Abstract
In epithelial cells, myosin-II-dependent forces regulate many aspects of animal morphogenesis, such as apical constriction, cell intercalation, cell sorting, and the formation and maintenance of the adherens junction. These forces are mainly generated by the circumferential actomyosin belt, which is composed of F-actin–myosin II bundles located along apical cell–cell junctions. Although several of the molecular pathways regulating the belt have been identified, the precise mechanisms underlying its function are largely unknown. Our recent studies identified Lulu proteins (Lulu1 and Lulu2), FERM-domain-containing molecules, as the regulators of the belt. Lulus activate the circumferential actomyosin belt and thereby induce apical constriction in epithelial cells; conversely, RNAi-mediated Lulu-knockdown results in the severe disorganization of the circumferential actomyosin belt. We also showed that p114RhoGEF is a downstream molecule of Lulu2 in its regulation of the belt; Lulu2 enhances the catalytic activity of p114RhoGEF through a direct interaction and thereby activates the circumferential actomyosin belt. We further identified aPKC and Patj as regulators of Lulu2-p114RhoGEF. In this commentary, we discuss current knowledge of the circumferential actomyosin belt's regulation, focusing on the Lulu2-p114RhoGEF system.
Keywords: :
Introduction
Epithelial cell sheets dynamically change their shape as part of normal animal morphogenesis. This shape change is thought to be performed by individual epithelial cells under the regulation of myosin-II-driven mechanical forces, which are mainly generated by the circumferential actomyosin belt ().Citation1-Citation3 The F-actin–myosin II bundles that comprise the belt are positioned at the apical portion of epithelial cells, along apical cell–cell boundaries ().Citation4-Citation6 Through its connections to apical junctional complexes, namely the zonula adherens (ZA) and tight junctions (TJs), the circumferential actomyosin belt serves as a force-transmitter between neighboring cells and thereby integrates individual cell behaviors into overall shape changes of epithelial cell sheets during animal morphogenesis.
Apical constriction, a well-studied shape change process of epithelial cells, triggers the invagination and folding of epithelial cell sheets by altering the morphology of individual cells from cuboidal to bottle-shaped.Citation7-Citation11 One of the mechanisms inducing apical constriction is the myosin-II-dependent contractile force generated by the circumferential actomyosin belt (), in which Rho-associated kinase (Rock), an effecter of RhoA, plays a central role.Citation8-Citation14 Rock is thought to regulate the phosphorylation state of myosin regulatory light chains, thereby activating myosin-II-driven contractile forces. Regulation of the Rock–myosin II pathway in apical constriction is not completely understood; but recent studies identified Shroom3, an actin-binding protein, as an upstream regulator.Citation10-Citation13 Shroom3 was shown to recruit Rock to apical cell–cell junctions, thereby inducing apical constriction.Citation13 However, Shroom3 is obviously not a sole molecule capable of inducing apical constriction in epithelial cells; and we found that Lulu molecules also have an ability to induce apical constriction in epithelial cells.Citation14
In vertebrates, there are two Lulus, Lulu1 and Lulu2 (also known as Epb41l5 and Epb41l4b/Ehm2, respectively), which have well conserved FERM and FERM adjacent domains, although regions beyond these domains do not resemble each other (). Lulu1 mutant mice exhibit embryonic lethality, with defects in gastrulation and folding of the neural plate.Citation15-Citation17 Yurt, a counterpart of the Lulu proteins in Drosophila (), also regulates several morphogenetic processes, including head involution and germ band elongation, which have been attributed to negative regulation of apical membrane size in epithelial cells via the inhibition of Crumbs activity.Citation18,Citation19 We showed however, that mammalian Lulus regulate the circumferential actomyosin belt.Citation14 Specifically, the overexpression of Lulus in epithelial cells results in strong upregulation of the belt, thereby inducing apical constriction.
