Abstract
Small GTPases are molecular switches at the hub of many signaling pathways and the expansion of this protein family is interwoven with the origin of unique eukaryotic cell features. We have previously reported on the evolution of CDC25 Homology Domain containing proteins, which act as guanine nucleotide exchange factors (GEFs) for Ras-like proteins. We now report on the evolution of both the Ras-like small GTPases as well as the GTPase activating proteins (GAPs) for Ras-like small GTPases. We performed an in depth phylogenetic analysis in 64 genomes of diverse eukaryotic species. These analyses revealed that multiple ancestral Ras-like GTPases and GAPs were already present in the Last Eukaryotic Common Ancestor (LECA), compatible with the presence of RasGEFs in LECA. Furthermore, we endeavor to reconstruct in which order the different Ras-like GTPases diverged from each other. We identified striking differences between the expansion of the various types of Ras-like GTPases and their respective GAPs and GEFs. Altogether, our analysis forms an extensive evolutionary framework for Ras-like signaling pathways and provides specific predictions for molecular biologists and biochemists.
Introduction
The Ras-like GTPases perform central regulatory functions in a myriad of cellular processes, such as cell division, differentiation, cell-cell adhesion, growth, apoptosis. The well-studied oncogenes H-, N- and K-Ras belong to this family. The Ras-like GTPase family belong to the small GTPase superfamily of which all members share high sequence similarity and structural features.Citation1 Other well-known members of this superfamily are the Rab, Ran, Rho and Arf GTPases.
The expansion and evolution of the small GTPases have been related to the evolution of unique eukaryotic cellular features, such as phagocytosisCitation2,Citation3 and specific studies on the evolution of the Rab and Rho GTPases have been published in reference Citation4–Citation6. However, to our knowledge no large scale and broad phylogenetic analysis has been reported for the Ras-like GTPases.
The small GTPases are inefficient GTPases that require GTPase Activating Proteins (GAPs) to enhance the intrinsic GTPase activity and Guanine Exchange Factors (GEFs) to efficiently exchange bound GDP for GTP. Regulation of GAP and GEF activity allows for small GTPases to function as an efficient, highly regulated molecular switch that can quickly alternate between its active GTP bound state and its inactive GDP bound state. Although virtually all small GTPases share this common principle of regulation, each subfamily of GTPases has its own set of evolutionary unrelated GAPs and GEFs.Citation7 Importantly, whereas Ras-like small GTPase are almost uniquely regulated by CDC25 HD domain containing proteins as GEF, the GAP proteins belong to two unrelated families, the RasGAPs and the RapGAPs.Citation7
Specific members of the Ras-like GTPase family regulate a multitude of diverse cellular processes. Therefore analyzing the phylogeny of the Ras-like GTPases and their GAPs and GEFs not only provides an in depth evolutionary history of one of the most enigmatic families belonging to the small GTPase superfamily, but also provides an evolutionary framework for researchers to clarify and explain the observed complexity.
We have previously shown a surprisingly tight evolutionary relationship between Ras-like GTPases and their Guanine Exchange Factors.Citation8 The CDC25 Homology Domain (CDC25HD) or RasGEF domain, was found in animal, fungal, Chromalveolate and Excavate species. The CDC25HD co-occurs perfectly with the Ras, Rap and Ral GTPases (i.e., either both GTPase and CDC25HD are present or both are absent in all species). We observed that the significant differences in the RasGEF repertoire between fungi and animals has been the result of differential loss of RasGEFs in the animal and fungal ancestor (see ). The tight evolutionary relationship between Ras-like GTPases and their RasGEFs raises the possibility that the evolution of the regulatory domains likely holds information on the evolution of the Ras-like GTPases as well.
We now report on the evolutionary history both of Ras- and RapGAPs as well of Ras-like small GTPases. We found that the origin of the RapGAP and RasGAP domains can reliably be traced back to four ancestral RapGAPs and five ancestral RasGAPs in the Last Eukaryotic Common Ancestor (LECA). We identified a previously unrecognized RapGAP domain in RalGAPB, a subunit of the RalGAPA/B dimer and we are able to show that this RalGAPA/B dimer was already present in LECA. The domain architecture of the RasGAP protein family is extremely well conserved throughout the eukaryotic kingdom. Phylogenetic analysis of the Ras-like GTPase protein sequences reveal that multiple Ras-like GTPases were present in LECA. We reconstruct the early evolution of the Ras-like GTPases and its regulatory domains and show in which order the Ras, Rap, Ral and Rheb GTPases diverged from each other. Lastly we combine the analyses of the phylogenies of the Ras-like GTPases, their GAPs and GEFs and examine the evolutionary dynamics of the whole Ras regulatory system (i.e., the GTPase and its GAPs and GEFs). We show that Rap and Ras display strikingly different behaviour in evolution with respect to the expansion of their respective GEFs and GAPs.
Results and Discussion
Evolution of the rap Gtpase activating protein domain; conservation of Gtpase specificity.
We collected RapGAP domain sequences from our genome dataset containing 64 different eukaryotic genomes using a custom RapGAP hidden markov model based on the original Pfam Rap_GAP model (Pfam: PF02145). For a full overview of all species included in this set please see Table S1 and Figure S3. This genome set was also used for the phylogenetic analysis of RasGAP and Ras-like GTPases described below.
The sequences of only the RapGAP domain were considered for the phylogenetic analysis as the variation of additional non homologous protein domains in RapGAP domain containing proteins make it impossible to make reliable full length alignments. In addition it allows us to identify domain acquisitions relative to the RapGAP domain. The gathered sequences were aligned using MAFFTCitation9 and a phylogenetic tree was constructed using RAxML.Citation10 The resulting phylogenetic tree clearly shows a trifurcation into Rap, Rheb and Ral specific monophyletic groups of RapGAP domain proteins (). For a detailed phylogenetic tree including domain architecture of each protein see Figure S1.
Rheb specific RapGAP domain containing proteins.
The cluster of Rheb specific RapGAP domain containing proteins encompasses the TSC2 gene and its orthologs as described in previous work (van Dam TJP, et al. unpublished). The TSC2 protein contains a DUF3384 domain, a Tuberin domain and a RapGAP domain. This domain architecture is strongly conserved in animal and fungal TSC2-like proteins (see Fig. S1). Additionally, we find sequences containing the TSC2-like RapGAP domain in Dictyostelium discoideum (amoeba), Phytophthora species (oomycetes), Phaeodactylum tricornutum (diatom), Tetrahymena thermophila (ciliate) and the red algae C. merolae. The species distribution for the TSC2 GAP domain strongly suggests that the TSC2-like RapGAP domain originated in or before LECA. Except for T. thermophila the TSC2-like RapGAP domain can be found exclusively in species that also contain a Rheb ortholog (van Dam et al. unpublished and this study), indicating that there is an exceptionally strong evolutionary link between Rheb and the TSC2-like GAP domain.
The Ral specific RapGAP domain containing proteins; evidence for an ancient heterodimer complex.
The GAP specific for the Ral GTPases was already discovered earlier in reference Citation11, but has only been recently shown to specifically regulate Ral.Citation12 The RalGAP is a protein complex that consists of RalGAPB with either RalGAPA1 or RalGAPA2. RalGAPA1 and 2 have been shown to harbor the RapGAP domain and perform the GAP function towards Ral. The RalGAP complex has been described as a TSC1/2-like complex as RalGAPB initially seemed to have no protein domain or other obvious structural features, much like TSC1. However, we identified a putative RapGAP domain at the C-terminal part of RalGAPB by sequence analysis (custom HMM model, e-value 7.1e-08 for the human RalGAPB, see also Fig. S1). Although the RapGAP domain sequence is highly conserved, RalGAPB has an insertion/deletion where normally the catalytic loop can be found (positions 282 to 290 in the Rap1GAP protein sequence, see Scrima et al.Citation13). This raises questions about the specific function of RalGAPB in the protein complex as it has no sequence similarity to TSC1 but also apparently lacks the ability to provide GAP function towards Rap, Ral or Rheb.
