream post-transcriptional steps e.g., in NMD. The formation of the EJC core establishes a platform that is used by additional factors for dynamic interactions (reviewed in ref. Citation4) ().
It is presently still unclear whether BTZ is part of the nuclear core of the EJC or if it joins the EJC only after the nuclear export of the spliced mRNA. Cellular localization studies would point toward a predominantly cytoplasmic sub-cellular localization and/or a shuttling activity of BTZ.Citation8,Citation10,Citation35 BTZ has been found associated to nuclear mRNPs in proteomic analysis of spliceosomal complexes from DrosophilaCitation37 but not from human cells.Citation38 Stable EJC assembly intermediates lacking BTZ can also be formed in human splicing extractsCitation33 suggesting that BTZ recruitment might not be required for EJC stability in the nucleus. It is puzzling, however, that BTZ can be loaded on the mRNA during in vitro splicing reactionsCitation39 and the recombinant EJC is unstable without BTZ.Citation12 Furthermore, an overlapping region on the surface of eIF4AIII is used for its interaction with the spliceosome and with BTZ.Citation33 Here, we will consider the EJC core as a tetramer, including BTZ.
Interestingly, the interaction of the EJC does not appear to be dependent on specific RNA sequences.Citation3,Citation32 The crystal structure of the EJC, in complex with RNA, has revealed how the EJC holds a tight sequence-independent grip on the mRNA.Citation40,Citation41 The major RNA binding component is eIF4AIII, an ATPase from the DEAD box family that binds RNA in an ATP-dependent manner (reviewed in ref. Citation42 and Citation43). BTZ (found in the structure as the minimal interacting domain of the protein, also known as SELOR domainCitation8) also contributes to RNA binding, and MAGO-Y14 stabilizes the core by inhibiting eIF4AIII ATPase activityCitation12,Citation40,Citation41 (). eIF4AIII has a conserved helicase core with two flexibly linked RecA-like domains. The N-terminal (domain 1) and the C-terminal (domain 2) domains of eIF4AIII are arranged to form a deep interdomain cleft where ATP binds. The RNA binds with a bent conformation in a shallow cleft across the two domains and is flanked by BTZ ( and D). One residue of BTZ stacks against the first base, increasing the affinity for the RNA.Citation12,Citation44 RNA binds mainly with the hydroxyls of the riboses and the phosphate moieties, while the RNA bases are exposed to the solvent. This explains the sequence independent binding and RNA specificity of the EJC.Citation12,Citation40,Citation41 Structural analysis revealed that the ATPase inhibition is not achieved from disrupting the ATP-binding-site. Instead eIF4AIII is locked into the closed conformation on the RNA by MAGO-Y14, which prevents the release of inorganic phosphate rather than the actual hydrolysis reaction.Citation45 Several loops from MAGO-Y14 grab domain 2 of eIF4AIII and stabilize the linker between domain1 and 2 of eIF4AIII. This prevents the transition to an open conformation that would result in the disruption of the RNA binding site and RNA release ( and C).
Among the more peripherally associated EJC proteins that remain bound to the spliced mRNA in the cytoplasm is the central NMD effector, UPF3b.Citation13 UPF3b associates with the EJC core in the nucleus and provides the physical link between the EJC core and the NMD pathway.Citation46,Citation47 The structure of the EJC core with UPF3b shows that the C-terminal region of UPF3b binds as an elongated peptide to a composite interaction patch involving the tip of the helices of MAGO, the conserved loop of Y14 and also a conserved patch on eIF4AIIICitation48 (). The structural analysis explains why UPF3b can associate only with the assembled core of the E°C but shows no interaction with the individual E°C proteins.Citation48
EJC Disassembly
In the cytoplasm, translation alters mRNP protein composition. It has been proposed that translating ribosomes at the first round of translation, strip off bound proteins from their binding sites on the mRNA.Citation49,Citation50 The EJC can be divided both biochemically and structurally, into two stable binary complexes: the MAGO-Y14 heterodimer and the eIF4AIII-BTZ subcomplex ( and and C) that could represent an intermediate step in the assembly/disassembly of the EJC.