Lulu2 Interacts with p114RhoGEF
As the activity of Lulu2 is much stronger than that of Lulu1 in the induction of apical constriction, we studied Lulu2 in further detail.Citation20 Lulu2 is localized along apical cell–cell boundaries in polarized epithelial cells and, as expected, its depletion by RNAi results in disorganization of the circumferential actomyosin belt. However, Lulu2 depletion did not apparently affect the accumulation of TJ components to apical cell–cell boundaries, suggesting that Lulu2 does not regulate TJ integrity.
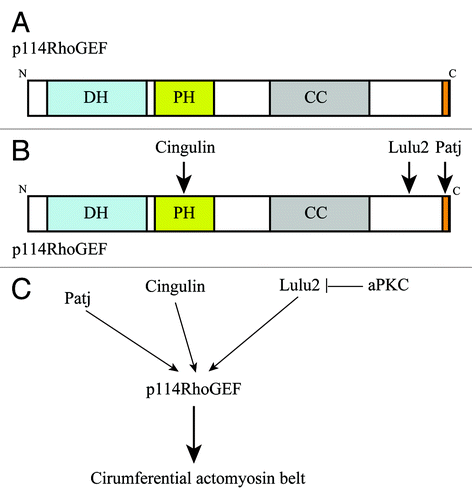
We previously determined that Shroom3 is dispensable for Lulus-induced apical constriction.Citation14 Those studies suggested that Lulus regulate apical constriction independently of Shroom3 through as yet unidentified molecular pathways. To uncover these pathways, we searched for interacting molecules of Lulu2 by a GST pull-down assay followed by MS/MS analysis, resulting in the identification of p114RhoGEF, a Rho-specific GEF, as a candidate (). p114RhoGEF was recently shown to regulate RhoA activity at apical cell–cell boundaries, thereby regulating the circumferential actomyosin belt in epithelial cells.Citation21 p114RhoGEF was also reported to play a role in neurite extension, by interacting with disheveled and DAAM1 in neuroblastoma cells,Citation22 as well as stress-fiber formation and reactive oxygen species production, through binding to Septin9 or the Gβγ subunit of heterotrimeric G proteins in fibroblasts.Citation23,Citation24 We confirmed the interaction between Lulu2 and p114RhoGEF by GST pull-down and co-immunoprecipitation assays. Lulu2 was co-localized with p114RhoGEF along apical cell–cell boundaries in epithelial cells, where, according to an in situ proximity assay, they appear to interact with each other. Furthermore, p114RhoGEF-depletion by RNAi caused severe disorganization of the circumferential actomyosin belt. As a functional link between Lulu2 and p114RhoGEF, it was found that p114RhoGEFis necessary for apical constriction driven by Lulu2 overexpression, suggesting that Lulu2 regulates the belt through p114RhoGEF.
Activation Mechanism of p114RhoGEF by Lulu2
As Lulu2 depletion by RNAi in epithelial cells did not markedly change the cellular localization of p114RhoGEF, Lulu2 is unlikely to serve as an anchoring molecule of p114RhoGEF at apical cell–cell boundaries. We thus postulated that Lulu2 directly activates p114RhoGEF. In experiments examining whether purified Lulu2 could enhance the catalytic activity of purified p114RhoGEF in vitro, we found that the guanine nucleotide exchange reaction between p114RhoGEF and RhoA was enhanced in the presence of Lulu2. In its N-terminal half, p114RhoGEF has Dbl homology (DH) and Pleckstrin homology (PH) domains, which are necessary for its catalytic activity, while its C-terminal half contains a coiled-coil region and a PDZ domain-binding motif (PBM). We identified the Lulu2-binding site on p114RhoGEF in its C-terminal half, specifically, within the region C-terminal to the coiled-coil region without the PBM. Therefore, the C-terminal portion of p114RhoGEF may serve as an intramolecular inhibitory region, with Lulu2 attenuating inhibition; however, this might not be the case because, at least in vitro, the N-terminal catalytic domain of full-length p114RhoGEF exhibits comparable catalytic activity (unpublished data). Thus, at this point we can speculate that the binding of Lulu2 causes a conformational change in p114RhoGEF that enhances its catalytic activity.