Analysis of the phylogenetic tree as represented in indicates that the RapGAP domain of RalGAPA1/2 and RalGAPB are closely related and distinct from the Rap specific and Rheb specific RapGAP domain containing proteins. The close relation between RalGAPA1/2 and RalGAPB suggests that the RalGAP complex originated from a single gene, of which the gene product likely formed a homodimeric complex that after gene duplication became an heterodimeric complex. Interestingly, based on the species distribution in the gene tree we can reconstruct both RalGAPA1/2 (the result of a duplication in the vertebrate ancestor) and RalGAPB as ancestral genes in LECA. The species distribution of RalGAPA and RalGAPB are identical suggesting that the RalGAP complex is perfectly conserved in evolution and was already present as a heterodimeric complex in LECA.
Intriguingly, the species distribution of the RalGAP complex matches the species distribution of the Ral subtype GTPases (animals + chytrid and zygomycota fungi), but can additionally be found in the amoeba D. discoideum, the oomycetes Phytophthora sojae and Phytophthora infestans and the excavate N. gruberi. We have not identified Ral-like GTPases nor putative RalGEFsCitation8 in these organisms. It could therefore be possible that the RalGAP complexes in these species perform GAP functions towards other small GTPases. Accordingly, this then raises the question if the RalGAP complex might also still perform GAP activity to other GTPases in human since RalGAP complex activity has been tested only on the Ras-like GTPases RalA and B, H-Ras, Rap1 and Rheb.Citation12 This leaves many Ras-like GTPases still to be tested.
The Rap subtype specific RapGAP domain containing proteins; the odd ones.
As the Rap specific RapGAPs form a well defined cluster outside of the TSC2 and RalGAP clusters (100% bootstrap support), they too can be reconstructed into the LECA gene repertoire. The Rap specific RapGAPs are by far the most expanded RapGAP domain containing proteins in mammals. In this group we find SIPA and the SIPA-like RapGAPs as well as the Rap1GAPs. We also find an uncharacterized RapGAP domain containing protein called GARNL3 (bootstrap support of 100%). GARNL3 contains in addition to a RapGAP domain a CNH domain (InterPro IPR001180, Pfam PF00780) which is present in a number of proteins that interact with Rho, Rac and or CDC42.Citation14 GARNL3 therefore could possibly link Rho GTPase activity to RapGAP activity as GARNL3 clearly clusters together with known RapGAPs.
The Rap1GAPs and SIPA-like RapGAPs each occur in all animals and likely originated from a duplication in the animal ancestor. The GARNL3 cluster also contains protein sequences from the chytrid fungus B. dendrobaditis and the amoeba D. discoideum. Sequences belonging to the fungi B. dendrobaditis, R. oryzae and P. blackleeanus cluster outside of the RapGAPs including GARNL3. These sequences potentially belong to the SIPA-like and Rap1GAP cluster. The species distribution suggests that GARNL3 and the SIPA/Rap1GAP genes originated in the Unikont or Opisthokont ancestor from a gene duplication. Interestingly the Rap specific RapGAP group of proteins lack sequences from a large group of fungal species (i.e., basidiomycetes and ascomycetes) while these species do contain a Rap ortholog (e.g., BUD1 in S. cerevisiae). It is likely that the Rap specific RapGAP domain containing proteins have been replaced by the C2 domain containing RasGAPs (the RASA-like RasGAPs, BUD5 in S. cerevisiae, see below).
Evolution of the Ras GTPase activating protein domain; conservation of domain architecture.
We collected RasGAP domain sequences from our genome dataset using the original Pfam Ras_GAP model (Pfam: PF00616). The sequences of only the RasGAP domain were considered similar to the RapGAP domain sequences described previously. The gathered sequences were aligned using MAFFTCitation9 and a phylogenetic tree was constructed using RAxML.Citation10
The phylogenetic tree of the Ras GTPase Activating Proteins or RasGAPs shows a similar behavior as the RapGAP tree. We observe five clusters which each represent an ancestral RasGAP gene in LECA (). For two of the five ancestral groups there is evidence to support a common origin predating LECA (GAP1 and IQGAP-like RasGAPs). However in contrast to the RapGAP domain phylogeny, these five ancestral groups are not only supported by the gene tree, but also by their domain architectures (i.e., the composition and order of protein domains in a protein). The domain architectures of the RasGAP domain containing proteins are highly conserved and can, in part, be reconstructed to the ancestral RasGAPs in LECA. Below we discuss the phylogenetic reconstruction and ancestral domain architectures of the RasGAP domain containing proteins.
The GAPVD1-like RasGAPs; an ancestral link between Rab and Ras signaling.
GAPVD1 (also known as RAP6, Gapex-5 or RME-6) represents a conserved type of RasGAP that can be found in animals, the amoeba D. discoideum, the oomycete P. infestans and the ciliates T. tetrahymena and P. tetraurelia. Interestingly we identified the RasGAP domain followed by the VPS9 domain in both the animal sequences and in the ciliate sequences. Even though there are no sequences found in representative species of the other phyla (e.g., plants and excavata), the identification of the GAPVD1 orthologs in the Chromalveolata (i.e., the ciliates and oomycetes) indicates that GAPVD1 is ancient and perhaps originated from LECA.
GAPVD1 has been characterized in C. elegans, mouse and human.Citation15–Citation17 The VSP9 domain exhibits Guanine Exchange Factor activity towards Rab5-like GTPases and the RasGAP domain has been shown to bind to H-Ras and stimulate its GTPase hydrolysis in human.Citation17 GAPVD1 therefore forms a conserved bridge between Ras and Rab signaling.
The GAP1 and IQGAP-like RasGAPs; diversification between the animal and fungal RasGAP repertoire.
The human IQGAPs and the S. pombe GAP1 [genedb: SPBC646.12c] each represent an ancestral RasGAP gene in LECA. Both families are characterized by a highly conserved RasGAP-RasGAP_C domain architecture, which does not occur in other GAP proteins. We identified IQGAP-like genes in animals, fungi (e.g., IQG1 in S. cerevisiae) including Enchephalitozoon cuniculi, the amoeba D. discoideum and the excavate N. gruberi. The N-terminal CH domain of the IQGAPs is highly conserved in all species, eventhough the IQ repeats, characteristic for this RasGAP family, is only conserved in animals and fungi. The CH-RasGAP-RasGAP_C architecture therefore represents the ancestral domain architecture of the IQGAP ancestral gene in LECA.
Interestingly the IQGAPs have no reported GAP activity to any Ras-like GTPase. Instead IQGAP1 and IQGAP2 inhibit the intrisinc GTP hydrolysis of Rac and CDC42.Citation18,Citation19 In S. cerevisiae the interaction between the IQGAP ortholog IQG1 and CDC42 is conserved.Citation20 It would be interesting to speculate as to why the IQGAPs do not have any observed GAP activity towards Ras-like GTPases while retaining the conserved RasGAP domain. However, we have not identified any obvious reason for the loss of GAP function from the sequences. Based on experimental descriptions of the IQGAPs in fungi and animals, it appears that the RasGAP function within the IQGAP family has been lost early in evolution and likely occurred before the Opisthokont ancestor leading up to animals and fungi.
The GAP1-like RasGAP does not occur in higher animals but can be found in the excavate N. gruberi, the amoeba D. discoideum, the choanoflagellate Monosiga brevicollis, the primitive animal Trichoplax adhearens and fungi. In contrast to the IQGAPs S. pombe GAP1 has been reported to regulate Ras1.Citation21 In fungi however the GAP1 gene appears to have been lost in the Saccharomyceae (S. cerevisiae, Candida glabrata, Kluyveromyces lactis, Eremothecium gossypii, Candida guilliermondii and Debaryomyces hansenii). The presence of a GAP1 ortholog in the placozoan T. adhearens and choanoflagellate M. brevicollis indicates that GAP1 has been lost early in the animal lineage. The conserved RasGAP-RasGAP_C domain architecture and position in the gene tree next to the IQGAP-like sequences indicates that both groups originated from a single ancestral gene that predates LECA and already had the RasGAP-RasGAP_C domain architecture. This particular domain architecture is not shared with any of the other RasGAP types.