The structures of the MAGO-Y14 heterodimer in isolation have shown that unexpectedly, the Y14 RNA-recognition motif (RRM) is involved in protein-protein interaction and does not bind RNACitation51–Citation53 (). MAGO-Y14 structures in different complexes reveal that the heterodimer behaves like a rigid unit and likely represents a constitutive complex (, B, D and E). In contrast, eIF4AIII undergoes a dramatic conformational change upon assembly/disassembly ( and C). On disassembly, the RNA binding site is disrupted and the surfaces involved in ATP and RNA binding are exposed to solvent. In the structures of apo eIF4AIII and in complex with BTZ, domain 1 and domain 2 are connected by an elongated linker and make few contacts with each otherCitation40,Citation41 (). It is at present not known whether, in the cytoplasm, the dissociation between eIF4AIII and BTZ takes places as a further intermediate step. The interaction surface of eIF4AIII and BTZ in the structure is specific and conserved but involves only few contacts, raising the possibility that the two proteins could be easily dissociated ( and ).
Although the processivity of the ribosome is likely sufficient to completely remove RNA-binding proteins and therefore to remodel mRNPs, complexes like the EJC that bind with high affinity could perhaps significantly slow down the progress of the ribosome. The removal of all EJC core components from the mRNA should involve a release of the inhibition by MAGO-Y14 on the ATPase activity followed by a decrease in the affinity of eIF4AIII for RNA. This would result in RNA release and a change in conformation to the open state of eIF4AIII. The N-terminal part of the cytoplasmic protein PYM associates with MAGO-YCitation14,Citation54,Citation55 while the C-terminal region of the protein binds to the 40S ribosomal subunit.Citation26 As suggested by the structural studies, PYM represents an EJC disassembly factor that equips the ribosome with the ability to remove EJCs as it processes along the mRNA.Citation41,Citation55,Citation56 PYM is able to dismantle EJCs that are formed by in vitro splicing reactions, whereas EJCs assembled from recombinant subunits are not efficiently targeted by PYM.Citation56 In cells, overexpression of PYM leads to a decreased association of EJCs with spliced mRNAs, while PYM inhibition by RNAi leads to an accumulation of EJC components in cytoplasmic cell fractions. This last observation is compatible with a recycling defect of EJC proteins under these conditions. Together, these data demonstrate that PYM is an EJC disassembly factor both in vitro and in vivo. PYM function is regulated by its association with ribosomes, which restricts the disassembly activity to actively translated mRNAs. This mechanism prevents the uncontrolled disassembly of EJCs in the cytoplasm before they have fulfilled their functions (e.g., as marker for NMD).
The structure of the MAGO-Y14-PYM (N-terminal part) ternary complex shows that the binding mode of PYM to MAGO-Y14 would not be compatible in the EJC due to steric hindrance with other components of the complex. Interestingly, the same interaction surface on MAGO-Y14 is involved in the binding of UPF3b and of PYM, although in the first case the binding occurs only in the presence of all EJC components and in the second case, it competes with eIF4AIII and BTZ. Therefore, it is likely that PYM has to displace UPF3b to bind to MAGO-Y14 and disrupt the EJC. Residues in this patch on the surface of MAGO were shown to affect NMD when mutated.Citation51,Citation55
Recycling of EJC Components
The efficient disassembly and recycling of EJC proteins is essential for their reassembly in the nucleus. Quantifications of endogenous proteins in cell lysates show that at steady state there are significantly fewer EJC components present than exonexon junctions (12,000 eIF4AIII, 40,000 MAGO-Y14 vs. 400,000 exon-exon junctions).Citation56 This underlines the requirement for an efficient system to disassemble, recycle and reuse EJC components. In order to be reloaded onto mRNA, the EJC components have to be re-imported into the nucleus.