Cingulin and Patj Anchor p114RhoGEF at Apical Cell–Cell Boundaries
The targeting mechanism of p114RhoGEF to apical cell–cell boundaries was also investigated. Cingulin, a TJ-associated protein known to interact with zona occludens (ZO) proteins, F-actin, and myosin II, was reported to bind to the PH domain of p114RhoGEF and to be necessary for p114RhoGEF localization at apical cell–cell boundaries.Citation21 Depletion of cingulin by RNAi resulted in the delocalization of p114RhoGEF from apical cell–cell boundaries and concomitant disorganization of the circumferential actomyosin belt. On the other hand, we found that Patj, an apical cell polarity regulator, binds to p114RhoGEF via a PDZ domain-mediated interaction.Citation20 As was the case with cingulin, depletion of Patj resulted in delocalization of p114RhoGEF from apical cell–cell boundaries and disorganization of the circumferential actomyosin belt, although cingulin remained localized at TJs in these cells. These results suggest that both cingulin and Patj are necessary for the localization of p114RhoGEF at apical cell–cell boundaries. Since their binding sites on p114RhoGEF are different, we can speculate that cingulin and Patj simultaneously bind to p114RhoGEF at apical cell–cell boundaries and thus form a ternary molecular complex (). However, whereas the C-terminal half of p114RhoGEF including the Patj binding site was efficiently targeted to apical cell–cell boundaries, the N-terminal half of p114RhoGEF including the cingulin-binding site was not. Thus, the role of cingulin might be different from that of Patj in terms of recruiting p114RhoGEF to apical cell–cell boundaries. As a cue to understand this difference, we found that full-length p114RhoGEF bound less efficiently than its C-terminal half to Patj (unpublished observations). We can thus speculate that the binding of cingulin to the N-terminal portion of p114RhoGEF causes a conformational change in p114RhoGEF that allows it to be more easily accessed by Patj at the C-terminal tail, thereby accelerating junctional accumulation of p114RhoGEF. However, more detailed studies are needed to understand the relationship among p114RhoGEF, cingulin, and Patj.
Patj has 10 PDZ domains and is known to bind to various molecules, including Pals1, angiomotin, JAM-1, ZO-3, nectins, Claudin, and Par6 at apical cell–cell boundaries.Citation25,Citation26 In addition, p114RhoGEF was reported to interact with Patj, myosin II, RockII, cingulin and Lulu2.Citation20,Citation21 Cingulin also interacts with several molecules, including GEF-H1, myosin II, F-actin, and ZO proteins at apical cell–cell boundaries.Citation21,Citation27 Therefore, at apical cell–cell boundaries, p114RhoGEF might be a component of a large multi-molecular complex composed of the above molecules. The possible existence of this multi-molecular complex would imply a more complicated interaction between Lulu2 and p114RhoGEF, i.e., Lulu2 might activate p114RhoGEF not only through a direct interaction, as we proposed, but also by collaborating with other molecules of the complex. To fully understand the activation mechanisms of p114RhoGEF by Lulu2 will require much deeper functional analyses, including reconstitution of this putative multi-molecular complex in vitro followed by studies of its regulation.
Atypical PKC Regulates the Circumferential Actomyosin Belt
Recent work showed that aPKC, a well-known cell polarity regulator, regulates the circumferential actomyosin belt.Citation20,Citation28,Citation29 RNAi-mediated knockdown of aPKC, or expression of a dominant negative form of aPKC, induced apical constriction in epithelial cells, suggesting that aPKC counteracts the contractility of the circumferential actomyosin belt. The molecular mechanism of aPKC action on the belt involves its anchoring by Par3 and Willin at the apical junctions.Citation28 In turn, anchored aPKC phosphorylates Rock I, which prevents it from accumulating at the apical junctions, thereby negatively regulating the belt's myosin II activity. We found that aPKC phosphorylates and negatively regulates Lulu2 by reducing its ability to interact with and activate p114RhoGEF ().Citation20 Where and how these two molecular system, aPKC-Lulu2-p114RhoGEF and Willin-aPKC-Rock I are utilized during morphogenesis are interesting questions that remain to be addressed in future studies.
