Abstract
3’ processing of mRNA precursors is frequently coupled to transcription by RNA polymerase II (RNAP II). This coupling is well known to involve the C-terminal domain of the RNAP II largest subunit, but a variety of other transcription-associated factors have also been suggested to mediate coupling. Our recent studies have provided direct evidence that transcriptional activators can enhance the efficiency of transcription-coupled 3’ processing. In this point-of-view, we discuss the mechanisms that underlie coupling, and their implications for control of alternative polyadenylation, which is emerging as a significant regulator of cell growth control.
3′ end formation of nearly all mRNA precursors in eukaryotic cells involves an endonucleolytic cleavage followed by synthesis of the poly(A) tail.Citation1 This is achieved by a macromolecular complex assembled onto the nascent pre-mRNA through the recognition of RNA sequences that constitute the poly(A) site.Citation2 This complex contains not only core polyadenylation factors such as cleavage-polyadenylation specificity factor (CPSF), cleavage stimulatory factor (CstF), cleavage factor I and II (CFI and CFII) and poly(A) polymerase, but also protein factors involved in transcription, splicing and translation.Citation2
Considerable evidence now indicates that polyadenylation occurs cotranscriptionally.Citation3–Citation6 The resulting coupling of transcription and 3′ processing is believed to ensure accurate, efficient and rapid processing of nascent pre-mRNAs. An important factor in this coupling is the carboxy-terminal domain of the RNA polymerase II (RNAP II) largest subunit (CTD), which has a significant role in enhancing the efficiency of 3′ processing in both in vivo and in vitro assays.Citation7,Citation8 In addition to the CTD, general transcription factors (GTFs) also function in transcription/polyadenylation coupling. For example, TFIID associates with and recruits CPSF to the preinitiation complex. After transcription initiates, CPSF is loaded to the elongating RNAP II, likely via the CTD.Citation9 Furthermore, the connection between transcription initiation and 3′ processing is not unidirectional, as 3′ processing has been shown to stimulate transcription initiation in a manner that involves the GTFs TFIIB and TBP.Citation10
Transcription elongation factors also help to coordinate cotranscriptional polyadenylation. Positive transcription elongation factor b (P-TEFb, similar to Ctk1 in yeast) is a CTD kinase responsible for phosphorylation of ser2 residues in the CTD heptad repeats.Citation11,Citation12 Ser2 phosphorylation has been shown to be important for CTD recruitment of 3′ processing factors to the site of processing on many genes.Citation13,Citation14 Another example is the PAF1 complex (PAF1c), which functions during elongation in a number of ways. For example, on chromatin templates in vitro it acts synergistically to enhance elongation with another elongation factor, TFIIS.Citation15 We showed previously that PAF1c associates with 3′ processing factors and functions in transcription-coupled polyadenylation. Depletion of PAF1c from cell extracts in vitro inhibited polyadenylation but not transcription or splicing, and from cells in vivo reduced expression and extended transcription of a target gene.Citation16 The interaction between 3′ processing factors and PAF1c is also observed in yeast, and poly(A) tail lengthCitation17 and poly(A) site selectionCitation18 were found to be altered in PAF mutant cells. These findings all support a role for PAF1c in helping to couple transcription to 3′ processing.
Interconnections between transcription termination and pre-mRNA 3′ processing have also been described.Citation19 The CFII component Pcf11 is capable of dismantling the RNAP II elongation complex in vitro via the CTD,Citation20–Citation23 consistent with a role in termination. The multifunctional p54nrb/PSF dimer promotes recruitment of the exonuclease XRN2, which participates in 3′ processing and termination.Citation24 Our recent work has also revealed that the CTD phosphatase Ssu72, which dephosphorylates Ser5 and has a role in efficient transcription termination of some genes in yeast,Citation25,Citation26 is specifically necessary for transcription-coupled polyadenylation in HeLa cell extracts.Citation27 An interaction between catalytically active Ssu72 and the core 3′ processing factor symplekin was found to be required for transcription-coupled (but not uncoupled) polyadenylation.