Among the proteins that associate with the EJC or its components there are transport receptors that recycle them into the nucleus (reviewed in ref. Citation57). Importin13 (Imp13) is the karyopherin nuclear receptor that specifically imports MAGO-Y14 as a cargo into the nucleus.Citation58 In the cytoplasm, MAGO-Y14 associates with Imp13, which imports it into the nucleus and releases it there in a RanGTP-dependent manner. The crystal structures of Imp13 both in the cytoplasmic state (in a ternary complex with MAGO and Y14) and in the nuclear state (in a binary complex with RanGTP) give insights into how MAGO-Y14 is imported into the nucleus and how it is then released.Citation59 Imp13 forms a ring-like structure that encircles the MAGO-Y14 cargo with a conserved surface (). The structure shows that Imp13 binding blocks the surface of MAGO-Y14 that is used to bind other proteins including PYM and UPF3b (). This explains why PYM is left behind in the cytoplasm after EJC dissociation. The comparison of the Imp13-Mago-Y14 structure and the Imp13-RanGTP structure shows that upon RanGTP binding, Imp13 assumes a more closed conformation. Mago-Y14 and RanGTP are bound on opposite sides of the molecule. Superposition of the two complexes reveals that MAGO-Y14 and RanGTP are almost in a side-by-side position when bound to Imp13, but concomitant binding is not possible because of steric clashes. This provides a rationale for how RanGTP could dock to the N-terminus of Imp13 first and then release the MAGO-Y14 cargo in the nucleus via a steric hindrance mechanism.Citation59
Perspectives
At the global level it is still largely unknown how extensive the EJC's occupancy is on exon-exon junctions. Nor it is understood what the deciding factors are that determine at which junction an EJC should be positioned by the splicing machinery. In this context, the identification of additional spliceosomal components that are involved in the deposition of EJCs will be of importance.
Structural studies will be required to understand how the ribosome, the mRNA localization machinery and the surveillance machinery interplay at the mechanistic level. How eIF4AIII is imported back into the nucleus to be reincorporated into the EJC is still unknown and the specific import receptor involved in this process remains to be identified. The function of BTZ in the context of the EJC is also not fully understood and it is not clear whether this protein could shuttle across the nuclear envelope.
Dissecting the complex network of interactions stemming from the EJC will ultimately be required to fully understand the regulatory role of this complex in mRNA metabolism. How the EJC mediates downstream interactions is still partly an open question. This is of particular interest since the EJC peripherally associated proteins are involved in several aspects of mRNA metabolism. Biochemical and structural studies have revealed that exclusive and concomitant interactions are central to mRNP dynamics. Future investigations will no doubt unravel the sequentiality with which multifunctional surfaces are adopted (such as the Y14 interface that binds PYM, Upf3b and Imp13).
Figures and Tables
Figure 1 Interactions in the EJC cycle. The scheme shows the sequence of either simultaneous or mutually exclusive interactions underlying function of the EJC and its sub-complexes. The EJC is assembled during splicing onto the mRNA. After the mRNP is exported into the cytoplasm to function in translational regulation (ribosome in gray), the EJC is disassembled with the aid of a ribosomal-binding protein, PYM (in orange). The MAGo-Y14 heterodimer (in blue and cyan, respectively) is re-imported into the nucleus without PYM by Imp13 (in yellow). Here, it is released by RanGTP binding (gray/black rectangle) and incorporated into a spliced mRNP with eIF4AIII and BTZ (in pink and red, respectively) in the form of the EJC (other mRNP proteins are shown in gray with different shapes and sizes). mRNP composition varies dynamically as well as in the composition of the periphery of the EJC. UPF3b (in purple) associates to the EJC in the nucleus and travels with the complex to the cytoplasm. The question marks indicate open issues in the cycle: it is unclear whether eIF4AIII and BTZ are imported into the nucleus separately or as a single unit and the putative import receptor(s) has not been identified yet.