It was recently reported that aPKC is downregulated by pro-inflammatory TNF-α signaling in intestinal epithelial cells in vitro and during intestinal inflammation in mice.Citation30,Citation31 In these cases, aPKC downregulation correlated well with apical myosin II accumulation and MRLC phosphorylation, hallmarks of the pathogenesis of inflammatory bowel disease (IBD). In intestinal cells, apical myosin II activation is thought to be a cause of TJ disruption, an important pathophysiologic factor of IBD.Citation32,Citation33 Furthermore, in IBD patients, as inflammation worsens, active aPKC decreases while apical myosin IIA accumulation increases.Citation31 Therefore, it would be interesting to examine whether aPKC-Lulu2-p114RhoGEF and/or Willin-aPKC-Rock I systems participate in TJ disruption in IBD.
Future Perspectives
The circumferential actomyosin belt regulates several morphogenetic processes. Our understanding of the molecular mechanisms regulating the circumferential actomyosin belt has been improved by recent studies, but is still incomplete. Several questions remain unanswered. For example, it is not yet known how the belt is activated locally and how it is able to induce the changes in apical membrane shape that lead to the desired morphological configurations. It might be the case that cells are able to adopt the desired shape by uneven activation or focal inactivation of the circumferential actomyosin belt. Also, depending on contraction of the belt, changes in the area of the respective apical membrane regions would be expected, perhaps regulated by intracellular membrane-traffic pathways; however, a relationship between such pathways and the contraction forces of the belt remains to be confirmed. Another question relates to the mechanism that allows tension generated by the circumferential actomyosin belt to be transmitted to neighboring cells. Finally, recent studies show that cell shape change is involved in cell proliferation and differentiation, but how the circumferential actomyosin belt regulates these cellular processes remains to be determined. These questions and many others regarding the circumferential actomyosin belt will be the focus of future studies.
Figure 1. The circumferential actomyosin belt regulates apical constriction. (A) The circumferential actomyosin belt is positioned along apical cell-cell junctions (tight and adherens junctions) in polarized epithelial cells. (B) Myosin II-dependent contractile forces of the circumferential actomyosin belt induce apical constriction in epithelial cells.
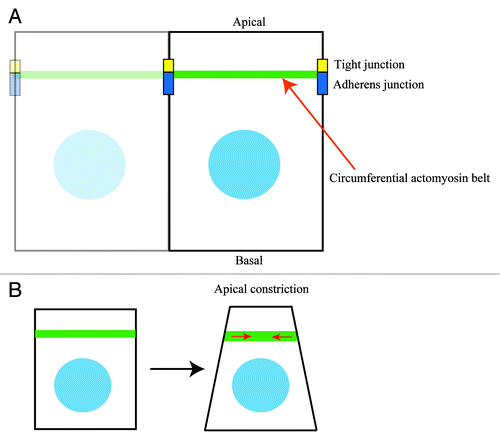
Figure 2. Lulus commonly have FERM and FERM-adjacent domains. (A) Both Lulu1 and Lulu2 have two alternative spliced transcripts that share FERM and FERM-adjacent domains in their N-terminal portion. The C-terminal portions of the isoforms do not obvious homology among them. (B) FERM and FERM-adjacent domains are conserved not only between Lulu1 and Lulu2, but also across species. The numbers indicate similarities or identities of the domains compared with Lulu1 (left or right, respectively).
