Abstract
Cellular signaling by small GTPases is critically dependent on proper spatio-temporal orchestration of activation and output. In addition to their core G (guanine nucleotide binding)-domain, small GTPases comprise a hypervariable region (HVR) and a lipid anchor that are generally accepted to control subcellullar localization. The HVR encodes in many small GTPases a polybasic region (PBR) that permits charge-mediated association to the inner leaflet of the plasma membrane or to intracellular organelles. Over the past 15–20 years, evidence has accumulated for specific protein–protein interactions, mediated by the HVR, that control both targeting and signaling specificity of small GTPases. Using the RhoGTPase Rac1 as a paradigm we here review a series of protein partners that require the Rac1 HVR for association and that control various aspects of localized Rac1 signaling. Some of these proteins represent Rac1 activators, whereas others mediate Rac1 inactivation and degradation and yet others potentiate Rac1 downstream signaling. Finally, evidence is discussed which shows that the HVR of Rac1 also contributes to effector interactions, co-operating with the N-terminal effector domain. The complexity of localized Rac1 signaling, reviewed here, is most likely exemplary for many other small GTPases as well, representing a challenge to identify and define similar mechanisms controlling the specific signaling induced by small GTPases.
Keywords: :
Introduction
Cellular signaling employs complex networks that are composed of cell surface receptors, adaptor proteins, molecular switches, and a host of downstream effector pathways that are all spatially organized by the actin and microtubule cytoskeleton as well as by vesicle transport and membrane dynamics. These effector pathways may require various types of enzyme that either feedback positively or negatively into surface receptors, modulate actin dynamics and adhesion molecules, or transfer their signal into the nucleus, resulting in transcriptional regulation, cell differentiation or growth. The nature of a cells’ intracellular response to an outside stimulus is determined early in the signaling cascade, at or near the plasma membrane. This is the site at which small GTPases of the Ras and Rho families trigger pathways that control cell growth and differentiation, cytoskeletal dynamics, adhesion and migration.
The molecular mechanisms that govern GTPase activation, inactivation and downstream signaling have been extensively studied over the past 25 years. The model describing small GTPases as molecular switches that can cycle between an “off” and an “on” state is in good agreement with models for other guanine nucleotide binding proteins, including elongation factor Tu and heterotrimeric G-proteins, that share sequence homology and biochemical properties with small GTPases.Citation1,Citation2 The GTPase “switch” is represented by the binding to either GTP (“on”) or GDP (“off”). Upstream activation of GTPases requires GEFs (Guanine Nucleotide Exchange Factors) that promote the exchange of bound GDP for GTP, which is in excess in the cytosol. Inactivation requires GTP hydrolysis, an intrinsic but low activity that is stimulated by GAPs (GTPase Activating Proteins). GTPases of the Rho and Rab families are, in the inactive, GDP-bound form, associated to cytosolic chaperones called GDIs (Guanine nucleotide Dissociation Inhibitors). Other regulators include the GDS (Guanine nucleotide Dissociation Stimulator) proteins, such as Smg (Small G-protein) GDS or Ral-GDS, that also acts as GEFs, activating subclasses of the superfamily of small GTPases.Citation3
In the active, GTP-bound, conformation, small GTPases bind to effector molecules, translating the upstream signal into downstream responses. The regions required for effector binding are primarily located in the GTPase N-terminus, from amino acid ~25–40.Citation4 These interactions are quite specific, as single point mutations can generate GTPase mutants that bind to one and not another effector protein, as was shown for Rac1.Citation5 Additional regions located more toward the C-terminus have been implicated in effector binding as well.Citation6 Rho GTPases encode a unique “insert region,” i.e., amino acids 123–135 (Rac1 numbering), which is not found in other small GTPases. This region has been implicated in regulating a subset of effector pathways such as actin dynamicsCitation7 and activation of the NADPH oxidase,Citation8 albeit that the relevance of the insert region for oxidase activation was not confirmed in other studies.Citation9,Citation10
Most families of small G-proteins comprise several members that can be highly homologous in sequence, in particular in the G (Guanine nucleotide-binding)-domain, the core structure of small GTPases. In this overview, we will focus on the Rho-family of small GTPases (22 members), of which some, such as the Rac1 and Rac2 GTPases, share over 90% sequence identity.Citation11 This identity also includes the effector domains, suggesting that other regions determine signaling specificity of the Rac GTPases. The most likely region controlling such specificity is the hypervariable region (HVR) in the GTPase C-terminus. This small region shows striking sequence diversity among related GTPases, which has been linked to their differential localization to intracellular membranes, nuclear translocation as well as binding to regulatory- and effector- proteins.Citation12-Citation15
In the early 90s a series of studies established the functional importance of the HVR of Rac GTPases by focusing on its role in the activation of the NADPH oxidase complex. This complex was, at the time, already identified as an established Rac effector, responsible for production of reactive oxygen species in granulocytes.Citation16 Using purified proteins and membrane extracts in a cell-free assay, the activity of the oxidase complex could be quantified, which allowed the testing of Rac1 and Rac2 mutants as well as interfering peptides. Whereas GTP-bound Rac1 binds through its N-terminal effector domain to the principal oxidase effector component, p67phox, and to a region more toward the C-terminus,Citation17 several studies showed that the hypervariable C-terminal domain of Rac proteins was also critical for oxidase activation.Citation18-Citation21 This could relate to the fact that the HVR is required for proper targeting of Rac1 in the cell-free assay. Targeting of most small GTPases to membranes also requires modification of the C-terminus by covalent attachment of a lipid anchor.Citation22 However, in the cell-free oxidase assays, most Rac proteins used were made in E. coli and thus not prenylated. Peptides, encoding the HVR could interfere with oxidase activation by the full-length Rac protein, further underscoring the notion that the HVR of Rac1 was responsible for this effect. However, whether this result was sequence-specific or due to the highly charged nature of the HVR has been subject of conflicting findings.Citation18,Citation19 Differential binding of GTPases to cellular membranes and compartments has been demonstrated using GFP (Green Fluorescent Protein)-fusion proteins of different HVRs of a subset of RhoGTPases. This shows that the HVRs are not only required but also sufficient for such targeting. Moreover, differential localization correlated with the number of basic residues in the C-terminus, with highly basic domains targeting preferentially to the plasma membrane.Citation12
Also nuclear localization of Rac1 has been linked, in several studies, to its HVR. The Rac1 C-terminus harbors a nuclear localization signal (NLS) represented by its PBR.Citation13,Citation23 A fusion protein in which the Rac1 C-terminus is coupled to GFP shows a nuclear localization, indicating that the Rac1 C-terminus is sufficient to drive proteins into the nucleus of MDCK, COS-1, porcine aortic endothelial (PAE) cells as well as ECV304 human bladder carcinoma, HeLa and NIH 3T3 cells.Citation24 The NLS is inactive when Rac1 is bound to RhoGDI, which explains its cytosolic localization. Nuclear translocation of Rac GTPases has been linked to their degradation.Citation13,Citation25 Rac1 degradation, in turn, requires its activation and can be induced by the CNF1 (Cytotoxic Necrotizing Factor1) toxin from E. coli. CNF1 de-amidates Rac/Rho GTPases at position 61/63, thereby generating a constitutively active GTPase.Citation26 Degradation of CNF-activated Rac1, but not Rac2 or Rac3, is dependent on the HVR, since the Rac1 HVR, swapped for the Rac2 HVR, promotes degradation of a full length Rac2/Rac1 HVR chimera. This finding is in good agreement with the absence of a NLS in the Rac2 and Rac3 C-termini and the notion that nuclear translocation of activated Rac1 is required for proteasome-mediated degradation.Citation13,Citation25
Whereas the targeting of small GTPases by a C-terminal lipid anchor and charge-dependent association of the PBR to the plasma membrane is clearly a key mechanism that controls differential localization and signaling, evidence has been accumulating over the past 15–20 years for supplementary modes of regulation, based on specific protein–protein interactions. In all this work, the HVR of Rac1 has been the most extensively studied. Here, we will review a series of protein partners of Rac1 that require the HVR for their association (). This represents a diverse group of proteins that reside in various locations, in line with the notion that Rac1 exerts distinct functions in different cellular compartments.