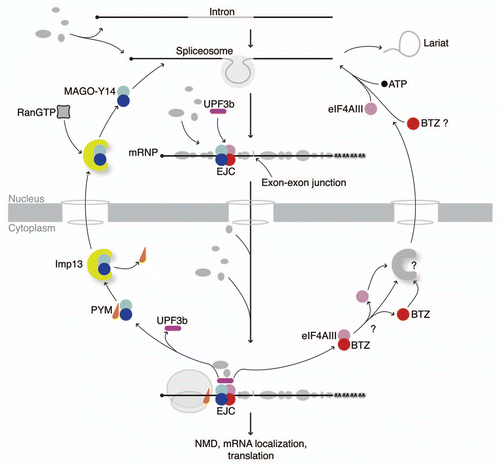
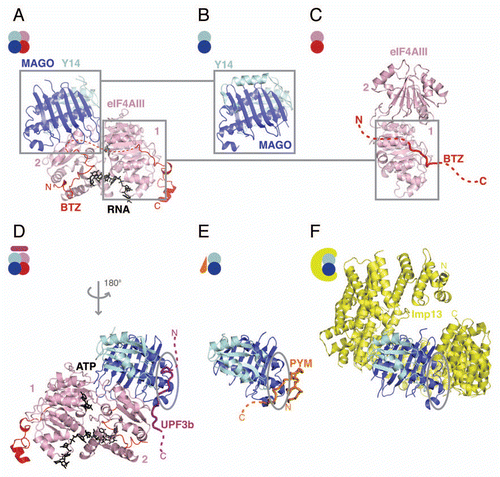
Figure 2 Structures in the EJC cycle. (A) Structure of the human EJC (pdb id. 2J0Q). eIF4AIII is shown in pink and BTZ in red, with ATP and RNA shown in black. The N- and C-terminal domains of eIF4AIII are labeled as 1 and 2, respectively. MAGO is shown in blue and Y14 in cyan. The disordered loop of BTZ is represented as a dashed red line. The N- and C-termini of the protein are labeled. The same color-coding is applied to all the panels and is consistent with the scheme in ; (B) Structure of the MAGO-Y14 heterodimer in isolation from Drosophila (pdb id. 1HL6) in a similar view as in (A); (C) Structure of the eIF4AIII-BTZ binary complex (pdb id. 2J0U in a similar view as in (A). The disordered portions of BTZ that could not be modeled in the structure are shown as a red dashed line; (D) Crystal structure of the EJC-UPF3b complex (pdb id. 2XB2). The complex is shown with a 180° rotation around the y-axis as compared with the orientation of the complex in (A). The UPF3b peptide is rendered as a purple cartoon tube. The dashed line represents parts of the protein that were not modeled in the structure. Highlighted with a gray circle is a conserved patch on MAGO-Y14 that is involved in protein-protein interactions also in (E and F). (E) Structure of the Drosophila MAGO-Y14-PYM complex (pdb id. 2RK8). PYM is rendered as a cartoon tube in orange. The N- and C-termini of the protein are labeled. (F) Structure of the Imp13-MAGO-Y14 complex with Imp13 in yellow (pdb id. 2X1G). The Imp13 N- and C-termini are indicated. This and all other protein structure figures were generated using PyMOL (www.pymol.org).
Acknowledgements
We would like to thank Atlanta Cook, Elisa Izaurralde and Gáspár Jékely, for critical reading of the manuscript. The authors are funded by the Max Planck Gesellshaft (F.B.) and by the Deutsche Forschungsgemeinschaft (BO3588/1-1 and GE2014/2-1). We apologize, in advance, to those investigators whose work was inadvertently overlooked.