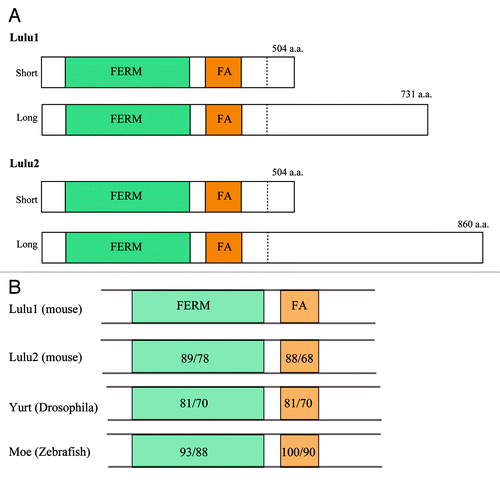
Figure 3. p114RhoGEF is activated by Lulu2. (A) p114RhoGEF has Dbl homology and pleckstrin homology (PH) domains, which are necessary for its catalytic activity, followed by a coiled-coil region and a PDZ domain-binding motif in its C-terminal tail. (B) p114RhoGEF interacts with cingulin via the PH domain, Lulu2 via C-terminal portion, and Patj via the PDZ domain-binding motif in its C-terminal tail. (C) Cingulin and Patj anchor p114RhoGEF at apical cell-cell boundaries. Lulu2 interacts with and activates p114RhoGEF. aPKC inhibits Lulu2. This molecular system regulates the circumferential actomyosin belt.
Acknowledgments
This work was supported by grants from the G-COE Program, a Grant-in-Aid for Scientific Research on Innovative Areas, the Inamori Foundation and Daiichi Sankyo Foundation of Life Science (to T.T.).
References
- Lecuit T, Lenne PF. Cell surface mechanics and the control of cell shape, tissue patterns and morphogenesis. Nat Rev Mol Cell Biol 2007; 8:633 - 44; http://dx.doi.org/10.1038/nrm2222; PMID: 17643125
- Nishimura T, Takeichi M. Remodeling of the adherens junctions during morphogenesis. Curr Top Dev Biol 2009; 89:33 - 54; http://dx.doi.org/10.1016/S0070-2153(09)89002-9; PMID: 19737641
- Kasza KE, Zallen JA. Dynamics and regulation of contractile actin-myosin networks in morphogenesis. Curr Opin Cell Biol 2011; 23:30 - 8; http://dx.doi.org/10.1016/j.ceb.2010.10.014; PMID: 21130639
- Yonemura S, Wada Y, Watanabe T, Nagafuchi A, Shibata M. alpha-Catenin as a tension transducer that induces adherens junction development. Nat Cell Biol 2010; 12:533 - 42; http://dx.doi.org/10.1038/ncb2055; PMID: 20453849
- Yamazaki Y, Umeda K, Wada M, Nada S, Okada M, Tsukita S, et al. ZO-1- and ZO-2-dependent integration of myosin-2 to epithelial zonula adherens. Mol Biol Cell 2008; 19:3801 - 11; http://dx.doi.org/10.1091/mbc.E08-04-0352; PMID: 18596233
- Smutny M, Cox HL, Leerberg JM, Kovacs EM, Conti MA, Ferguson C, et al. Myosin II isoforms identify distinct functional modules that support integrity of the epithelial zonula adherens. Nat Cell Biol 2010; 12:696 - 702; http://dx.doi.org/10.1038/ncb2072; PMID: 20543839
- Sawyer JM, Harrell JR, Shemer G, Sullivan-Brown J, Roh-Johnson M, Goldstein B. Apical constriction: a cell shape change that can drive morphogenesis. Dev Biol 2010; 341:5 - 19; http://dx.doi.org/10.1016/j.ydbio.2009.09.009; PMID: 19751720
- Lee JY, Harland RM. Actomyosin contractility and microtubules drive apical constriction in Xenopus bottle cells. Dev Biol 2007; 311:40 - 52; http://dx.doi.org/10.1016/j.ydbio.2007.08.010; PMID: 17868669
- Martin AC, Kaschube M, Wieschaus EF. Pulsed contractions of an actin-myosin network drive apical constriction. Nature 2009; 457:495 - 9; http://dx.doi.org/10.1038/nature07522; PMID: 19029882
- Hildebrand JD, Soriano P. Shroom, a PDZ domain-containing actin-binding protein, is required for neural tube morphogenesis in mice. Cell 1999; 99:485 - 97; http://dx.doi.org/10.1016/S0092-8674(00)81537-8; PMID: 10589677