Figure 1. Overview of Rac1-interacting proteins. In the left part of the figure, proteins known to bind to the effector domain (ED) of Rac1 via their CRIB domain or not are listed. In the right part of the figure, proteins known to bind to the hypervariable (HV) region are listed. These proteins are clustered dependent on their known interactions. See text for details.
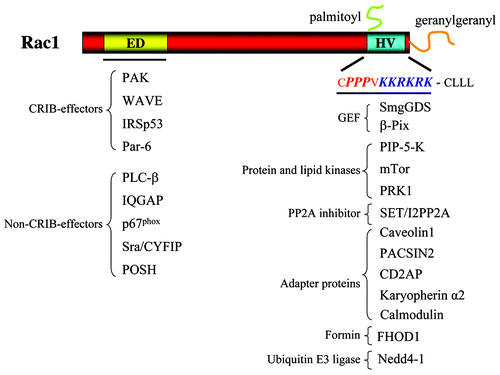
Protein–Protein Interactions Mediated by the Rac1 C-Terminus
Activation by guanine nucleotide exchange factors
Because the C-terminal HVR of RhoGTPases is one of the main features discriminating these highly homologous family members, it represents a prime determinant for specific interaction with one of the many (~80) RhoGEFsCitation3 that have been identified. However, GTPase-GEF interactions are mediated by the switch I and switch II regions, rather than by the C-terminus, as was shown, for example, for the GEFs Tiam1 as well as Vav1.Citation27 Many GEFs can activate several different RhoGTPases,Citation3 that are sometimes highly homologous in their N-terminal Switch I/II regions. Specificity of GEF-mediated activation is therefore the result of cell-specific expression of the GEFs as well as of the GTPases.Citation28 As exceptions to this, there are at least two GEFs for which there is evidence that they also bind to the Rac1 HVR, i.e., smgGDS (small G-protein dissociation stimulator) and β-PIX (Pak-Interacting eXchange factor).
SmgGDS
The armadillo protein smgGDS is an atypical GEF that shows selectivity toward some, but not all RhoGTPases.Citation29 SmgGDS also binds to the inactive N17 mutants of H- and N-Ras, but does not promote nucleotide exchange. Further experiments showed that the PBR present in the C-terminus of RhoA and Rac1 but not in H- or N-Ras, is necessary but not sufficient for smgGDS-mediated nucleotide exchange.Citation29 A more recent study further confirmed the requirement for the RhoA C-terminus for nucleotide exchange by smgGDS.Citation30 This work showed that smgGDS is a bona fide GEF for RhoA and RhoC, but, in contrast to the study by Vikis et al.,Citation29 does not activate other RhoGTPases including Rac1, Rac2 and Cdc42.
The laboratory of Williams and colleagues has studied the interaction between smgGDS and Rac1, focusing on the Rac1 C-terminal PBR.Citation23 Rac1 binds smgGDS through this region and this interaction controls the nucleo-cytoplasmic shuttling of the Rac1-smgGDS complex. The Rac1 PBR comprises, in contrast to that of RhoA, a nuclear localization signal and Rac1 mutants lacking the PBR, do not localize to the nucleus.Citation13 These authors further proposed that smgGDS-promoted Rac1 activation stimulated nuclear translocation of the complex, whereas smgGDS binding to and activating RhoA would promote its cytoplasmic accumulation, due to the absence of a NLS in RhoA.
The functional consequences of the nuclear shuttling of the Rac1-smgGDS complex are not clear. Lanning et al. proposed that the complex would dissociate upon nuclear entry releasing Rac1 from smgGDS to allow local signaling.Citation13 Later studies have linked nuclear translocation of activated Rac1, dependent on its PBR and its interaction with the armadillo protein karyopherin α2, to Rac1 degradation (See below; refs 13 and 25 ).
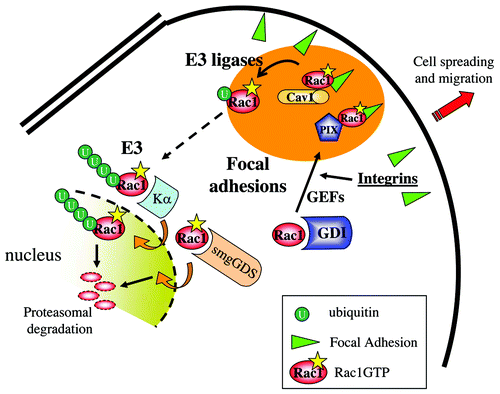
Figure 2. Rac1 interactions at Focal Adhesions and in the nucleus. Figure provides a model of the different local interactions of Rac1 with the various C-terminal-interacting proteins. The various regions that focus Rac1 activity and regulation are indicated by the different colors. Integrin-mediated Rac1 activation, possibly by PIX, leads to its targeting to FAs and recruitment of Caveolin1. Caveolin1 subsequently regulates Rac1 ubiquitylation and internalization. Internalized Rac1 may associate to Karyopherin α, translocate to the nucleus for further ubiquitylation and proteasomal degradation.