The NF1-like RasGAPs; an ancient link between Ras and Rheb signaling?
Neurofibromin 1 (NF1) is RasGAP and a known tumor suppressor. NF1 mediates Ras dependent mTOR activation via TSC2.Citation22 Dysfunction of NF1 in mammals results in a disorder called neurofibromatosis which is characterized by the development of neurofibromas, that are benign tumors found on and around the peripheral nerves. We find NF1 orthologs in all animals and in fungi where the gene has duplicated in the yeasts S. cerevisiae and C. glabrata (IRA1 and IRA2 in S. cerevisiae). We also find multiple NF1-like RasGAP sequences in N. gruberi, D. discoideum, P. sojae and P. infestans. The sequences of one gene in P. infestans [PITG_09962] and P. sojae [131833] and two genes in the excavate N. gruberi [80941, 73399] are conserved over the full length of NF1. This strongly indicates that the ancestral gene in LECA likely coded for a NF1-like protein.
The RASA-like RasGAPs; origins of the C2-RasGAP domain architecture, linking Ras and Rap signaling in evolution.
The RASA-like RasGAPs contain by far the most human RasGAPs (10 of 17, excluding the Plexin genes, see below) and includes mammalian RASA1-4, SynGAP1 and yeast Bud2. While the other classes of RasGAPs contain only 1 group of animal genes, the RASA-like RasGAPs have undergone an animal specific expansion. Additionally, this class has undergone extensive protein domain rearrangements and domain acquisitions. However, nearly all sequences contain at least the C2 domain followed by the RasGAP domain. This indicates that although the overall bootstrap values for this class is low, the clustering itself is correct.
Proteins belonging to the RASA-like RasGAPs have been reported to be dual specific towards Ras- and Rap GTPases.Citation23–Citation25 Pena et al. has shown that the individual RasGAP domain of SynGAP1 specifically deactivates Ras, while the full length protein is a Rap specific GAP. They also showed that the C2 domain is necessary for its Rap specificity. The phylogenetic distribution of the C2—RasGAP domain architecture matches the phylogenetic distribution of the Rap GTPase subtype suggesting that the C2—RasGAP architecture may represent conservation of specificity towards the Rap GTPases and therefore represents an evolutionary link between the RASA-like RasGAPs and Rap signaling.
We note above that Rap specific RapGAP domain containing proteins do not occur in fungal species belonging to the basidiomycetes and ascomycetes. A known GAP for the fungal Rap ortholog BUD1 in the yeast S. cerevisiae is the BUD2 gene.Citation23 BUD2 belongs to the RASA-like GTPases and contains the C2-RasGAP domain architecture. Therefore, BUD2 and its fungal orthologs fill the gap left by the loss of the RapGAP domain containing proteins in fungal Rap signaling.
The plexin type RasGAPs; unclear origin.
The Plexin type RasGAPs cluster in the RASA-like RasGAPs but do not share the C2 domain or any other domain except for the RasGAP domain. The Plexin type RasGAPs from an animal specific group of genes but show a particularly long branch indicating a period of fast sequence evolution of the RasGAP domain sequence early in or just before the emergence of animals in evolution. Therefore it is uncertain if the Plexin type RasGAPs indeed belongs to the RASA-like RasGAPs or if its placement is a Long Branch Attraction artifact (LBA) of the Maximum Likelihood inference.
Localized signaling and cross-signaling in Ras signaling network.
An interesting observation is that although both the RasGAP and RapGAP domains had multiple copies in LECA we observe conservation of protein domain architecture only in the RasGAP family. Two of three protein domains that are conserved in LECA together with the RasGAP domain are involved in localization (CH for binding to the actin skeleton, C2 for Ca2+ dependent membrane targeting). This strengthens the idea that sub-localization is an crucial factor in Ras-like GTPase signaling.Citation27 The conservation of the VPS9 domain (a GEF domain for Rab5-like GTPases) and the C2 domain with the RasGAP domain across the eukaryotic kingdom is indicative for the interconnectivity of Ras signaling pathways with other signaling pathways. This interconnectivity or cross-signaling, is one of the hallmarks of Ras signal transduction.
A high resolution phylogeny of the Ras-like subfamily of small GTPases.
In order to classify the Ras-like GTPases, sequence comparison and phylogenetic analysis have often been employed.Citation3,Citation28–Citation33 However most focused solely on (a subset of) human GTPase sequences or common model species (e.g., the yeasts S. cerevisiae and S. pombe and animal models such as C. elegans and D. melanogaster). The small species selection does not provide insights into how old the subtypes (e.g., Ras, Rap, Ral, etc.,) are and when duplications and losses actually occurred. Interestingly the relationships between the various subtypes seem to differ in the phylogenetic trees reported in these publications. This suggests that the phylogenetic relationships between the Ras-like GTPases is still unresolved. Below we report a detailed phylogenetic analysis of the Ras-like GTPase subfamily and a reconstruction of the early evolution of Ras-like GTPases.
We identified small GTPase protein sequences in the genomes of 64 divergent eukaryotic genomes using the PfamCitation34 Ras HMM model (Pfam PF00071.15). The small GTPase super family members all share a high degree of sequence similarity and the Pfam Ras model therefore also matches significantly to hits belonging to the other small GTPase subfamilies, such as Rho, Rab and Ran. In order to identify Ras-like subfamily members we aligned all small GTPase sequences using MAFFT and constructed a phylogenetic tree using the Neighbor Joining algorithm. We identified a cluster that contained all currently classified mammalian and yeast Ras-like GTPase subfamily members but did not contain small GTPase members belonging to the other subfamilies. Therefore we can confidently classify all GTPases in this cluster, mostly belonging to species for which there is no gene function information available, as belonging to the Ras-like subfamily of GTPases.
A phylogeny of the sequences belonging to the Ras-like subfamily cluster was constructed by using RAxMLCitation10 and PhyML.Citation35 Bayesian methods did provide any results as the MCM chains would not converge. The evolutionary relationships between the Ras-like GTPases were determined as well as the timing of the duplications based on species distribution in each phylogenetic branch (see and Sup. data).
The phylogenetic trees as produced by PhyML and RAxML are not in full agreement. This is likely due to the relatively short sequences of the Ras-like GTPases (average of 159 aa) and the high sequence similarity which limits the amount of information present to make a reliable distinction between related sequences. This is reflected in the low bootstrap support, specifically in the deep branches. We address the uncertainty in the phylogenetic trees case by case below.
We define Ras-like GTPase subtypes based on their relatedness as observed in clustering in the phylogenetic trees. We can trace back the origin of four subtypes to in or before the LECA (i.e., Ras, Ral, Rheb and Rap), but for other subtypes we are unable to do so. These other subtypes do not affiliate specifically to any other subtypes and encompasses only metazoan species implicating a more recent origin. The many isoforms of each subtype observed in vertebrates, for instance Rap1A and Rap1B, are likely the results of multiple whole genome duplications (WGD) that occurred in the vertebrate ancestor. Below we will discuss the evolution of each Ras-like GTPase subtype in more detail.
The Ras subtype (canonical).