The N-terminal region of symplekin may provide a scaffold that mediates the coupling between transcription and polyadenylation. The region has the ability to associate with RNAP II,Citation27 and has been shown to function in HSF1-dependent expression of heat shock responsive genes.Citation28 Heat shock of human K562 cells induces the interaction between the transcriptional activator HSF1 and the N-terminal region of symplekin, and overexpression of a mutant form of HSF1 defective in binding to DNA templates inhibits 3′ processing of Hsp70 mRNA in heat-shocked cells. This observation suggests that sequence-specific transcription factors (TFs) can function in transcription-coupled polyadenylation. In support of this, transient cotransfection assays showed that TFs can indeed increase not only transcription but also 3′-end processing.Citation29
Recently, we provided evidence that TFs can directly stimulate the efficiency of transcription-coupled 3′ processing, and that this stimulation requires the PAF1c.Citation30 We showed that the prototypical transcription activator GAL4-VP16 (as well as a similar GAL4-p53 fusion protein) strongly induced transcription-coupled polyadenylation in HeLa nuclear extract (NE) using naked DNA templates, and that depletion of PAF1c markedly reduced VP16-mediated polyadenylation, but not transcription, in vitro. Furthermore, depletion of PAF subunits by RNAi in vivo resulted in decreased 3′ end cleavage, and nuclear export, of a reporter transcript. Finally, the VP16 activation domain was shown to interact directly with PAF1c and to recruit it to DNA templates in NE, thereby enhancing transcription-coupled polyadenylation.Citation30 These findings provide yet another mechanism for coupling transcription and 3′ processing, further integrating these two processes to enhance the efficiency of gene expression. And, as we discuss below, the ability of TFs to affect the efficiency of 3′ processing offers additional mechanisms for gene control via alternative mRNA processing.
Utilization of alternative polyadenylation (APA) sites is emerging as a widespread mechanism for control of gene expression during cell proliferation and differentiation. Several recent global studies have provided evidence that rapidly proliferating cells, including cancer cells, tend to use promoter-proximal poly(A) sites, while non-proliferating or slowly growing cells favor distal sites.Citation31–Citation33 Importantly, this provides cells a mechanism to alter gene expression either qualitatively (if the APA sites affect coding sequences) or quantitatively (by the inclusion or exclusion of repressive sites in affected 3′ UTR sequences). Thus, the use of proximal sites that occurs in rapidly growing cells will increase protein expression.
Multiple mechanisms are likely to control APA. One established mechanism involves changes in the concentrations of core 3′ processing factors. For example, we showed previously that changes in the levels of CstF influence APA of the IgM heavy chain pre-mRNA during B-cell activation.Citation34,Citation35 Specifically, CstF levels were found to increase dramatically during activation, and this was sufficient to switch APA from the stronger, distal site to the weaker, proximal site. Importantly, the much more recent bioinformaic analyses have also observed changes in levels of 3′ processing factors, and that proximal poly(A) sites tend to be weaker than distal sites. Additionally, levels of CFI have also been shown to alter poly(A) site use of several genes.Citation36,Citation37 Therefore, it may be that changes in the levels or activity of polyadenylation factors constitute a general mechanism for regulating APA.
RNA binding proteins that function as splicing factors can also regulate APA. For example, Nova2, a neuron specific splicing factor, has been shown to bind not only to coding region but also to 3′ UTR of mRNAs, and knockout of Nova2 leads to APA in brain.Citation38 PGC-1, a transcriptional coactivator that contains RS domains characteristic of SR proteins, is another good candidate to regulate APA. PGC-1 is recruited to gene promoters through direct interactions with sequence-specific TFs, and enhances transcription by associating with the histone acetyltransferase p300.Citation39 PGC-1 also associates with RNAP II and splicing factors, and modulates alternative pre-mRNA splicing when loaded onto the promoter region of a target gene.Citation40 Since PGC-1 binds not only to splicing factors but also to polyadenylation factor CPSF100,Citation40 it may well also function to modulate APA in a transcription-coupled manner.
Extending the link between transcription and APA, expression levels of the transcription elongation factor ELL2 have been found to contribute to APA of the IgM heavy chain mRNA, in conjunction with CstF.Citation41 Overexpressed ELL2 stimulates CstF loading to the IgM promoter, which enhances processing at the proximal poly(A) site, while lowering ELL2 expression decreased use of this site. ChIP analysis showed that ELL2 and CstF track together with RNAP II across the gene locus. These results indicate that APA of IgM pre-mRNA can be controlled in part by a transcription-coupled mechanism.