Beta-PIX
The Rac1/Cdc42 GEF β-PIX (also known as ArhGEF7) binds to the Rac1 C-terminal proline-rich region, directly N-terminal of the PBR.Citation31 This finding was based on the notion that this region is similar to an earlier defined PIX-binding motif in PAK, which is in fact an atypical SH3 domain binding motif.Citation32 We could show that the SH3 domain of PIX was required for efficient binding to the proline stretch in the Rac1 C-terminus, and that in HEK293 cells this sequence was required for targeting the activated V12Rac1 mutant to Focal Adhesions (FAs) (). In addition, β-PIX also serves as a genuine Rac1-targeting protein, since a Rac1C186S mutant, that cannot be prenylated and localizes primarily to the nucleus, shows increased cytosolic and membrane localization upon co-transfection with β-PIX.Citation31
Since PAK also uses a proline-rich domain for β-PIX binding, these interactions will result in competition between PAK1 and Rac1. This suggests that, following β-PIX-mediated activation of Rac1, Rac1 will bind through its effector domain to PAK, which may then replace Rac1 from binding to the β-PIX-SH3 domain, allowing Rac1 signaling and subsequent inactivation. Interestingly, an earlier study had shown that the Rac1 PBR is also required for PAK1 binding.Citation33 This suggests that, perhaps transiently, a trimeric complex can form comprising Rac1, PAK and β-PIX. However, formal proof will require additional biochemical studies using the appropriate mutants.
Previously, PAK was proposed to function upstream of Rac1, mediating CDC42-dependent Rac1 signaling. Thus, the PIX-PAK complex may well be part of a positive feedback loop, potentiating Rac1 signaling at FAs. The notion of such a signaling pathway is supported by work from Romer and colleagues, who showed that Focal Adhesion Kinase (FAK) can phosphorylate βPIX, promoting its binding to Rac1 as well as the targeting of Rac1 to Fas.Citation34
Downstream effectors: Protein- and lipid-kinases
Although most protein kinases in Rac1 signaling pathways act as effectors that require the GTP-induced conformational switch to become activated (), for some of these an interaction with the Rac1 HVR was shown. This suggests that co-operativity exists between this region and the N-terminal effector domain in Rac1.
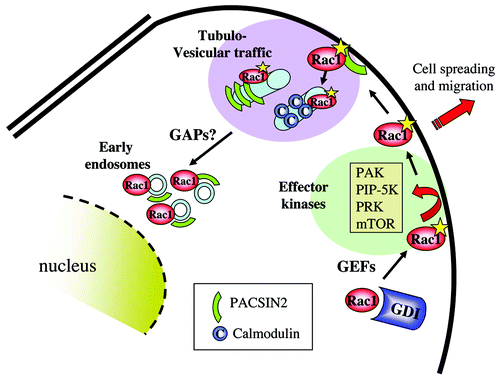
Figure 3. Internalization of activated Rac1 and binding to effector kinases. Activated Rac1 may interact with a series of protein kinases at the plasma membrane. These represent primarily effector kinases such as PAK, PRK, PIP-5-K and mTor. Active Rac1 at the membrane also associates with PACSIN2, which regulates Rac1 internalization toward early endosomes, paralleled by GAP-mediated Rac1 inactivation.
PAK1
Using Far-Western blotting, Knaus et al.,Citation33 showed that the HVR is required for efficient binding of GTPγS-loaded Rac1 to PAK1. In line with this, stimulation of PAK1 auto-phosphorylation was severely reduced by a PBRQ mutant of Rac1, in which all basic residues in the PBR are mutated to Gln (Q). This suggests that the Rac1 PBR is also involved in PAK1 activation. Use of chimeras showed that Rac1 with the intact Rac2 C-terminus was less efficient in activating PAK1, whereas a Rac2 chimera with the Rac1 C-terminus gained capacity to activate PAK1.Citation33 In this study, the PBR was proposed to act as a novel “effector domain.” Whereas this is true for Rac1 effectors in general remains to be established, but there are other examples, such as for the kinase PRK (see below), that suggest that the PBR in Rac1 is more than just a membrane-targeting sequence.
PIP-5-K
In search for a link between actin-regulating RhoGTPases and actin-regulating lipid kinases, Tolias et al.Citation35,Citation36 showed that Rac1 binds to PIP-5-K (phosphatidylinositol-4-phosphate 5-kinase) as well as to DGK (diacylglycerol kinase) in a nucleotide-independent manner. In contrast, the association of Rac1 with PI3-Kinase was found to be GTP-dependent.Citation35 In a subsequent study it was shown that Rac1, but not RhoA or Cdc42, associated, through its HVR, to PIP-5-K and DGK. This association was subsequently linked to K186 in the Rac1 HVR.Citation37
The interaction of Rac1 with these lipid kinases was stimulated in vitro by a subset of phosopholipids such as PS (phosphatidylserine), PA (phosphatidic acid) and PI-3,4-P2, but not PC (phosphatidylcholine) or PI-3,4,5,-P3. Moreover, PIP-5-K and DGK were found to complex both with Rac1 and with RhoGDI. Rac1 associates to RhoGDI through a series of protein-interface-based interactions, as well as through its geranylgeranyl anchor.Citation38,Citation39 In this complex, the HVR appears to be relatively exposed, allowing interactions with other proteins, such as the lipid kinases discussed here. In fact, our lab has also shown that a Rac1 C-terminal peptide does not bind to RhoGDI, but does compete for an association between PIX and RhoGDI.Citation31 This suggests that Rac1 simultaneously binds to PIX, through its C-terminus, and to RhoGDI, using both its lipid anchor and regions in the Rac1 N-terminus.
The model proposed by Tolias et al. suggested that the DGK-PIP-5-K axis generates, once at the membrane, the phospholipids that would promote dissociation of RhoGDI allowing Rac1 activation.Citation36 Later studies suggested various lipid-based feed-back loops in the complex. Tolias et al.Citation37 showed that PIP-5-K acts as a Rac1 effector, producing PIP2 that promotes actin uncapping and local actin polymerization.Citation37 More recently, Chae et al.Citation40 presented a model in which phosphatidic acid, derived from integrin-regulated phospholipase D but which can also be synthesized by DGK, binds to the HVR of Rac1, leading to GDI dissociation and recruitment of GTP-bound Rac1 to the plasma membrane.