- Haigo SL, Hildebrand JD, Harland RM, Wallingford JB. Shroom induces apical constriction and is required for hingepoint formation during neural tube closure. Curr Biol 2003; 13:2125 - 37; http://dx.doi.org/10.1016/j.cub.2003.11.054; PMID: 14680628
- Hildebrand JD. Shroom regulates epithelial cell shape via the apical positioning of an actomyosin network. J Cell Sci 2005; 118:5191 - 203; http://dx.doi.org/10.1242/jcs.02626; PMID: 16249236
- Nishimura T, Takeichi M. Shroom3-mediated recruitment of Rho kinases to the apical cell junctions regulates epithelial and neuroepithelial planar remodeling. Development 2008; 135:1493 - 502; http://dx.doi.org/10.1242/dev.019646; PMID: 18339671
- Nakajima H, Tanoue T. Epithelial cell shape is regulated by Lulu proteins via myosin-II. J Cell Sci 2010; 123:555 - 66; http://dx.doi.org/10.1242/jcs.057752; PMID: 20103536
- García-García MJ, Eggenschwiler JT, Caspary T, Alcorn HL, Wyler MR, Huangfu D, et al. Analysis of mouse embryonic patterning and morphogenesis by forward genetics. Proc Natl Acad Sci U S A 2005; 102:5913 - 9; http://dx.doi.org/10.1073/pnas.0501071102; PMID: 15755804
- Hirano M, Hashimoto S, Yonemura S, Sabe H, Aizawa S. EPB41L5 functions to post-transcriptionally regulate cadherin and integrin during epithelial-mesenchymal transition. J Cell Biol 2008; 182:1217 - 30; http://dx.doi.org/10.1083/jcb.200712086; PMID: 18794329
- Lee JD, Silva-Gagliardi NF, Tepass U, McGlade CJ, Anderson KV. The FERM protein Epb4.1l5 is required for organization of the neural plate and for the epithelial-mesenchymal transition at the primitive streak of the mouse embryo. Development 2007; 134:2007 - 16; http://dx.doi.org/10.1242/dev.000885; PMID: 17507402
- Laprise P, Beronja S, Silva-Gagliardi NF, Pellikka M, Jensen AM, McGlade CJ, et al. The FERM protein Yurt is a negative regulatory component of the Crumbs complex that controls epithelial polarity and apical membrane size. Dev Cell 2006; 11:363 - 74; http://dx.doi.org/10.1016/j.devcel.2006.06.001; PMID: 16950127
- Laprise P, Lau KM, Harris KP, Silva-Gagliardi NF, Paul SM, Beronja S, et al. Yurt, Coracle, Neurexin IV and the Na(+),K(+)-ATPase form a novel group of epithelial polarity proteins. Nature 2009; 459:1141 - 5; http://dx.doi.org/10.1038/nature08067; PMID: 19553998
- Nakajima H, Tanoue T. Lulu2 regulates the circumferential actomyosin tensile system in epithelial cells through p114RhoGEF. J Cell Biol 2011; 195:245 - 61; http://dx.doi.org/10.1083/jcb.201104118; PMID: 22006950
- Terry SJ, Zihni C, Elbediwy A, Vitiello E, Leefa Chong San IV, Balda MS, et al. Spatially restricted activation of RhoA signalling at epithelial junctions by p114RhoGEF drives junction formation and morphogenesis. Nat Cell Biol 2011; 13:159 - 66; http://dx.doi.org/10.1038/ncb2156; PMID: 21258369
- Tsuji T, Ohta Y, Kanno Y, Hirose K, Ohashi K, Mizuno K. Involvement of p114-RhoGEF and Lfc in Wnt-3a- and dishevelled-induced RhoA activation and neurite retraction in N1E-115 mouse neuroblastoma cells. Mol Biol Cell 2010; 21:3590 - 600; http://dx.doi.org/10.1091/mbc.E10-02-0095; PMID: 20810787