The increased polyadenylation efficiency brought about TFs also has the potential to influence APA (). Specifically, the increased efficiency would be predicted to favor use of proximal poly(A) sites. Indeed, a recent bioinformatics analysis provides important support for this idea: Upregulated genes tend to use promoter-proximal poly(A) sites, while downregulated ones prefer distal poly(A) sites (B. Tian, personal communication). Additionally, it is likely that PAF1c participates in APA control. PAF1c mutants in yeast are known to alter poly(A) site utilization for certain genes,Citation18 while in humans siRNA-mediated depletion of the Cdc73 subunit was found to reduce expression and extend transcription of a target gene, Ints6, likely favoring utilization of downstream poly(A) sites.Citation16 Intriguingly, low expression of PAF1c and its translocation to the cytoplasm were observed in differentiating cells,Citation42,Citation43 which might explain at least in part why distal poly(A) sites are favored in these cells.
Finally, we speculate that the network coupling transcription and 3′ processing can be extended by taking into consideration the role of chromatin. For example, PAF1c is required for histone lysine methylation, such as H3-K4, -K36, -K79,Citation44,Citation45 and immunopurified PAF1c contains H3K4 methyltransferase subunits in addition to polyadenylation factors.Citation16 Furthermore, the H3K4 methyltransferase ALL1 assembles into protein complexes that include polyadenylation factors,Citation46 suggesting a possible link between H3K4 methylation and polyadenylation. Since PAF1c is necessary for H3K4 methylation and expression of the ALL1-target gene HoxA9,Citation47,Citation48 an intriguing possibility is that H3K4 methylation directed by PAF1c contributes to transcription-coupled polyadenylation of HoxA9 pre-mRNA. Establishing in vitro transcription-coupled polyadenylation assays with chromatin templates should provide further insight into the mechanism by which transcriptional activators direct 3′ processing. In any event, it is now clear that numerous distinct factors contribute to the coupling of transcription and polyadenylation, reflecting the important role that regulated 3′ end formation plays in the control of gene expression.
Figures and Tables
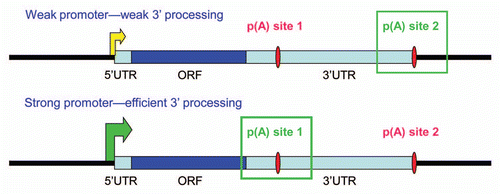
Figure 1 Transcriptional activation and poly(A) site use. In many mRNAs containing multiple poly(A) sites, promoter-proximal sites are weaker than distal ones. Weakly transcribed genes, or genes on which transcription and 3′ processing are poorly coupled, will tend to polyadenylate less efficiently, leading to preferential use of distal poly(A) sites. This will result in long 3′ UTRs containing repressive sites, thus lowering protein expression. In contrast, strong transcriptional activation and the resulting enhanced 3′ processing efficiency will lead to use of proximal poly(A) sites, shorter 3′ UTRS and increased protein expression.
Acknowledgments
Work from the authors' lab is supported by grants from the NIH (J.L.M.).
References
- Colgan DF, Manley JL. Mechanism and regulation of mRNA polyadenylation. Genes Dev 1997; 11:2755 - 2766
- Shi Y, Di Giammartino DC, Taylor D, Sarkeshik A, Rice WJ, Yates JR 3rd, et al. Molecular architecture of the human pre-mRNA 3′ processing complex. Mol Cell 2009; 33:365 - 376
- Hirose Y, Manley JL. RNA polymerase II and the integration of nuclear events. Genes Dev 2000; 14:1415 - 1429
- Bentley D. The mRNA assembly line: transcription and processing machines in the same factory. Curr Opin Cell Biol 2002; 14:336 - 342
- Maniatis T, Reed R. An extensive network of coupling among gene expression machines. Nature 2002; 416:499 - 506
- Proudfoot NJ, Furger A, Dye MJ. Integrating mRNA processing with transcription. Cell 2002; 108:501 - 512