Our own lab later confirmed the PIP-5-K binding of Rac1 through its HVR and identified the proline-rich region in the HVR, as a region in Rac1 that can bind to SH3 domains (as in Crk, PIX, CD2AP and PACSIN2).Citation31,Citation41-Citation43 We showed that protein–protein interactions that required the PBR did not necessarily require the proline-rich domain but that, conversely, the proline-dependent interactions did require the PBR. This is in agreement with other SH3 domain-based interactions wherein one or more downstream basic residues are required to stabilize the binding to the proline-rich sequence.Citation44,Citation45
PRK
The PRK kinases (Protein Kinase C-related kinase, also known as Protein Kinase N, PKN) are established effectors of both RhoA and Rac1.Citation46,Citation47 These kinases regulate cytoskeletal dynamics, as well as transcriptional activity and mitogenesis.Citation48 The analysis by scintillation proximity assays and NMR of the interaction between Rac1 and PRK1 supported a model in which the Rac1 PBR mediates specific interactions with effector proteins.Citation15,Citation49 In these analyses, the relevance of any of the basic residues for the interaction were underscored and a model depicting the specific contribution of the Rac1 C-terminus in PRK1 binding was presented. Intriguingly, the analysis by Modha et al. suggested that the Rac1 C-terminus has a rather fixed structure comprising a proline-based α-helix, followed by an extended stretch of large basic residues.Citation15 Importantly, the C-terminal portion of this region (Lys 183-Arg185) was proposed to loop back to interact with the core G-domain, in particular with portions of the switch II region and the region C-terminal to helix 3. In addition, several of the basic residues in the PBR contact the cognate binding region in PRK1.Citation15 Since the Rac1 C-terminus is predicted to interact with its switch II region (i.e., Pro73, Gln74), it is likely that GTP binding does affect the relative position of the C-terminus, and thereby the affinity for effectors such as PRK1. Reminiscent of this is our own finding that Rac1 binding to the ubiquitin ligase Nedd4, for which the Rac1 C-terminus is sufficient, is more efficient for activated Rac1 (Q61 mutant) as compared with inactive Rac1 (N17 mutant).
mTor
The mTor kinase is part of the mTORC1 and mTORC2 complexes that relay growth factor signaling, nutrient status and oxygen concentration into cellular responses such as gene translation and protein synthesis.Citation50 The mTORC2 complex signals toward the Akt kinase and regulates the actin cytoskeleton through RhoGTPases.Citation51 Recent work showed that RhoGTPases, in particular Rac1, can also function upstream of mTOR.Citation52 These authors showed that Rac1 binds in a nucleotide-independent fashion to mTOR though the Rac1 HVR, specifically through the PBR, but not the proline-rich region. Importantly, it was shown that Rac1 controls the subcellullar localization of mTOR, recruiting the kinase to the plasma membrane and, in parallel, activating the kinase. These data suggest a model in which activated Rac1, which localizes to membranes, recruits and activates mTOR and thereby both mTORC1 and mTORC2 complexes, through the binding via the Rac1 PBR. Rac1 thus acts both upstream and downstream of mTOR and is among a growing group of small GTPases, including Rheb and RalA, that regulate different aspects of TOR-mediated signaling.Citation53 It is likely that this novel interaction between Rac1 and mTOR is of relevance for many biological systems, including platelet activation as well as cancer cell motility and metastasis.Citation54,Citation55
Regulation of Rac1 by adaptor proteins
CD2-associated protein
CD2-associated proteinCitation56 (CD2AP, also known as CMS: Cas ligand with multiple Src homology 3 (SH3) domains) is an adaptor protein implicated actin dynamics, cell adhesion and receptor traffic.Citation57-Citation59 CD2AP harbors three SH3 domains in its N-terminus and an actin binding region, proline-rich domains and a coiled-coil region in its C-terminus. CD2AP is also closely related to CIN85 (Cbl-interacting protein, also known as Ruk, SETA, SH3KBP1).Citation56 CD2AP is best known for its role in the kidney, where it links nephrins, homotypic cell-adhesion proteins that are critical for renal filtration, to the actin cytoskeleton. As a result, mice lacking CD2AP show kidney failure and die at 6 weeks of age.Citation60,Citation61 Interestingly, nephrin was previously shown to bind to the Rac1 effector IQGAP1 suggesting Rac1 signaling affects podocyte cell–cell contacts.Citation62 This is further supported by the analysis of the Rac1GAP ArhGAP24, which is mutated in glomerulosclerosis, in line with the notion that the balance between Rac1 and RhoA signaling is important for proper kidney function.Citation63
We found CD2AP in a screen for proteins binding to the peptide encoding the Rac1 C-terminus.Citation42 This binding is very efficient, most likely because the proline-rich region in the Rac1 peptide can bind to all three SH3 domains. Mutating each of these individually in the context of full-length CD2AP did not dramatically block binding of Rac1, indicating that Rac1 can bind to either SH3 domain in CD2AP. Interestingly, we found that in epithelial cells expressing activated Rac1, CD2AP co-localizes with active Rac1 in membrane ruffles in single cells, and at cell-cell contacts in confluent cells. The latter effect promotes cell–cell adhesion, since knockdown of CD2AP resulted in a reduction in transepithelial resistanceCitation42 (). These findings are in line with the role of CD2AP in podocytes and are in good agreement with findings in Drosophila, where the CD2AP ortholog Cindr promotes E-cadherin localization to junctions and stimulates junctional stability.Citation64 CD2AP may, in conjunction with Rac1 activity, induce actin rearrangements, as CD2AP binds to the actin capping proteins CapZ and the cortical actin regulator cortactin.Citation58,Citation59 In fact, we could show that CD2AP links Rac1 to CapZ and cortactin.Citation42 Finally, E-cadherin itself could promote CD2AP recruitment by acting as a junctional “anchor,” since it was shown to bind to CD2AP in gastric epithelial cells.Citation65
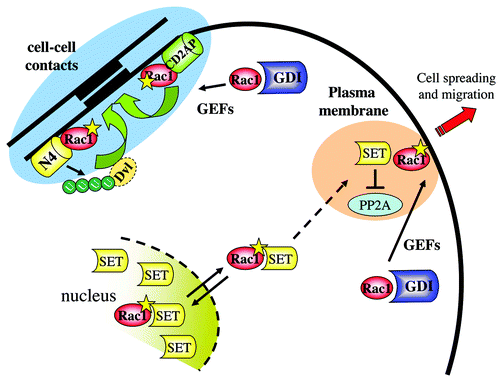
Figure 4. The nuclear connection for Rac1 and its role in cell–cell contacts. Activated Rac1 recruits the nuclear protein SET and/or a Rac1-SET complex from the nucleus to the plasma membrane to regulate cell migration. Conversely, at cell–cell contacts, activated Rac1 recruits CD2AP as well as Nedd4 (N4) to promote intercellular adhesion. The latter pathways also involves Nedd4-mediated ubiquitylation of the adaptor protein disheveled (Dvl1).