The canonimal Raslike GTPases N-, H-, K-Ras and the closely related TC21, R- and M-Ras regulate diverse cellular functions such as cell differentiation, apoptosis, cell proliferation, cell division and functions as input into many other signaling pathways. All canonical Ras GTPases require many of the same GEF and GAP proteins for regulation.Citation7 Indeed we find that all six are closely related, but form a bi-partitioned group: one consisting out of N-, H-, K-Ras and the other consisting out of TC21, R- and M-Ras (). We find that E-Ras, generally not considered to be closely related to the Ras subtypeCitation29 is directly related to N-, H-, K-Ras (bootstrap support of 82%). We find that the fungal orthologs of Ras (Ras1 and Ras2 in Saccharomyces cerevisiae, Ras1 in Schizosaccaromyces pombe) are orthologous (i.e., sharing common ancestry) to TC21, R- and M-Ras but are paralogous to N-, H-, K- and E-Ras, whom appear to have diverged earlier. However, the phylogenetic signal is weak (0% and 1% bootstrap support for the PhyML and RAxML trees respectively) and therefore an alternative evolutionary scenario could be that the N-, H-, K-, E-Ras and TC21, M- and R-Ras have arisen from animal specific duplications and all are orthologous to the fungal Ras genes. Nonetheless, GTPase genes from the slime mould Dictyostelium discoideum are found in both the N-, H-, K-, E-Ras partition and the TC21, M- and R-Ras partition, supporting the first scenario. Because of the uncertainty concerning the placement of the fungal Ras orthologs, Ras1 & Ras2 are colored gray in .
The Ral subtype; opisthokont origin or much older?
The Ral GTPases (RalA and RalB) function in a number of different cellular processes, including the regulation of exocytosis.Citation36,Citation37 Ral has its own complement of GEFs (RalGDS- and RalGPS-like proteins) and GAPs (RalGAPA/B complexCitation12). Previously we observed that the RalGDS and RalGPS genes form a single evolutionary gene family with fungal orthologs in the Chytrid and Zygomycetae fungi.Citation8 We also identified putative Ral orthologs in the same fungi indicating a strong evolutionary link between Ral and the RalGEFs.
It is unclear whether Ral clusters into the Ras subtype or forms a separate cluster, since different phylogenetic methods (e.g., maximum likelihood, neighbor joining) and programs (RAxML or PhyML) reached different conclusions (see Sup. material). This raises the question of how old the Ral subtype truly is. The species that contain a clear Ral ortholog all belong to the Opisthokonta (animals and fungi), so clear evidence is lacking that Ral may be older than the Opisthokonta. However, the fact that Ral has a GAP which is distinct from Rap- and RhebGAPs (see section RalGAPs) may indicate that Ral form their own evolutionary subtype and that Ral emerged earlier in eukaryotic evolution and subsequently has been lost in all other phyla.
The Rap subtype.
Rap has a suppressing effect on the Ras oncogenesCitation38 and have therefore attracted much attention in the GTPase field. Rap1 (Rap1A and Rap1B) is known to regulate many aspects of cell-cell adhesion, development of focal adhesions and junction formation. Although similar functions for Rap2 (Rap2A/B/C) are observed it is currently still unclear what the distinct cellular function of Rap2 is relative to Rap1.Citation27
We identify Rap orthologs in animals, fungi, the amoeba D. discoideum and the excavate N. gruberi. The Naegleria gruberi Rap ortholog (55569) indicates that Rap might be older than the Unikonta and might even have originated in or before LECA. This is supported by the presence of RapGEF orthologs and a RASA RasGAP ortholog in N. gruberi (van Dam et al.Citation8 and this manuscript). Rap must have subsequently been lost in the Archaeplastida, Chromalveolates and specific lineages within the Excavata. It has been observed that Naegleria gruberi contains many genes belonging to orthologous groups of genes previously thought to be Unikont specific.Citation39 Therefore our observation of the phylogenetic distribution of the Rap-like GTPases is not uncommon.
The Rap1 and Rap2 genes from two separate groups within the Rap subtype and have arisen from a duplication in the Unikont or Opisthokont ancestor, depending on the placement of orthologous genes from the amoeba D. discoideum (). Fungal Rap orthologs (e.g., BUD1 in S. cerevisiae) consistently cluster to Rap1 but not Rap2, indicating that fungi have lost the Rap2 gene. S. pombe appears to also have lost the Rap1 ortholog as well.
The Rheb subtype.
Rheb is an integral part of the TOR pathway and plays a central role in the regulation of the TOR complex 1 (TORC1). Activation of Rheb results in the activation of TORC1 which leads to increased translation and growth.Citation40 Via its GAP, the TSC1/2 complex, Rheb integrates many signals including nutrient availability and growth factor signaling.Citation41–Citation43 The TSC2 protein harbors a RapGAP domain that is needed for its catalytic activity towards Rheb.
Previously, the phylogeny of Rheb has been extensively discussed in light of the TOR pathway by us (van Dam TJP, et al. unpublished). We found that the Rheb subtype is well conserved in all eukaryotic lineages and we have been able to identify clear orthologs in animals, fungi, amoebazoa, stramenophila (Chromalveolates), excavates and even Archaeplastida (specifically in the red algae Cyanidioschyzon merolae). The Rheb-like GTPases are generally found as a single copy in most eukaryotic species, except vertebrates in which Rheb has duplicated giving rise to the Rheb and RhebL1 genes. The Rheb subtype is well established and consistent in the phylogenetic trees. The Rheb subtype represents the least ambiguous orthologous group of Ras-like GTPases spanning all major eukaryotic phyla and very likely represents a single ancestral GTPase in LECA (see ).
The identification of a Rheb ortholog in the red algae C. merolae is substantiated by the discovery of a TSC2 ortholog. The position of sequences of the putative Rheb and TSC2 orthologs in the phylogenetics trees indicates that their presence in C. merolae is not the result of horizontal gene transfer. The presence of Rheb and TSC2-like proteins in the red algae suggests that part of the Rheb-TOR pathway is conserved in red algae but has subsequently been lost in green algae and plants. To our knowledge the Rheb ortholog in C. merolae represents the only Ras-like GTPase family member that is conserved in a species belonging to plant lineage. This indicates that the Ras-like GTPases and their pathways have been selectively lost early in the plant lineage.
Other Ras-like GTPase subtypes.
The Ras, Ral, Rap and Rheb subtypes represent the most well characterized and frequently studied Ras-like GTPases. However, there are more Ras-like GTPases (18 out of 34 Ras-like GTPases in human) including the RIT, RERG, GEM and DiRAS GTPases. To the best of our knowledge there has been no (confirmed) report on the characterization of any GAP or GEF proteins specific for these Ras-like GTPases considerably hampering biochemical characterization of the pathways in which these GTPases are involved.
Interestingly for the Ras-like GTPases other than the Ras, Rap, Ral and Rheb subtypes we cannot identify any orthologs in S. cerevisiae, S. pombe or other fungi. In fact we only find well defined animal specific clusters, but very quickly the resolution in the trees is lost due to weak bootstrap support and/or absence of sequences from non-animal species (see ). The subtypes rarely cluster together (except the RERG and REM subtypes in the NJ tree and RERG and RIT subtypes in the RAxML tree, but both with a bootstrap value of 0). In fact none of the Ras-like GTPase subtypes cluster together consistently, for which the low bootstrap support is indicative. It is therefore impossible to determine with any confidence the origin of groups we have termed the REM, RasD, RERG and the RIT subtypes.
The number of ancestral GTPases that gave rise to the REM, RasD, RERG and the RIT subtypes may vary between one and four. Either these subtypes are the result of multiple rounds of duplications from a single ancestral gene in the animal ancestor and were lost only once in the other major phyla (i.e., a single ancestral gene in LECA) or all four subtypes emerged in or before LECA and all subtypes were individually lost in the major phyla, except the Metazoa. The first possibility requires the least evolutionary events to describe the observed tree and must therefore be considered as the most likely scenario.
The early emergence of the Ras, Rap, Ral and Rheb GTPases and their regulatory domains.