References
- Dreyfuss G, Matunis MJ, Piñol-Roma S, Burd CG. hnRNP proteins and the biogenesis of mRNA. Annu Rev Biochem 1993; 62:289 - 321
- Moore MJ. From birth to death: the complex lives of eukaryotic mRNAs. Science 2005; 309:1514 - 1518
- Le Hir H, Izaurralde E, Maquat LE, Moore MJ. The spliceosome deposits multiple proteins 20–24 nucleotides upstream of mRNA exon-exon junctions. EMBO J 2000; 19:6860 - 6869
- Tange TØ, Nott A, Moore MJ. The ever-increasing complexities of the exon junction complex. Curr Opin Cell Biol 2004; 16:279 - 284
- Kim VN, Yong J, Kataoka N, Abel L, Diem MD, Dreyfuss G. The Y14 protein communicates to the cytoplasm the position of exon-exon junctions. The EMBO J 2001; 20:2062 - 2068
- Le Hir H, Gatfield D, Braun IC, Forler D, Izaurralde E. The protein Mago provides a link between splicing and mRNA localization. EMBO Rep 2001; 2:1119 - 1124
- Chan CC, Dostie J, Diem MD, Feng W, Mann M, Rappsilber J, et al. eIF4A3 is a novel component of the exon junction complex. RNA 2004; 10:200 - 209
- Degot S, Le Hir H, Alpy F, Kedinger V, Stoll I, Wendling C, et al. Association of the breast cancer protein MLN51 with the exon junction complex via its speckle localizer and RNA binding module. J Biol Chem 2004; 279:33702 - 33715
- Ferraiuolo MA, Lee CS, Ler LW, Hsu JL, Costa-Mattioli M, Luo MJ, et al. A nuclear translation-like factor eIF4AIII is recruited to the mRNA during splicing and functions in nonsense-mediated decay. Proc Natl Acad Sci USA 2004; 101:4118 - 4123
- Palacios IM, Gatfield D, St. Johnston D, Izaurralde E. An eIF4AIII-containing complex required for mRNA localization and nonsense-mediated mRNA decay. Nature 2004; 427:753 - 757
- Shibuya T, Tange TØ, Sonenberg N, Moore MJ. eIF4AIII binds spliced mRNA in the exon junction complex and is essential for nonsense-mediated decay. Nat Struct Mol Biol 2004; 11:346 - 351
- Ballut L, Marchadier B, Baguet A, Tomasetto C, Séraphin B, Le Hir H. The exon junction core complex is locked onto RNA by inhibition of eIF4AIII ATPase activity. Nat Struct Mol Biol 2005; 12:861 - 869
- Tange TØ, Shibuya T, Jurica MS, Moore MJ. Biochemical analysis of the E°C reveals two new factors and a stable tetrameric protein core. RNA 2005; 11:1869 - 1883
- Conti E, Izaurralde E. Nonsense-mediated mRNA decay: molecular insights and mechanistic variations across species. Curr Opin Cell Biol 2005; 17:316 - 325
- Chang YF, Imam JS, Wilkinson MF. The nonsense-mediated decay RNA surveillance pathway. Annu Rev Biochem 2007; 76:51 - 74
- Newmark PA, Boswell RE. The mago nashi locus encodes an essential product required for germ plasm assembly in Drosophila. Development 1994; 120:1303 - 1313
- Micklem DR, Dasgupta R, Elliott H, Gergely F, Davidson C, Brand A, et al. The mago nashi gene is required for the polarisation of the oocyte and the formation of perpendicular axes in Drosophila. Curr Biol 1997; 7:468 - 478
- Hachet O, Ephrussi A. Drosophila Y14 shuttles to the posterior of the oocyte and is required for oskar mRNA transport. Curr Biol 2001; 11:1666 - 1674
- Mohr SE, Dillon ST, Boswell RE. The RNA-binding protein Tsunagi interacts with Mago Nashi to establish polarity and localize oskar mRNA during Drosophila oogenesis. Genes Dev 2001; 15:2886 - 2899