PACSIN2
The BAR (Bin/Amphiphysin/Rvs)-domain containing adaptor protein PACSIN2 is a member of a large family of regulatory proteins (adaptor proteins, GAPs and GEFs) that comprise various forms of BAR domain.Citation66,Citation67 BAR domains bind to phospholipids in intracellular membranes and sense and promote membrane curvature. BAR domain proteins reside on the inner leaflet of the plasma membrane and on intracellular vesicles and regulate protein traffic and local cell signaling.Citation68,Citation69 The PACSIN proteins (i.e., PACSIN1,2,3) encode a so-called F-BAR (FCH-BAR e.g., Fes/CIP4 homology BAR) domain that regulates membrane curvature such that internalization of the plasma membrane is induced. Expression of F-BAR domains induces the formation of tubulo-vesicular organelles that arise at the plasma membrane and move toward the cell center.Citation66,Citation70
Like CD2AP, PACSIN2 binds to the Rac1 C-terminus through a SH3 domain in its C-terminus. PACSIN2 binds to Rac1 in a nucleotide-independent fashion.Citation43 We showed that PACSIN2 co-localizes with Rac1 in membrane ruffles, on tubules and on early endosomes (). We found an interesting bi-directional regulatory crosstalk between PACSIN2 and Rac1. An activated mutant of Rac1, Rac1Q61, inhibited formation of PACSIN2-positive tubulo-vesicular structures which was promoted by an inactive mutant of Rac1, Rac1N17. Based on live-cell imaging using GFP-tagged PACSIN2, we concluded that these tubules, upon inward movement, break and form early endosomes. Conversely, we found that expression of PACSIN2 in cells reduced the levels of endogenous GTP-bound Rac1 and Rac1-mediated spreading of epithelial cells. These effects were dependent on an intact SH3 domain in PACSIN2, suggesting that its binding to Rac1 was required. In contrast, siRNA-mediated knockdown of PACSIN2, or expression of a PACSIN2 mutant with a mutated, inactive BAR domain, promoted RacGTP loading, epithelial cell spreading and migration in a wound-healing assay. These findings suggest a model in which PACSIN2 becomes recruited to areas of Rac1 activity, such as membrane ruffles, where PACSIN2 promotes membrane internalization, transporting membranes and activated Rac1 toward early endosomes. Subsequent inactivation of Rac1 may occur either on these early endosomes, or on another intracellular compartment. Intriguingly, there are several RhoGAP proteins that encode BAR domains, such as srGAP3/MEGAPCitation71,Citation72 and these may well reside on similar membrane compartments as PACSIN2, regulating GAP-mediated inactivation of Rac1.
Calmodulin
A recent study by Vidal-Quadras et al.Citation73 showed that Rac1 binds to calmodulin in a calcium-dependent manner. This interaction required both the PBR in the Rac1 C-terminus, as well as the lipid anchor. Binding appears to be selective for activated, membrane-associated Rac1, since an activated Rac1V12 mutant, lacking the C-terminal prenylation site (Cys 189), did not associate to calmodulin.Citation73 This interaction of Rac1 and calmodulin is reminiscent of the interaction between calmodulin and K-Ras 4B, which also requires the K-Ras4B HVR.Citation74 Notably, efficient interaction of K-Ras4B with calmodulin was nucleotide-dependent and required the catalytic region of the GTPase. This co-operativity between the hypervariable C-terminus and the N-terminal effector domains in K-Ras4B is again similar to the interactions described for Rac1 and PRK,Citation15 for Rac1 and Nedd4Citation75 and may also apply to the interaction Rac1 and calmodulin.
The functional consequences of Rac1 interacting with calmodulin were analyzed for the Rac1-PIP5K interaction. Inhibiting calmodulin function with an antagonist (W13) enhanced the binding of Rac1 to PIP5K in co-immunoprecipitation experiments.Citation73 Moreover, W13 induced the formation of Arf6-positive and Rac1-positive membrane tubules in COS cells, which is regulated by Rac1 activity, PIP-5-K and microtubules. Expression of an inactive Rac1N17 mutant promoted tubule formation, whereas an activated Rac1V12 mutant reduced tubule formation. These findings are identical to results from our group on PACSIN2-positive, microtubule-dependent tubules in HeLa cells.Citation43 This suggests that the Rac1-PACSIN2 interaction, and the Rac1-Calmodulin interaction are components of the same internalization pathway. Moreover, both PACSIN2 and calmodulin act as negative regulators of Rac1 signaling, further linking pathways driving membrane internalization to the inhibition of Rac1 ().
Caveolin1
The membrane-associated adaptor Caveolin1 is a key regulator of cell signaling, due to its role in membrane domain organization and traffic of cell surface proteins, including integrins, growth factor receptors and cadherins.Citation76 Caveolin1 has been implicated in cell spreading and migration, cell growth, EMT (epithelial-mesenchymal transition), tumor cell metastasis, inflammation and cardiovascular disease.Citation14,Citation76-Citation81 Caveolin1 connects integrin function with Rac1 traffic and activity since integrin-mediated adhesion reduces Caveolin1-dependent membrane internalization. Conversely, cell detachment promotes Caveolin1-mediated membrane internalization, leading to the inhibition of Rac1.Citation82,Citation83
We found Rac1 binding to Caveolin1 serendipitously in the context of our work on the actin-binding protein filamin, which associates to ICAM-1 in endothelial cells.Citation84 Filamin associates with and is regulated by Caveolin1Citation84-Citation87 and we tested binding to the Rac1 C-terminus as a control for Rac1 or ICAM-1 binding to Filamin. Caveolin1 is in many cells a polarity marker, which accumulates in the back of migrating cells. This is in good agreement with the notion that Caveolin1 positively regulates RhoA activity.Citation83 However, in cells expressing activated Rac1, cell polarity is lost and Caveolin1 becomes more diffusely localized.Citation14,Citation83 Detailed confocal analysis showed that endogenous Caveolin1 is recruited to FAs in cells expressing active Rac1, which localizes to these FAs as well. In line with earlier findings,Citation82,Citation83 we found that Caveolin1 is a negative regulator of Rac1 as siRNA-mediated downregulation of Caveolin1 increases basal levels of Rac1GTP. Surprisingly however, we found that this correlated with an increase in total Rac1 protein levels. In other words, Caveolin1 regulates Rac1 expression, which is reflected in concomitant changes in the levels of GTP-bound Rac1Citation14 ().
Because of this latter finding, regulation of Rac1 ubiquitylation was investigated. Rac1 is known to be ubiquitylated at Lys147Citation88 and this occurs primarily on activated Rac1Citation89,Citation90 Analysis of Rac1 ubiquitylation showed that loss of Caveolin1 promotes mono-ubiquitylation of Rac1.Citation14 Poly-ubiquitylation was, however, reduced under these conditions, resulting in an accumulation of mono-ubiquitylated and non-ubiquitylated, activated and non-activated Rac1 protein. It is not clear how Caveolin1 regulates the poly-ubiquitylation of mono-ubiquitylated Rac1, but this may require Rac1 internalization or targeting toward a Rac1 E3 ligase. Recently, the HACE1 and the cIAP/XIAP ubiquitin ligases were found to target Rac1.Citation91-Citation93 Interestingly, Caveolin1 binds to XIAP in the context of VEGF-induced cell survival of endothelial cells.Citation94 XIAP also binds to Rac1, in a nucleotide-independent fashionCitation91 but the domain in Rac1 that is required for this association was not identified. Importantly, Caveolin1 links XIAP to integrins and FAK (Focal Adhesion Kinase), regulating integrin function and adhesion in endothelial cells.Citation95 Given that Rac1 binding to Caveolin1 is stimulated by cell adhesion, it may well be that integrin-mediated Rac1 activation is accompanied by the recruitment of both Caveolin-1 and XIAP to Focal Adhesions, resulting in the local ubiquitylation of active Rac1.