Many of the Ras-like GTPase subtypes emerged before the final radiation of all extent eukaryotic species, placing the duplication events and subsequent divergence of Ras, Rap, Rheb and other Ras-like subtypes somewhere in the large time span between the First Eukaryotic Common Ancestor (FECA) and LECA. Because all extant eukaryotic species radiated after LECA we initially have only the phylogenetic tree topology to describe the relationships between the Ras-like GTPase subtypes. However, the short sequence length of the small GTPases (average sequence length of 159 aa) is restricting the reliability and reconstruction of the phylogenetic relationships between the GTPase subtypes. Altogether our initial observation in literature that the phylogenetic relationships between Ras-like GTPases vary greatly between publications is confirmed by our own analysis. We may however be able to at least refine the phylogenetic relationships between the Ras, Rap, Ral and Rheb GTPase subtypes using additional information. We previously observed a strong evolutionary relation between Ras-like GTPases Ras, Rap and Ral and the CDC25HDCitation8 and also between Rheb and the TSC2 GAP domain (van Dam et al. unpublished and this manuscript) and Rap and RASA-like RasGAPs. The RasGEF domain (the CDC25HD) and the GAP domains (RasGAP and RapGAP) provide catalytic activity to two or more of the Ras, Ral, Rap and Rheb subtypes and therefore could possibly be used to infer the order of divergence into these four subtypes. Below we discuss our results on the GAPs and our previous study on the evolution of the CDC25HD (i.e., the RasGEF domain) in light of the Ras-like GTPase subtypes.
In we have condensed the specificity of the regulatory domains to their GTPase subtypes into a matrix. For each possible evolutionary scenario for the Ras-like GTPase subtypes we have reconstructed the point of invention and or loss of subtype specific regulatory domains while trying to minimize the number of events (i.e., the number of inventions and losses, see ). Of the 14 possible scenario's there is only one scenario () that requires the least amount of events (four) to describe the matrix in . Five scenarios require five events ( and D, H, M and N) and the remaining eight scenarios require 6 or 7 events. The scenario depicted in therefore represents the most parsimonious scenario of how the Ras-like subtypes could have differentiated from each other before the LECA. It also depicts clearly when and in which order the regulatory domains emerged that regulate current day Raslike GTPases.
The parsimonious scenario in describes a chain of successive inventions of regulatory domains and the duplications of the Ras-like GTPases in the eukaryotic ancestors before the radiation in all the current eukaryotic phyla. According to this scenario the invention of the RapGAP domain preceded the emergence of the Ras, Rap, Ral and Rheb GTPases in an early eukaryotic ancestor predating LECA. Subsequently Rheb diverged from the Ras/Rap/Ral ancestral GTPase and the RasGEF domain was invented. The early divergence of Rheb explains the lack of observed Rheb specific RasGAP and RasGEFs. After the addition of the RasGEF domain the Ral subtype diverged from the Ras/Rap ancestral GTPase. At this point all regulatory domains for Ral already existed (i.e., the RasGEF and RapGAP domains). Finally the RasGAP protein domain was invented after which the Rap and Ras subtypes diverged from each other. The Ras subtype then lost regulation by RapGAP domain containing regulatory proteins. Only after the divergence of all four Ras-like GTPase subtypes did the radiative expansion of eukaryotes take place.
Expansion of the GTPases and their regulatory domains in animals.
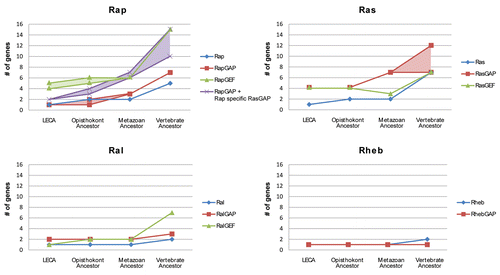
We have shown that the Ras-like GTPases and their regulatory domains (i.e., the RasGEF, RasGAP and RapGAP domain) are highly conserved in the eukaryotic lineages and that there is compelling evidence that all protein families occurred in multiple copies in the LECA. We have described in detail the phylogeny of all domains and protein families involved. Here we will discuss the evolutionary dynamics (i.e., expansion of the protein families over time) that these protein families display. In we show the expansion of the GTPases and their specific GEF and GAP proteins for each of the Ras, Ral, Rap and Rheb subtypes.
The number of GTPases and regulatory proteins remained relatively equal for a long time in evolution (LECA leading up to the Metazoa) but have greatly expanded (doubled) in the vertebrate ancestor (a period of roughly 500 million yearsCitation44) see –. The Ras and Rap GTPase subtypes and their regulators contributed the most to this expansion. The Rheb and Ral subtypes and regulators remained mostly constant in numbers with only a significant increase of the Ral specific GEFs in the vertebrate ancestor.
Interestingly the Ras and Rap subtype exhibit different dynamics compared to each other in regards to their regulatory domains. In the early expansion of the eukaryotic lineage the Ras subtype supported more GAP and GEF regulatory genes than there were Ras GTPases (four GEFs and four GAPs to one ancestral Ras GTPase). However, leading up to the vertebrate lineages the number of Ras specific GEFs remained equal as duplications were balanced by multiple loss events ( and ). In contrast, the RasGAP domain greatly increased in numbers, hence indicating a relevant importance of regulation via GAP proteins over regulation by GEF proteins in the expansion of the Ras subtype.
In contrast to the Ras subtype, the Rap subtype shows a dependency on GEF regulation over GAP regulation in the early eukaryote as GTPase and GAP numbers are similar (one and two genes respectively), but already supported seven Rap specific GEFs. Interestingly, the number of GAPs increased consistently in time and matches the number of Ras specific GEFs in the vertebrate ancestor (14 RapGEFs and 14 RapGAPs of which seven contain the RapGAP domain and another seven contain the C2—RasGAP domain architecture). The Rap subtype therefore displays an ancestral dependency on regulation by its GEFs in strong contrast to Ras. However, regulation by GAP activity at some point in the Opisthokont lineage acquired importance and the Rap specific GAPs started to expand. The slow expansion of the Ral and Rheb subtype regulatory systems is most likely linked to their specialized role in cellular signaling and regulation. The Rheb GTPase has a singular role in the eukaryotic TOR pathway, namely to regulating TOR activity based on nutrient availability and growth signals.Citation45 The Ral GTPase plays a critical role in exocytosis in animals.Citation36,Citation37 The Rap and Ras subtypes play important roles in multiple pathways and provide many cross-signaling routes between distinct signaling pathways. These multiple roles and a suspected need to separate the cellular role of activated Ras and Rap in the multitude of cross-signaling pathways might provide a reason for the expansion of the Ras and Rap regulatory systems compared to Ral and Rheb.
Evolution of Ras-like GTPase regulation.
We have discussed in detail the evolution of the Ras-like GTPases and their GAPs and GEFs and focused mainly on the co-evolution of the GTPases and their regulatory domains (i.e., the RasGAP, RapGAP and RasGEF domains). However, protein domains other than the GAP and GEF domains may also play an important role in the cellular functions of RasGEFs, RasGAPs and RapGAPs (for instants localization and regulation of catalytic activity by the GAP or GEF domains). We find that the domain architectures of these regulatory protein families are generally not conserved beyond the Metazoa (RasGEFs see van Dam et al.Citation8 RapGAPs see Fig. S1), suggesting intrinsic adaptability of Ras-like GTPase regulation via acquisition and loss of regulatory domains in RasGEFs and RapGAPs. However RasGAPs clearly show strong conservation of ancestral domain architectures. This indicates that the adaptability of Ras regulation via GAPs is constrained. Additionally, the expansion of the Ras-like GTPases and their regulatory proteins in evolution show that Ras GTPases are preferably regulated by GAPs while Rap GTPases are preferably regulated by GEFs. Together, the conservation of RasGAP domain architecture and an evolutionary preference for regulation via GAPs indicates a necessity for strict downregulation of canonical Ras GTPases.
By analyzing the evolution of not only the Ras-like GTPases but also the evolution of their regulatory domains we are able to paint a complete evolutionary scenario of the Ras-like GTPase protein family and it regulation. We have shown that the RasGAP, RapGAP and RasGEF domains co-evolved coherently with their respective GTPases indicating robustness in the way Ras-like GTPase activity are regulated throughout evolution. But on the other hand we observe flexibility in how the GAPs and GEFs themselves are regulated as observed in the variability of the domain architectures observed in the specific subtypes of GAPs and GEFs. We believe that this aspect, the regulation of the regulators, makes the Ras-like GTPase family a most versatile molecular switch in eukaryotic evolution. Altogether, our analysis provides a detailed evolutionary framework but also provides specific predictions for molecular biologists and biochemists working on Ras-like GTPases and their signaling pathways.