- van Eeden FJ, Palacios IM, Petronczki M, Weston MJ, St. Johnston D. Barentsz is essential for the posterior localization of oskar mRNA and colocalizes with it to the posterior pole.. J Cell Biol 2001; 154:511 - 523
- Hachet O, Ephrussi A. Splicing of oskar RNA in the nucleus is coupled to its cytoplasmic localization. Nature 2004; 428:959 - 963
- Gatfield D, Unterholzner L, Ciccarelli FD, Bork P, Izaurralde E. Nonsense-mediated mRNA decay in Drosophila: at the intersection of the yeast and mammalian pathways. EMBO J 2003; 22:3960 - 3970
- Wiegand HL, Lu S, Cullen BR. Exon junction complexes mediate the enhancing effect of splicing on mRNA expression. Proc Natl Acad Sci USA 2003; 100:11327 - 11332
- Nott A, Le Hir H, Moore MJ. Splicing enhances translation in mammalian cells: an additional function of the exon junction complex. Genes Dev 2004; 18:210 - 222
- Gudikote JP, Imam JS, Garcia RF, Wilkinson MF. RNA splicing promotes translation and RNA surveillance. Nat Struct Mol Biol 2005; 12:801 - 809
- Diem MD, Chan CC, Younis I, Dreyfuss G. PYM binds the cytoplasmic exon-junction complex and ribosomes to enhance translation of spliced mRNAs. Nat Struct Mol Biol 2007; 14:1173 - 1179
- Ma XM, Yoon SO, Richardson CJ, Jülich K, Blenis J. SKAR links pre-mRNA splicing to mTOR/S6K1-mediated enhanced translation efficiency of spliced mRNAs. Cell 2008; 133:303 - 313
- Le Hir H, Nott A, Moore MJ. How introns influence and enhance eukaryotic gene expression. Trends Biochem Sci 2003; 28:215 - 220
- Giorgi C, Moore MJ. The nuclear nurture and cytoplasmic nature of localized mRNPs. Semin Cell Dev Biol 2007; 18:186 - 193
- Ideue, Sasaki, Hagiwara, Hirose. Introns play an essential role in splicing-dependent formation of the exon junction complex. Genes Dev 2007; 21:1993 - 1998
- Bessonov S, Anokhina M, Will CL, Urlaub H, Lührmann R. Isolation of an active step I spliceosome and composition of its RNP core. Nature 2008; 452:846 - 850
- Mishler DM, Christ AB, Steitz JA. Flexibility in the site of exon junction complex deposition revealed by functional group and RNA secondary structure alterations in the splicing substrate. RNA 2008; 14:2657 - 2670
- Gehring NH, Lamprinaki S, Hentze MW, Kulozik AE. The hierarchy of exon-junction complex assembly by the spliceosome explains key features of mammalian nonsense-mediated mRNA decay. PLoS Biol 2009; 7:1000120
- Deckert J, Hartmuth K, Boehringer D, Behzadnia N, Will CL, Kastner B, et al. Protein composition and electron microscopy structure of affinity-purified human spliceosomal B complexes isolated under physiological conditions. Mol Cell Biol 2006; 26:5528 - 5543
- Macchi P, Kroening S, Palacios IM, Baldassa S, Grunewald B, Ambrosino C, et al. Barentsz, a new component of the Staufen-containing ribonucleoprotein particles in mammalian cells, interacts with Staufen in an RNA-dependent manner. J Neurosci 2003; 23:5778 - 5788
- Herold N, Will CL, Wolf E, Kastner B, Urlaub H, Lührmann R. Conservation of the protein composition and electron microscopy structure of Drosophila melanogaster and human spliceosomal complexes. Mol Cell Biol 2009; 29:281 - 301
- Merz C, Urlaub H, Will CL, Lührmann R. Protein composition of human mRNPs spliced in vitro and differential requirements for mRNP protein recruitment. RNA 2007; 13:116 - 128