FHOD1
The Formin Homology (FH) domain proteins are established regulators of cytoskeletal dynamics. This family includes the diaphanous proteins, effectors of active RhoA (seeCitation96 for a review on formins), that promote actin nucleation and formation of short, unbranched actin fibers. Rac1, but not Rac2, Rac3 and RhoA, associates to FHOD1 (formin homology-2 domain containing protein) and frl (formin-related gene in leukocytes) in a nucleotide-independent manner.Citation97-Citation99 In addition, the interaction required the Rac1 C-terminal HVR.Citation97 Moreover, an activated mutant of Rac1 recruites FHOD1 to membrane ruffles.Citation99 In the initial functional studies on this interaction, both active and inactive Rac1 mutants were found to reduce FHOD1-mediated activation of the SRE (Serum-Response Element), suggesting Rac1 signaling affects FHOD1 function in a complex fashion.Citation97 A subsequent studyCitation98 showed that FHOD1 expression stimulates migration of melanoma cells on different substrates, but did not affect integrin expression or integrin-mediated adhesion. Moreover, expression of Rac1N17 as well as inhibition of RhoA or ROCK, inhibited the induction of action stress fibers in NIH3T3 cells by an activated mutant of FHOD1.Citation98 Conversely, FHOD1 is also a substrate for ROCK, and its thrombin-induced phosphorylation in endothelial cells mediates the formation of actin stress fibers.Citation100 Together, these findings suggest that FHOD1 binds selectively to Rac1, but operates in Rac1 and RhoA signaling pathways and may function upstream as well as downstream of these GTPases.
The ubiquitin E3 ligases Nedd4
In addition to XIAP and HACE1, Rac1 also binds to another E3 ubiquitin ligase, Nedd4.Citation75 Nedd4 is, like HACE1, a member of the HECT (“Homologous to the E6-AP Carboxyl Terminus”)-domain family. However, Rac1 is not ubiquitylated by Nedd4.Citation75,Citation93 Nedd4 has been implicated in various physiological processes including heart development,Citation101 adaptive immunityCitation102 and development of the neuromuscular junction, a specialized form of cell–cell contact.Citation103 In line with this latter finding, our lab showed that Nedd4 is required for the formation of stable cell–cell contacts in epithelial HeLa cells, which is a Rac1-dependent process.Citation75 Nedd4 binds, via its WW domains, to the HVR of Rac1 and this interaction is largely nucleotide-independent, although we detected increased interaction of Nedd4 with an activated mutant of Rac1.
Subsequent functional studies showed that expression of Rac1 recruits Nedd4 to cell–cell contacts and that Nedd4, but not Nedd4-2, is activated by GTP-bound Rac1, resulting in the ubiquitylation and degradation of the adaptor protein Dishevelled (Dvl1)Citation75 (). Expression of Dvl1, and in particular a K-R Dvl1 mutant that is not ubiquitylated by Nedd4, reduced transepithelial resistance in HeLa cells. This is in good agreement with earlier reports that identified Dvl1 as a negative regulator of epithelial cell–cell contact in MDCK cellsCitation104 and in developing mouse embryos.Citation105 The mechanism by which Dvl1 regulates epithelial cell–cell contacts is currently unknown, but could involve cooperation with Rho signaling components, such as RhoKinase, as was recently shown for adherens junctions dynamics, required for neural plate closure.Citation106
Rac1 signaling and the nucleus
Whereas RhoGTPases are generally accepted to signal at the plasma membrane or on intracellular vesicles, there is increasing evidence for GTPase signaling in the nucleus. GTPases are known to regulate transcription and proliferation although this does not necessarily require their localization in the nucleus.Citation107-Citation110 Nuclear localization of endogenous GTPases has been difficult to establish rigourously, because the quality of available antibodies has been poorCitation24 and because some secondary antibodies induce nuclear background staining. Expression of tagged, active mutants of Rac1 has been observed by many to result in nuclear localization of a fraction of the expressed protein. The work by Michaelson and colleaguesCitation24 further confirmed that also endogenous Rac1 can be detected in the nucleus, as concluded from fractionation experiments. However, the exact location of Rac1 in the nucleus is unknown.
Nuclear translocation of proteins occurs through the nuclear pore complex (NPC), which acts as a gatekeeper. While small molecules and peptides (< ~40 kD) can pass the nuclear pore by diffusion, active nuclear import of protein complexes is mediated by karyopherins.Citation111 Proteins with a basic, “classical” NLS are imported by the karyopherins β1 and -α (also known as importin β and importin α). Most NLS-containing proteins bind to importin α and this complex binds to importin β. Once inside the nucleus, RanGTP binds to importin β and the complex dissociates.Citation112 The Rac1 HVR binds at least two proteins that are associated with nucleo-cytoplasmic shuttling. These are karyopherin α2 (importin α-1) and the PP2A inhibitor SET/I2PP2A.
Karyopherin α2
Using a two-hybrid-based interaction screen, Sandrock and colleagues found Rac1 to bind directly to the armadillo protein Karyopherin α2 in a nucleotide-independent fashion.Citation25 Mutational analysis showed that the interaction was specific for Rac1 over Rac2 and Rac3 and required the Rac1 HVR, which harbors the NLS. Interestingly, Sandrock et al. showed that nuclear translocation of Rac1 required its activation, even though the binding to Karyopherin α2 did not.Citation25 This suggests that the interaction is required but is not sufficient for nuclear import of Rac1. Moreover, siRNA-mediated loss of Karyopherin α2 blocked nuclear import and prevented the degradation of Rac1 that follows its activation by the E. coli toxin CNF. These data suggest that one of the functions of the nuclear import of activated Rac1 is to promote its degradation, which is in good agreement with the data from Lanning et al., who previously showed that the Rac1 PBR is required for nuclear translocation and degradationCitation13 (). Since nuclear import required binding of Karyopherin α2 to the Rac1 C-terminus as well as Rac1 activity, additional factors must control nuclear accumulation of activated Rac1. These may be Rac1 effector domains, but could also be the (mono-)ubiquitylation of Rac1. This will require analysis of the nuclear targeting of the Rac1Q61-K147R mutant, an activated Rac1 mutant that cannot be ubiquitylated.