Methods
Genome selection.
We acquired best model protein sequences of 64 divergent eukaryotic species from EnsEMBL,Citation46 JGI, the Broad institute or their respective genome project sites. We have selected a wide range of animal and fungal genomes as most research on Ras signaling is being done in either animal or fungal model organisms. We also included a wide range of genomes belonging to other major phyla, such as the archeaplastida, chromalveolates and excavates, to be able to accurately time the duplication and loss events of the Ras-like GTPases as well as for the RasGAP and RapGAP domains. For a full overview of genomes, source and version information see Table S1 and Figure S3.
Phylogenetic analyses.
Identification of Ras-like GTPases and phylogenetic analysis. The sequences of the selected genomes were searched using the PfamCitation34 HMM profile for the Ras family (Pfam accession PF00071.12, Pfam version 23) and hmmsearch of the HMMER package version 2.3.2.Citation47 All sequences with a bitscore larger than 0 were selected. Due to the high sequence similarity of Ras to other small GTPases many other small GTPases are included in this set. An alignment of all sequences was made using the MAFFT programCitation9 with the ginsi option. A neighborjoining tree was constructed using the Quicktree program.Citation48 A sub tree was selected which contained all Ras-like subfamily members but no other small GTPases. The sequences were gathered from the initial alignment as manual inspection of the alignment produced from the subset showed it was suboptimal to the initial alignment. Subsequently a phylogenetic tree was constructed over all Ras-like subfamily members using RAxMLCitation10 (-T 4 -x 488761235 -f a -N 1000 -m PROTGAMMAIWAG), PhyMLCitation35 (phyml <file> 1 i 1 0 WAG e 6 e BIONJ y y) and Quicktree. For the Quicktree and RAxML analyses a 1000 bootstrap runs were performed. However the bootstrapping method implemented by the PhyML program is very slow compared to the RAxML bootstrap algorithm. We therefore used the bootstrap data from the RAxML run to calculate bootstrap values for the PhyML tree. All phylogenetic trees were visualized using Dendroscope.Citation49
Identification of RapGAP domain containing proteins and phylogenetic analysis. We gathered RapGAP domain sequences from the sequence set by using a custom made HMM model and hmmsearch of the HMMER package version 2.3.2. The custom RapGAP HMM model is based on an edited alignment of RapGAP domain sequences from a previous hmmsearch run using the Pfam RapGAP model. We did not use HMMER version 3 as the latest HMMER version does not allow full length domain detection (ls) and only returned segments. The domain sequences were aligned using MAFFT ginsi. A phylogenetic tree was constructed using RAxML (-T 6 -x 23421421 -f a -N 1000 -m PROTGAMMAIWAG). The domain architectures of the RapGAP containing proteins were visualized using the iTOL webserver application.Citation50 The domain architecture datasets as needed by the iTOL server were build using a custom perl script and data from hmmscan (HMMER package version 3.0b3) and Pfam version 24 hmm models.
Identification of RasGAP domain containing proteins and phylogenetic analysis. We gathered RasGAP domain containing proteins sequences from the sequence set by using the Pfam version 24 HMM model with hmmsearch of the HMMER package version 3.0b3. A custom build RasGAP HMM model did not result in the discovery of novel RasGAP domain containing proteins. The full length sequences were aligned using MAFFT ginsi to include conserved sequence positions bordering the detected Pfam domain (positions 2,870–5,010). A phylogenetic tree was constructed using RAxML (-T 8 -x 57231793 -f a -N 1000-m PROTGAMMAIWAG). The domain architectures of the RasGAP containing proteins were visualized using the iTOL webserver application.
Figures and Tables
Figure 1 Graphical summary of the evolution of the CDC25 Homology Domain as described in van Dam et al. For each class of RasGEFs we have traced back their perceived and reconstructed time of origin. For the mammalian RasGEFs we reconstructed the time of duplications to specific points in eukaryotic evolution. Note the differential loss of ancestral RasGEFs between animals and fungi. All gene names of fungal RasGEFs are from orthologs in S. cerevisiae unless specified otherwise. The evolutionary time scale is based on Douzery et al. but note that molecular dating is highly inaccurateCitation51 and that these dates are therefore approximate at best.
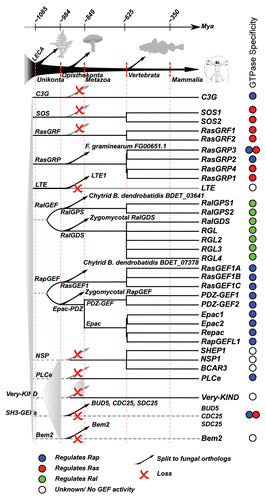
Figure 2 Representation of the phylogenetic tree of RapGAP domain containing proteins. We observe four clades, each of whicht represent a single ancestral gene in LECA . RalGAPB and RalGAPA each represent a single ancestral gene in LECA but cluster together indicating a common ancestral gene preceding LECA . The species present in each clade is depicted as a colored barcode. The differences in sequence between the Rap specific GAP sequences and the Rheb/Ral specific GAP sequences is well defined (100% bootstrap support) and a possible root may therefore lie between these two groups. *In the Rap specific clade we also observe GARNL3, a putative RapGAP, but it cellular function has not yet been reported. #It is unknown if RalGAPB harbors any GAP activity.
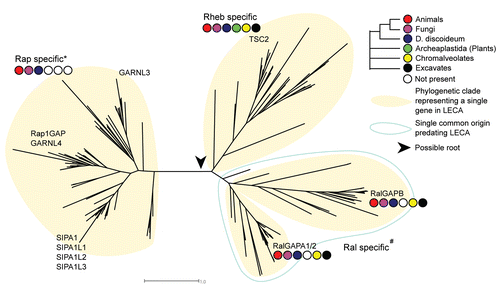
Figure 3 Simplified representation of the phylogenetic tree of RasGAP containing protein sequences. Gene names from human and S. cerevisiae are depicted as representative genes. Representative domain architectures for each clade are depicted in the tree and the five clades representing a single ancestral gene in or before LECA are numbered. The GAP1 and IQGAP clades from a combined clade in which a domain architecture is shared that predates LECA . The tree depicted has been modified to reflect proper grouping based on domain architecture: the clade containing RASA-like genes in D. discoideum, chromalveolates and N. gruberi is found between clusters 2 and (3,6) in the ML tree, see Figure S2.
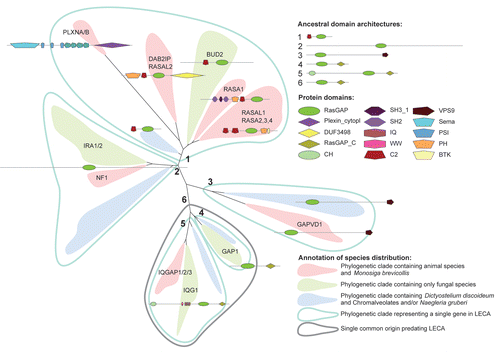
Figure 4 Evolutionary timeline of the Ras-like subfamily members. Many duplication events occurred in the common ancestor of the metazoa and vertebrates. The LECA likely contained multiple ancestral Ras-like GTPases although it is impossible to resolve how many there exactly were. Confident branches are depicted in black. Grey lines indicate possible alternative interpretations but are supported by circumstantial evidence. Gray dotted lines are based on a strict interpretation of the position of these GTPases in the phylogenetic tree but are otherwise unsupported and may originate from a more recent ancestral gene in the Unikont or Opisthokont ancestor. Fungal gene names are based on the orthologous genes in the yeast S. cerevisiae. The evolutionary time scale is based on Douzery et al. but note that molecular dating is highly inaccurateCitation51 and that these dates are therefore approximate at best.