- Niu J, Profirovic J, Pan H, Vaiskunaite R, Voyno-Yasenetskaya TG. G Protein betagamma subunits stimulate p114RhoGEF, a guanine nucleotide exchange factor for RhoA and Rac1: egulation of cell shape and reactive oxygen species production. Circ Res 2003; 93:848 - 56; http://dx.doi.org/10.1161/01.RES.0000097607.14733.0C; PMID: 14512443
- Nagata K, Inagaki M. Cytoskeletal modification of Rho guanine nucleotide exchange factor activity: identification of a Rho guanine nucleotide exchange factor as a binding partner for Sept9b, a mammalian septin. Oncogene 2005; 24:65 - 76; http://dx.doi.org/10.1038/sj.onc.1208101; PMID: 15558029
- Adachi M, Hamazaki Y, Kobayashi Y, Itoh M, Tsukita S, Furuse M, et al. Similar and distinct properties of MUPP1 and Patj, two homologous PDZ domain-containing tight-junction proteins. Mol Cell Biol 2009; 29:2372 - 89; http://dx.doi.org/10.1128/MCB.01505-08; PMID: 19255144
- Sugihara-Mizuno Y, Adachi M, Kobayashi Y, Hamazaki Y, Nishimura M, Imai T, et al. Molecular characterization of angiomotin/JEAP family proteins: interaction with MUPP1/Patj and their endogenous properties. Genes Cells 2007; 12:473 - 86; http://dx.doi.org/10.1111/j.1365-2443.2007.01066.x; PMID: 17397395
- Paschoud S, Yu D, Pulimeno P, Jond L, Turner JR, Citi S. Cingulin and paracingulin show similar dynamic behaviour, but are recruited independently to junctions. Mol Membr Biol 2011; 28:123 - 35; http://dx.doi.org/10.3109/09687688.2010.538937; PMID: 21166484
- Ishiuchi T, Takeichi M. Willin and Par3 cooperatively regulate epithelial apical constriction through aPKC-mediated ROCK phosphorylation. Nat Cell Biol 2011; 13:860 - 6; http://dx.doi.org/10.1038/ncb2274; PMID: 21685893
- Kishikawa M, Suzuki A, Ohno S. aPKC enables development of zonula adherens by antagonizing centripetal contraction of the circumferential actomyosin cables. J Cell Sci 2008; 121:2481 - 92; http://dx.doi.org/10.1242/jcs.024109; PMID: 18628308
- Mashukova A, Wald FA, Salas PJ. Tumor necrosis factor alpha and inflammation disrupt the polarity complex in intestinal epithelial cells by a posttranslational mechanism. Mol Cell Biol 2011; 31:756 - 65; http://dx.doi.org/10.1128/MCB.00811-10; PMID: 21135124
- Wald FA, Forteza R, Diwadkar-Watkins R, Mashukova A, Duncan R, Abreu MT, et al. Aberrant expression of the polarity complex atypical PKC and non-muscle myosin IIA in active and inactive inflammatory bowel disease. Virchows Arch 2011; 459:331 - 8; http://dx.doi.org/10.1007/s00428-011-1102-1; PMID: 21667320
- Ivanov AI, Parkos CA, Nusrat A. Cytoskeletal regulation of epithelial barrier function during inflammation. Am J Pathol 2010; 177:512 - 24; http://dx.doi.org/10.2353/ajpath.2010.100168; PMID: 20581053
- Turner JR. Intestinal mucosal barrier function in health and disease. Nat Rev Immunol 2009; 9:799 - 809; http://dx.doi.org/10.1038/nri2653; PMID: 19855405