SET/I2PP2A
A very abundant interactor of Rac1 is the PP2A inhibitor SET, also known as TAF1β (Template Activating Factor 1β).Citation113,Citation114 SET is a ubiquitously expressed, versatile protein that has been implicated in growth of myeloid leukemias, chronic lymphocytic leukemia and non-Hodgkin lymphoma and is thus characterized as a proto-oncogene, inhibiting the PP2A tumor suppressor.Citation115,Citation116 In addition, SET promotes granzyme B-expression in natural killer cells, thereby enhancing cytotoxicity,Citation117 inhibits granzyme A-activated DNaseCitation118 and plays a role in histone acetylation as part of the INHAT (Inhibitor of Histone AcetylTransferase) complex, thus regulating transcription.Citation114 Finally, SET is a substrate for Casein Kinase II (CKII)Citation119 and for PI-3-kinase γ.Citation120
Rac1 binds, through the PBR in the C-terminus, to the NAP (Nucleosome Assembly Protein) domain in SET.Citation121 This interaction is enhanced using a SET mutant that encodes a S9E mutation, which was used to mimic phosphorylation of SET.Citation120,Citation121 In addition, this mutant of SET does no longer dimerize, but still binds to PP2A suggesting that phosphorylation of SET on Ser9 promotes monomerization, binding to Rac1 and PP2A concomitant with shuttling toward the cytoplasm. Using a membrane-targeted SET mutant, we could show that increased plasma membrane localization of SET cooperated with active Rac1 to stimulate cell migrationCitation121 ().
SET encodes a bipartite, cryptic NLS in its C-terminal portion (amino acids 168–181.Citation122 A SET mutant lacking the NLS was found to promote cell migration in a wound-healing assay (B.D. Lam, unpublished data). Although the SET-regulated pathway that promotes cell migration is not yet identified, it is tempting to speculate that inhibition of PP2A plays an important role. PP2A is a regulatory phosphatase for several kinases, implicated in cell motility, including the Rac1 effector PAK1. Moreover, several studies have confirmed a role for PP2A in the control of cell migration.Citation123,Citation124 Finally, proteomic analyses have identified SET in complex with a host of cellular proteins including the Rac/Cdc42 GEF βPIX and the Rac/Cdc42 effector PAK1,Citation125 actin and dynamin-2119 and a β1-integrin-filamin complex.Citation126
A recent study by Switzer et al.Citation127 showed that a SET-binding peptide (COG112) does not only block the Rac1-SET interaction, but also reduces EGF-induced Rac1 activation and serum-induced cell migration and invasion. The same COG112 peptide, an apo-lipoprotein E mimetic, also inhibits inflammatory responses by blocking signaling through NFkappaB, the activity of which is regulated, in turn, by activated Rac1 as well as by PP2A.Citation128,Citation129 Thus, it may well be that the Rac1-SET complex outside the nucleus, in conjunction with PP2A, regulates cell motility and affects inflammatory responses by modulating NFkappaB activity.
Concluding Remarks
The above line-up of Rac1-interacting proteins discusses some unusual associations, but also reveals a few common themes. Most Rac1 C-terminal interactors identified so far are non-enzymatic adaptor proteins. Caveolin1, PACSIN2 and calmodulin all serve as “brakes” on Rac1 signaling, reducing the levels of RacGTP by different means. These proteins are also, in different ways, connected to membrane traffic, in good agreement with previous data that suggested that internalization of membrane domains is a prime event required for Rac1 inactivation.Citation130 This has previously been linked mainly to Caveolin1, which controls integrin-dependent Rac1 targeting and sustained Rac1 signaling.Citation83 Since Caveolin1 co-localizes with Rac1 at FAs, this inactivation pathway may well be relevant for the pool of active Rac1 that is integrin-controlled. PACSIN2 is not in FAs, but is in membrane ruffles,Citation43 suggesting that PACSIN2 is involved in inactivation of a different pool of active Rac1. PACSIN2 and calmodulin, but not Caveolin1, are on internal tubular structures, further reflecting distinct internalization and inactivation pathways.
The association of Rac1 to FAs appears very similar to what was described for R-Ras, which contains a FA-targeting domain in its HVR.Citation131 R-Ras is, like Rac1, tightly linked to integrin-mediated adhesion, the stimulation of which requires the binding of R-Ras to SH3-domain-containing proteins such as Nck, through the proline-rich domain in its HVR.Citation132 Also Rac1 shows a number of proline-SH3 domain dependent interactions, even though the proline-rich region in Rac1 is an a-typical SH3 domain binding motif. Rac1 binds to Crk, CD2AP and PACSIN2 via their SH3 domains and also binds to the SH3 domain of β-PIX, which is one of the FA-targeting proteins for activated Rac1.Citation31,Citation34
Some of the proteins that bind the Rac1 C-terminus promote its downstream signaling. CD2AP and Nedd4 co-localize with Rac1 at cell–cell contacts in epithelial cells and appear to stimulate Rac1-induced cell–cell adhesion. This involves different pathways, with Nedd4 controlling stability of the junction-inhibiting protein Dvl1 and CD2AP acting by promoting the remodelling of junctional F-actin through its association to CapZCitation42 and its regulation of surface protein internalization.Citation57 The interaction of Rac1 with the nuclear/cytoplasmic PP2A inhibitor SET/I2PP2A appears to be unique but also serves to amplify Rac1 signaling, since SET is a positive regulator of cell migration.
Protein–protein interactions through the Rac1 C-terminus were initially suggested to be independent from nucleotide binding, since the HVR was not considered a stable portion of the Rac1 3D-structure. Moreover, analysis of Rac1 in complex with RhoGDI suggested that the HVR is not part of the interaction surface and may be available for additional interactions with regulatory proteins. Yet, signaling specificity has been, in several studies, clearly linked to the C-terminus of RhoGTPases, both in vitro as well as in vivoCitation133-Citation135 More recently, several studies have shown for Rac1 that there is a functional connection between the C-terminus and the effector domain in the N-terminusCitation15 and that some C-terminal interactions are influenced by nucleotide binding. This shows that the functional relevance of the C-terminal targeting domain in activated Rac1 lies in part in its binding to membrane domains but also in its specific binding to a subset of effector proteins. The recent study by Abankwa and colleagues showed that the orientation of the G-domain of active Ras is determined in part by the C-terminal HVR, contributing to effector interactions.Citation136 This further supports the notion that the HVR in small GTPases is an important aspect of the overall GTPase structure.