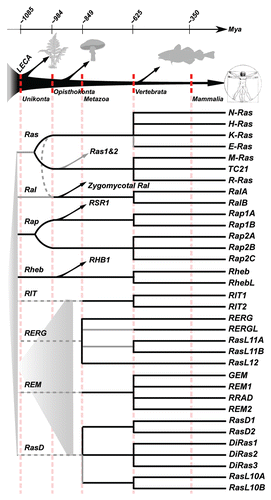
Figure 5 Evolutionary reconstruction of the Ras, Ral, Rap and Rheb GTPases based on co-evolution with their regulatory domains. (A) Matrix describing for each GTPase subtype which regulatory domain is active. (B–O) All scenarios possible for the order of duplications that gave rise to Ras, Rap, Ral and Rheb. For each scenario we reconstructed the order of invention and loss of regulation by the regulatory domains to fit the observed regulation of the GTPases as depicted in (A). Parts that exhibit the minimal number of events needed to fit the matrix in (A), (B) (4) and (C, D, H, M and N) (5 events) are marked.
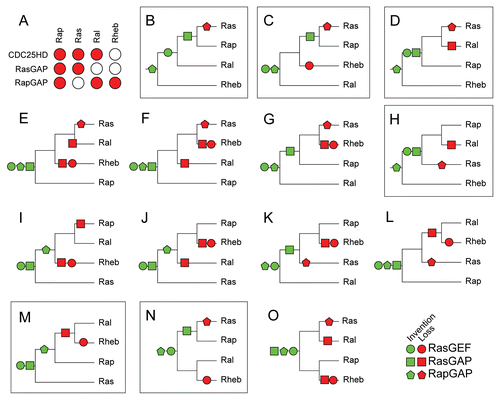
Figure 6 Expansion of Ras-like GTPase subtypes and their respective GEFs and GAPs (Ras-like regulatory system) in time leading up to mammalian organisms. We incorporated upper and lower estimates for GEFs and GAPs based on how the phylogenetic trees can be interpreted. The difference between higher and lower estimates for RapGAPs and RasGAPs is mainly caused by the RASA1 RasGAP protein family as GTPase specificity shifts within this family (see main text). The Rap GTPase regulatory system display a many to one GEF regulation network. This schema is maintained throughout eukaryotic evolution leading up to vertebrates. However with the inclusion of the C2-RasGAPs, the GAP regulation of Rap shows a similar trend. In contrast, the Ras regulatory system displays a many to one GAP regulation, while maintaining a relatively equal amount of Ras GTPase specific GEFs. The difference in the rate of expansion of the regulatory proteins for the Ras and Rap GTPase subtype indicates that there is a fundamental difference between the regulatory networks of Ras and Rap. Where Rap relies on multiple GEF and GAP proteins for its signaling diversification, Ras seems to rely mostly on its GAPs. The Ral and Rheb GTPases display a relatively compact regulatory system although in the vertebrates the number of RalGEFs expands significantly.
Additional material
Download Zip (1.3 MB)Acknowledgements
We like to thank Jos Boekhorst, Gabino Sanchez-Perez, Like Fokkens, Michael Seidl for their help in performing the analyses and support. This work is part of the BioRange program of the Netherlands Bioinformatics Centre (NBIC), which is supported by a BSIK grant through the Netherlands Genomics Initiative (NGI). The sequence data of selected genomes were produced by the US Department of Energy Joint Genome Institute http://www.jgi.doe.gov/, in collaboration with the user community and the Fungal Genome Initiative of the Broad Institute. For a full overview of the genomes see Supplemental Table S1 and Figure S3.
References
- Wennerberg K, Rossman KL, Der CJ. The Ras superfamily at a glance. J Cell Sci 2005; 118:843 - 846; http://dx.doi.org/10.1242/jcs.01660
- Yutin N, Wolf MY, Wolf YI, Koonin EV. The origins of phagocytosis and eukaryogenesis. Biology direct 2009; 4:9; http://dx.doi.org/10.1186/1745-6150-4-9
- Jékely G. Small GTPases and the evolution of the eukaryotic cell. BioEssays 2003; 25:1129 - 1138; http://dx.doi.org/10.1002/bies.10353
- Brighouse A, Dacks JB, Field MC. Rab protein evolution and the history of the eukaryotic endomembrane system. Cell Mol Life Sci 2010; 1:17; http://dx.doi.org/10.1007/s00018-010-0436-1
- Boureux A, Vignal E, Faure S, Fort P. Evolution of the Rho family of ras-like GTPases in eukaryotes. Mol Biol Evol 2007; 24:203 - 216; http://dx.doi.org/10.1093/molbev/msl145
- Elias M, Patron NJ, Keeling PJ. The RAB family GTPase Rab1A from Plasmodium falciparum defines a unique paralog shared by chromalveolates and rhizaria. J Eukaryot Microbiol 56:348 - 356; http://dx.doi.org/10.1111/j.1550-7408.2009.00408.x
- Bos JL, Rehmann H, Wittinghofer A. GEFs and GAPs: critical elements in the control of small G proteins. Cell 2007; 129:865 - 877; http://dx.doi.org/10.1016/j.cell.2007.05.018
- Dam TJP van, Rehmann H, Bos JL, Snel B. Phylogeny of the CDC25 homology domain reveals rapid differentiation of Ras pathways between early animals and fungi. Cell Signal 2009; 21:1579 - 1585; http://dx.doi.org/10.1016/j.cellsig.2009.06.004
- Katoh K, Misawa K, Kuma K, Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res 2002; 30:3059 - 3066; http://dx.doi.org/10.1093/nar/gkf436
- Stamatakis A, Ludwig T, Meier H. RAxML-III: a fast program for maximum likelihood-based inference of large phylogenetic trees. Bioinformatics 2005; 21:456 - 463; http://dx.doi.org/10.1093/bioinformatics/bti191
- Gridley S, Chavez JA, Lane WS, Lienhard GE. Adipocytes contain a novel complex similar to the tuberous sclerosis complex. Cell Signal 2006; 18:1626 - 1632; http://dx.doi.org/10.1016/j.cellsig.2006.01.002
- Shirakawa R, Fukai S, Kawato M, Higashi T, Kondo H, Ikeda T, et al. Tuberous sclerosis tumor suppressor complex-like complexes act as GTPase-activating proteins for Ral GTPases. J Biol Chem 2009; 284:21580 - 21588; http://dx.doi.org/10.1074/jbc.M109.012112
- Scrima A, Thomas C, Deaconescu D, Wittinghofer A. The Rap-RapGAP complex: GTP hydrolysis without catalytic glutamine and arginine residues. EMBO J 2008; 27:1145 - 1153; http://dx.doi.org/10.1038/emboj.2008.30
- Chen XQ. The Myotonic Dystrophy Kinase-related Cdc42-binding Kinase Is Involved in the Regulation of Neurite Outgrowth in PC12 Cells. J Biol Chem 1999; 274:19901 - 19905; http://dx.doi.org/10.1074/jbc.274.28.19901
- Lodhi IJ, Chiang SH, Chang L, Vollenweider D, Watson RT, Inoue M, et al. Gapex-5, a Rab31 guanine nucleotide exchange factor that regulates Glut4 trafficking in adipocytes. Cell Metab 2007; 5:59 - 72; http://dx.doi.org/10.1016/j.cmet.2006.12.006
- Sato M, Sato K, Fonarev P, Huang CJ, Liou W, Grant BD. Caenorhabditis elegans RME-6 is a novel regulator of RAB-5 at the clathrin-coated pit. Nat Cell Biol 2005; 7:559 - 569; http://dx.doi.org/10.1038/ncb1261
- Hunker CM, Galvis A, Kruk I, Giambini H, Veisaga ML, Barbieri MA. Rab5-activating protein 6, a novel endosomal protein with a role in endocytosis. Biochem Bioph Res Co 2006; 340:967 - 975; http://dx.doi.org/10.1016/j.bbrc.2005.12.099