- McCracken S, Fong N, Yankulov K, Ballantyne S, Pan G, Greenblatt J, et al. The C-terminal domain of RNA polymerase II couples mRNA processing to transcription. Nature 1997; 385:357 - 561
- Hirose Y, Manley JL. RNA polymerase II is an essential mRNA polyadenylation factor. Nature 1998; 395:93 - 96
- Dantonel JC, Murthy KG, Manley JL, Tora L. Transcription factor TFIID recruits factor CPSF for formation of 3′ end of mRNA. Nature 1997; 389:399 - 402
- Mapendano CK, Lykke-Andersen S, Kjems J, Bertrand E, Jensen TH. Crosstalk between mRNA 3′ end processing and transcription initiation. Mol Cell 2010; 40:410 - 422
- Cho EJ, Kobor MS, Kim M, Greenblatt J, Buratowski S. Opposing effects of Ctk1 kinase and Fcp1 phosphatase at Ser 2 of the RNA polymerase II C-terminal domain. Genes Dev 2001; 15:3319 - 3329
- Shilatifard A, Conaway RC, Conaway JW. The RNA polymerase II elongation complex. Annu Rev Biochem 2003; 72:693 - 715
- Ahn SH, Kim M, Buratowski S. Phosphorylation of serine 2 within the RNA polymerase II C-terminal domain couples transcription and 3′ end processing. Mol Cell 2004; 13:67 - 76
- Ni Z, Schwartz BE, Werner J, Suarez JR, Lis JT. Coordination of transcription, RNA processing and surveillance by P-TEFb kinase on heat shock genes. Mol Cell 2004; 13:55 - 65
- Kim J, Guermah M, Roeder RG. The human PAF1 complex acts in chromatin transcription elongation both independently and cooperatively with SII/TFIIS. Cell 2010; 140:491 - 503
- Rozenblatt-Rosen O, Nagaike T, Francis JM, Kaneko S, Glatt KA, Hughes CM, et al. The tumor suppressor Cdc73 functionally associates with CPSF and CstF 3′ mRNA processing factors. Proc Natl Acad Sci USA 2009; 106:755 - 760
- Mueller CL, Porter SE, Hoffman MG, Jaehning JA. The Paf1 complex has functions independent of actively transcribing RNA polymerase II. Mol Cell 2004; 14:447 - 456
- Penheiter KL, Washburn TM, Porter SE, Hoffman MG, Jaehning JA. A posttranscriptional role for the yeast Paf1-RNA polymerase II complex is revealed by identification of primary targets. Mol Cell 2005; 20:213 - 223
- Richard P, Manley JL. Transcription termination by nuclear RNA polymerases. Genes Dev 2009; 23:1247 - 1269
- Birse CE, Minvielle-Sebastia L, Lee BA, Keller W, Proudfoot NJ. Coupling termination of transcription to messenger RNA maturation in yeast. Science 1998; 280:298 - 301
- Zhang Z, Fu J, Gilmour DS. CTD-dependent dismantling of the RNA polymerase II elongation complex by the pre-mRNA 3′-end processing factor, Pcf11. Genes Dev 2005; 19:1572 - 1580
- Zhang Z, Gilmour DS. Pcf11 is a termination factor in Drosophila that dismantles the elongation complex by bridging the CTD of RNA polymerase II to the nascent transcript. Mol Cell 2006; 21:65 - 74
- West S, Proudfoot NJ. Human Pcf11 enhances degradation of RNA polymerase II-associated nascent RNA and transcriptional termination. Nucleic Acids Res 2008; 36:905 - 914
- Kaneko S, Rozenblatt-Rosen O, Meyerson M, Manley JL. The multifunctional protein p54nrb/ PSF recruits the exonuclease XRN2 to facilitate premRNA 3′ processing and transcription termination. Genes Dev 2007; 21:1779 - 1789
- Ganem C, Devaux F, Torchet C, Jacq C, Quevillon-Cheruel S, Labesse G, et al. Ssu72 is a phosphatase essential for transcription termination of snoRNAs and specific mRNAs in yeast. EMBO J 2003; 22:1588 - 1598
- Steinmetz EJ, Brow DA. Ssu72 protein mediates both poly(A)-coupled and poly(A)-independent termination of RNA polymerase II transcription. Mol Cell Biol 2003; 23:6339 - 6349
- Xiang K, Nagaike T, Xiang S, Kilic T, Beh MM, Manley JL, et al. Crystal structure of the human symplekin-Ssu72-CTD phosphopeptide complex. Nature 2010; 467:729 - 733
- Xing H, Mayhew CN, Cullen KE, Park-Sarge OK, Sarge KD. HSF1 modulation of Hsp70 mRNA polyadenylation via interaction with symplekin. J Biol Chem 2004; 279:10551 - 10555