- Zhang Z, Krainer AR. Splicing remodels messenger ribonucleoprotein architecture via eIF4A3-dependent and -independent recruitment of exon junction complex components. Proc Natl Acad Sci USA 2007; 104:11574 - 11579
- Andersen CBF, Ballut L, Johansen JS, Chamieh H, Nielsen KH, Oliveira CLP, et al. Structure of the exon junction core complex with a trapped DEAD-box ATPase bound to RNA. Science 2006; 313:1968 - 1972
- Bono F, Ebert J, Lorentzen E, Conti E. The crystal structure of the exon junction complex reveals how it maintains a stable grip on mRNA. Cell 2006; 126:713 - 725
- Cordin O, Banroques J, Tanner NK, Linder P. The DEAD-box protein family of RNA helicases. Gene 2006; 367:17 - 37
- Hilbert M, Karow AR, Klostermeier D. The mechanism of ATP-dependent RNA unwinding by DEAD box proteins. Biol Chem 2009; 390:1237 - 1250
- Noble CG, Song H. MLN51 stimulates the RNA-helicase activity of eIF4AIII. PLoS ONE 2007; 2:303
- Nielsen KH, Chamieh H, Andersen CBF, Fredslund F, Hamborg K, Le Hir H, et al. Mechanism of ATP turnover inhibition in the EJC. RNA 2009; 15:67 - 75
- Gehring NH, Neu-Yilik G, Schell T, Hentze MW, Kulozik AE. Y14 and hUpf3b form an NMD-activating complex. Mol Cell 2003; 11:939 - 949
- Chamieh H, Ballut L, Bonneau F, Le Hir H. NMD factors UPF2 and UPF3 bridge UPF1 to the exon junction complex and stimulate its RNA helicase activity. Nat Struct Mol Biol 2008; 15:85 - 93
- Buchwald G, Ebert J, Basquin C, Sauliere J, Jayachandran U, Bono F, et al. Insights into the recruitment of the NMD machinery from the crystal structure of a core EJC-UPF3b complex. Proc Natl Acad Sci USA 2010; 107:10050 - 10055
- Dostie J, Dreyfuss G. Translation is required to remove Y14 from mRNAs in the cytoplasm. Curr Biol 2002; 12:1060 - 1067
- Lejeune F, Ishigaki Y, Li X, Maquat LE. The exon junction complex is detected on CBP80-bound but not eIF4E-bound mRNA in mammalian cells: dynamics of mRNP remodeling. EMBO 2002; 21:3536 - 3545
- Fribourg S, Gatfield D, Izaurralde E, Conti E. A novel mode of RBD-protein recognition in the Y14-Mago complex. Nat Struct Biol 2003; 10:433 - 439
- Lau C-K, Diem MD, Dreyfuss G, Van Duyne GD. Structure of the Y14-Magoh core of the exon junction complex. Curr Biol 2003; 13:933 - 941
- Shi H, Xu RM. Crystal structure of the Drosophila Mago nashi-Y14 complex. Genes Dev 2003; 17:971 - 976
- Forler D, Köcher T, Rode M, Gentzel M, Izaurralde E, Wilm M. An efficient protein complex purification method for functional proteomics in higher eukaryotes. Nat Biotechnol 2003; 21:89 - 92
- Bono F, Ebert J, Unterholzner L, Güttler T, Izaurralde E, Conti E. Molecular insights into the interaction of PYM with the Mago-Y14 core of the exon junction complex. EMBO Rep 2004; 5:304 - 310
- Gehring NH, Lamprinaki S, Kulozik AE, Hentze MW. Disassembly of exon junction complexes by PYM. Cell 2009; 137:536 - 548
- Cook A, Bono F, Jinek M, Conti E. Structural biology of nucleocytoplasmic transport. Annu Rev Biochem 2007; 76:647 - 671
- Mingot JM, Kostka S, Kraft R, Hartmann E, Görlich D. Importin 13: a novel mediator of nuclear import and export. EMBO J 2001; 20:3685 - 3694
- Bono F, Cook AG, Grunwald M, Ebert J, Conti E. Nuclear Import Mechanism of the EJC Component Mago-Y14 Revealed by Structural Studies of Importin 13. Mol Cell 2010; 37:211 - 222