- Hart MJ, Callow MG, Souza B, Polakis P. IQGAP1, a calmodulin-binding protein with a rasGAP-related domain, is a potential effector for cdc42Hs. EMBO J 1996; 15:2997 - 3005
- Brill S, Li S, Lyman C, Church D, Wasmuth J, Weissbach L, et al. The Ras GTPase-activating-protein-related human protein IQGAP2 harbors a potential actin binding domain and interacts with calmodulin and Rho family GTPases. Mol Cell Biol 1996; 16:4869 - 4878
- Osman MA. Iqg1p, a Yeast Homologue of the Mammalian IQGAPs, Mediates Cdc42p Effects on the Actin Cytoskeleton. J Cell Biol 1998; 142:443 - 455; http://dx.doi.org/10.1083/jcb.142.2.443
- Imai Y, Miyake S, Hughes DA, Yamamoto M. Identification of a GTPase-activating protein homolog in Schizosaccharomyces pombe. Mol Cell Biol 1991; 11:3088 - 3094
- Johannessen CM, Reczek EE, James MF, Brems H, Legius E, Cichowski K. The NF1 tumor suppressor critically regulates TSC2 and mTOR. P Natl Acad Sci USA 2005; 102:8573 - 8578; http://dx.doi.org/10.1073/pnas.0503224102
- Park HO, Chant J, Herskowitz I. BUD2 encodes a GTPase-activating protein for Bud1/Rsr1 necessary for proper bud-site selection in yeast. Nature 1993; 365:269 - 274; http://dx.doi.org/10.1038/365269a0
- Krapivinsky G, Medina I, Krapivinsky L, Gapon S, Clapham DE. SynGAP-MUPP1-CaMKII synaptic complexes regulate p38 MAP kinase activity and NMDA receptor-dependent synaptic AMPA receptor potentiation. Neuron 2004; 43:563 - 574; http://dx.doi.org/10.1016/j.neuron.2004.08.003
- Kupzig S, Deaconescu D, Bouyoucef D, Walker SA, Liu Q, Polte CL, et al. GAP1 family members constitute bifunctional Ras and Rap GTPase-activating proteins. J Biol Chem 2006; 281:9891 - 9900; http://dx.doi.org/10.1074/jbc.M512802200
- Pena V, Hothorn M, Eberth A, Kaschau N, Parret A, Gremer L, et al. The C2 domain of SynGAP is essential for stimulation of the Rap GTPase reaction. EMBO Rep 2008; 9:350 - 355; http://dx.doi.org/10.1038/embor.2008.20
- Bos JL. Linking Rap to cell adhesion. Curr Opin Cell Biol 2005; 17:123 - 128; http://dx.doi.org/10.1016/j.ceb.2005.02.009
- Bos JL. ras oncogenes in human cancer: a review. Cancer Res 1989; 49:4682 - 4689
- Colicelli J. Human RAS superfamily proteins and related GTPases. Science STKE 2004; 13; http://dx.doi.org/10.1126/stke.2502004re13
- Cox AD, Der CJ. Ras history. Small GTPases 2010; 1:2 - 27; http://dx.doi.org/10.4161/sgtp.1.1.12178
- Karnoub AE, Weinberg RA. Ras oncogenes: split personalities. Nat Rev Mol Cell Bio 2008; 9:517 - 531; http://dx.doi.org/10.1038/nrm2438
- Drivas GT, Palmieri S, D'Eustachio P, Rush MG. Evolutionary grouping of the RAS-protein family. Biochem Bioph Res Co 1991; 176:1130 - 1135; http://dx.doi.org/10.1016/0006-291X(91)90402-S
- Valencia A, Chardin P, Wittinghofer A, Sander C. The ras protein family: evolutionary tree and role of conserved amino acids. Biochemistry 1991; 30:4637 - 4648
- Finn RD, Tate J, Mistry J, Coggill PC, Sammut SJ, Hotz HR, et al. The Pfam protein families database. Nucleic Acids Res 2008; 36:281 - 288; http://dx.doi.org/10.1093/nar/gkm960
- Guindon S, Gascuel O. A simple, fast and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol 2003; 52:696 - 704; http://dx.doi.org/10.1080/10635150390235520
- Moskalenko S, Henry DO, Rosse C, Mirey G, Camonis JH, White MA. The exocyst is a Ral effector complex. Nat Cell Biol 2002; 4:66 - 72; http://dx.doi.org/10.1038/ncb728
- Lipschutz JH, Mostov KE. Exocytosis: the many masters of the exocyst. Curr Biol 2002; 12:212 - 214; http://dx.doi.org/10.1016/S0960-9822(02)00753-4
- Kitayama H, Sugimoto Y, Matsuzaki T, Ikawa Y, Noda M. A ras-related gene with transformation suppressor activity. Cell 1989; 56:77 - 84; http://dx.doi.org/10.1016/0092-8674(89)90985-9
- Fritz-Laylin LK, Prochnik SE, Ginger ML, Dacks JB, Carpenter ML, Field MC, et al. The Genome of Naegleria gruberi Illuminates Early Eukaryotic Versatility. Cell 2010; 140:631 - 642; http://dx.doi.org/10.1016/j.cell.2010.01.032
- Wullschleger S, Loewith R, Hall MN. TOR signaling in growth and metabolism. Cell 2006; 124:471 - 484; http://dx.doi.org/10.1016/j.cell.2006.01.016
- Inoki K, Ouyang H, Zhu T, Lindvall C, Wang Y, Zhang X, et al. TSC2 integrates Wnt and energy signals via a coordinated phosphorylation by AMPK and GSK3 to regulate cell growth. Cell 2006; 126:955 - 968; http://dx.doi.org/10.1016/j.cell.2006.06.055
- Inoki K, Zhu T, Guan KL. TSC2 Mediates Cellular Energy Response to Control Cell Growth and Survival. Cell 2003; 115:577 - 590; http://dx.doi.org/10.1016/S0092-8674(03)00929-2
- Garami A, Zwartkruis FJT, Nobukuni T, Joaquin M, Roccio M, Stocker H, et al. Insulin Activation of Rheb, a Mediator of mTOR/S6K/4E-BP Signaling, Is Inhibited by TSC1 and 2. Mol Cell 2003; 11:1457 - 1466; http://dx.doi.org/10.1016/S1097-2765(03)00220-X
- Douzery EJP, Snell EA, Bapteste E, Delsuc F, Philippe H. The timing of eukaryotic evolution: does a relaxed molecular clock reconcile proteins and fossils?. P Natl Acad Sci USA 2004; 101:15386 - 15391; http://dx.doi.org/10.1073/pnas.0403984101
- Aspuria PJ, Tamanoi F. The Rheb family of GTPbinding proteins. Cell Signal 2004; 16:1105 - 1112; http://dx.doi.org/10.1016/j.cellsig.2004.03.019
- Hubbard TJ, Aken BL, Ayling S, Ballester B, Beal K, Bragin E, et al. Ensembl 2009. Nucleic Acids Res 2009; 37:690 - 697; http://dx.doi.org/10.1093/nar/gkn828
- Eddy SR. Profile hidden Markov models. Bioinformatics 1998; 14:755 - 763; http://dx.doi.org/10.1093/bioinformatics/14.9.755
- Howe K, Bateman A, Durbin R. QuickTree: building huge Neighbour-Joining trees of protein sequences. Bioinformatics 2002; 18:1546 - 1547; http://dx.doi.org/10.1093/bioinformatics/18.11.1546
- Huson D, Richter D, Rausch C, Dezulian T, Franz M, Rupp R. Dendroscope: An interactive viewer for large phylogenetic trees. BMC bioinformatics 2007; 8:460; http://dx.doi.org/10.1186/1471-2105-8-460
- Letunic I, Bork P. Interactive Tree Of Life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics 2007; 23:127 - 128; http://dx.doi.org/10.1093/bioinformatics/btl529
- Roger AJ, Hug LA. The origin and diversification of eukaryotes: problems with molecular phylogenetics and molecular clock estimation. Philos T Roy Soc B 2006; 361:1039 - 1054; http://dx.doi.org/10.1098/rstb.2006.1845