- Rosonina E, Bakowski MA, McCracken S, Blencowe BJ. Transcriptional activators control splicing and 3′-end cleavage levels. J Biol Chem 2003; 278:43034 - 43040
- Nagaike T, Logan C, Hotta I, Rozenblatt-Rosen O, Meyerson M, Manley JL. Transcriptional activators enhance polyadenylation of mRNA precursors. Mol Cell 2011; 41:409 - 418
- Sandberg R, Neilson JR, Sarma A, Sharp PA, Burge CB. Proliferating cells express mRNAs with shortened 3′ untranslated regions and fewer microRNA target sites. Science 2008; 320:1643 - 1647
- Ji Z, Lee JY, Pan Z, Jiang B, Tian B. Progressive lengthening of 3′ untranslated regions of mRNAs by alternative polyadenylation during mouse embryonic development. Proc Natl Acad Sci USA 2009; 106:7028 - 7033
- Mayr C, Bartel DP. Widespread shortening of 3′UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell 2009; 138:673 - 684
- Takagaki Y, Seipelt RL, Peterson ML, Manley JL. The polyadenylation factor CstF-64 regulates alternative processing of IgM heavy chain pre-mRNA during B cell differentiation. Cell 1996; 87:941 - 952
- Takagaki Y, Manley JL. Levels of polyadenylation factor CstF-64 control IgM heavy chain mRNA accumulation and other events associated with B cell differentiation. Mol Cell 1998; 2:761 - 771
- Kubo T, Wada T, Yamaguchi Y, Shimizu A, Handa H. Knock-down of 25 kDa subunit of cleavage factor Im in Hela cells alters alternative polyadenylation within 3′-UTRs. Nucleic Acids Res 2006; 34:6264 - 6271
- Kim S, Yamamoto J, Chen Y, Aida M, Wada T, Handa H, et al. Evidence that cleavage factor Im is a heterotetrameric protein complex controlling alternative polyadenylation. Genes Cells 2010; 15:1003 - 1013
- Licatalosi DD, Mele A, Fak JJ, Ule J, Kayikci M, Chi SW, et al. HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature 2008; 456:464 - 469
- Handschin C, Spiegelman BM. Peroxisome proliferator-activated receptor gamma coactivator 1 coactivators, energy homeostasis and metabolism. Endocr Rev 2006; 27:728 - 735
- Monsalve M, Wu Z, Adelmant G, Puigserver P, Fan M, Spiegelman BM. Direct coupling of transcription and mRNA processing through the thermogenic coactivator PGC-1. Mol Cell 2000; 6:307 - 316
- Martincic K, Alkan SA, Cheatle A, Borghesi L, Milcarek C. Transcription elongation factor ELL2 directs immunoglobulin secretion in plasma cells by stimulating altered RNA processing. Nat Immunol 2009; 10:1102 - 1109
- Agarwal SK, Simonds WF, Marx SJ. The parafibromin tumor suppressor protein interacts with actinbinding proteins actinin-2 and actinin-3. Mol Cancer 2008; 7:65
- Ding L, Paszkowski-Rogacz M, Nitzsche A, Slabicki MM, Heninger AK, de Vries I, et al. A genome-scale RNAi screen for Oct4 modulators defines a role of the Paf1 complex for embryonic stem cell identity. Cell Stem Cell 2009; 4:403 - 415
- Krogan NJ, Kim M, Tong A, Golshani A, Cagney G, Canadien V, et al. Methylation of histone H3 by Set2 in Saccharomyces cerevisiae is linked to transcriptional elongation by RNA polymerase II. Mol Cell Biol 2003; 23:4207 - 4218
- Zhu B, Mandal SS, Pham AD, Zheng Y, Erdjument-Bromage H, Batra SK, et al. The human PAF complex coordinates transcription with events downstream of RNA synthesis. Genes Dev 2005; 19:1668 - 1673
- Nakamura T, Mori T, Tada S, Krajewski W, Rozovskaia T, Wassell R, et al. ALL-1 is a histone methyltransferase that assembles a supercomplex of proteins involved in transcriptional regulation. Mol Cell 2002; 10:1119 - 1128
- Zhu B, Zheng Y, Pham AD, Mandal SS, Erdjument-Bromage H, Tempst P, et al. Monoubiquitination of human histone H2B: the factors involved and their roles in HOX gene regulation. Mol Cell 2005; 20:601 - 611
- Muntean AG, Tan J, Sitwala K, Huang Y, Bronstein J, Connelly JA, et al. The PAF complex synergizes with MLL fusion proteins at HOX loci to promote leukemogenesis. Cancer Cell 17:609 - 621