Finally, recent studies have highlighted that additional regulatory mechanisms, controlling Rac1 function, impinge on its C-terminal region. Castillo-Lluva showed that Rac1 can be sumoylated on lysine residues in the PBR, promoting its activation and the induction of cell migration and invasion.Citation137 This post-translational modification is distinct from Rac1 ubiquitylation, which occurs on Lys147 and triggers its degradation.Citation88 Preventing Rac1 sumoylation did not alter its localization to membranes, nor the binding to GEFs and effector proteins, but did reduce GTP binding. Rac1 binding to GTP, as well as its partitioning in “liquid-ordered plasma membrane domains,” is also stimulated by palmitoylation of Cys178, which is immediately N-terminal of the HVR.Citation138 Moreover, a palmitoylation mutant of Rac1 cannot rescue the spreading and migration defects in Rac1-deficient fibroblasts, supporting the requirement of this modification for proper Rac1-mediated signaling toward the actin cytoskeleton.
The complexity of Rac1 regulation that is discussed here is reflected by its signaling at different locations in the cell. It is very likely that such complexity is not unique for Rac1, but is similar for other small GTPases. For some of these such as RhoA and Rap1, phosphorylation within the HVR has been shown, expanding the options for membrane- and effector-protein interactions.Citation139-Citation141 The same is true for modifications such as (mono-, di-, poly-) ubiquitylation and acetylation that will further expand the cells’ options to regulate the targeting, output and stability of active GTPases.
As alluded to in the introduction, small GTPases are key players in cellular decision-making following stimulatory or inhibitory events. The surprising range of options to tune the localization and signaling capacity of small GTPases, in addition to the surplus of GEFs and GAPs, presents us with an exceptional challenge to chart local GTPase signaling. Use of novel tools, such as dedicated biosensorsCitation142 may eventually allow us to visualize local activation, post-translational modifications as well as GTPase output in live cells.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- Barbacid M. ras genes. Annu Rev Biochem 1987; 56:779 - 827; http://dx.doi.org/10.1146/annurev.bi.56.070187.004023; PMID: 3304147
- Allende JE. GTP-mediated macromolecular interactions: the common features of different systems. FASEB J 1988; 2:2356 - 67; PMID: 2452111
- Bos JL, Rehmann H, Wittinghofer A. GEFs and GAPs: critical elements in the control of small G proteins. Cell 2007; 129:865 - 77; http://dx.doi.org/10.1016/j.cell.2007.05.018; PMID: 17540168
- Wennerberg K, Rossman KL, Der CJ. The Ras superfamily at a glance. J Cell Sci 2005; 118:843 - 6; http://dx.doi.org/10.1242/jcs.01660; PMID: 15731001
- Westwick JK, Lambert QT, Clark GJ, Symons M, Van Aelst L, Pestell RG, et al. Rac regulation of transformation, gene expression, and actin organization by multiple, PAK-independent pathways. Mol Cell Biol 1997; 17:1324 - 35; PMID: 9032259
- Diekmann D, Nobes CD, Burbelo PD, Abo A, Hall A. Rac GTPase interacts with GAPs and target proteins through multiple effector sites. EMBO J 1995; 14:5297 - 305; PMID: 7489719
- Karnoub AE, Der CJ, Campbell SL. The insert region of Rac1 is essential for membrane ruffling but not cellular transformation. Mol Cell Biol 2001; 21:2847 - 57; http://dx.doi.org/10.1128/MCB.21.8.2847-2857.2001; PMID: 11283263
- Nimnual AS, Taylor LJ, Bar-Sagi D. Redox-dependent downregulation of Rho by Rac. Nat Cell Biol 2003; 5:236 - 41; http://dx.doi.org/10.1038/ncb938; PMID: 12598902
- Alloul N, Gorzalczany Y, Itan M, Sigal N, Pick E. Activation of the superoxide-generating NADPH oxidase by chimeric proteins consisting of segments of the cytosolic component p67(phox) and the small GTPase Rac1. Biochemistry 2001; 40:14557 - 66; http://dx.doi.org/10.1021/bi0117347; PMID: 11724569
- Miyano K, Koga H, Minakami R, Sumimoto H. The insert region of the Rac GTPases is dispensable for activation of superoxide-producing NADPH oxidases. Biochem J 2009; 422:373 - 82; http://dx.doi.org/10.1042/BJ20082182; PMID: 19534724
- Bishop AL, Hall A. Rho GTPases and their effector proteins. Biochem J 2000; 348:241 - 55; http://dx.doi.org/10.1042/0264-6021:3480241; PMID: 10816416
- Michaelson D, Silletti J, Murphy G, D’Eustachio P, Rush M, Philips MR. Differential localization of Rho GTPases in live cells: regulation by hypervariable regions and RhoGDI binding. J Cell Biol 2001; 152:111 - 26; http://dx.doi.org/10.1083/jcb.152.1.111; PMID: 11149925
- Lanning CC, Daddona JL, Ruiz-Velasco R, Shafer SH, Williams CL. The Rac1 C-terminal polybasic region regulates the nuclear localization and protein degradation of Rac1. J Biol Chem 2004; 279:44197 - 210; http://dx.doi.org/10.1074/jbc.M404977200; PMID: 15304504
- Nethe M, Anthony EC, Fernandez-Borja M, Dee R, Geerts D, Hensbergen PJ, et al. Focal-adhesion targeting links caveolin-1 to a Rac1-degradation pathway. J Cell Sci 2010; 123:1948 - 58; http://dx.doi.org/10.1242/jcs.062919; PMID: 20460433
- Modha R, Campbell LJ, Nietlispach D, Buhecha HR, Owen D, Mott HR. The Rac1 polybasic region is required for interaction with its effector PRK1. J Biol Chem 2008; 283:1492 - 500; http://dx.doi.org/10.1074/jbc.M706760200; PMID: 18006505
- DeLeo FR, Quinn MT. Assembly of the phagocyte NADPH oxidase: molecular interaction of oxidase proteins. J Leukoc Biol 1996; 60:677 - 91; PMID: 8975869
- Diekmann D, Abo A, Johnston C, Segal AW, Hall A. Interaction of Rac with p67phox and regulation of phagocytic NADPH oxidase activity. Science 1994; 265:531 - 3; http://dx.doi.org/10.1126/science.8036496; PMID: 8036496
- Kreck ML, Uhlinger DJ, Tyagi SR, Inge KL, Lambeth JD. Participation of the small molecular weight GTP-binding protein Rac1 in cell-free activation and assembly of the respiratory burst oxidase. Inhibition by a carboxyl-terminal Rac peptide. J Biol Chem 1994; 269:4161 - 8; PMID: 8307977
- Joseph G, Gorzalczany Y, Koshkin V, Pick E. Inhibition of NADPH oxidase activation by synthetic peptides mapping within the carboxyl-terminal domain of small GTP-binding proteins. Lack of amino acid sequence specificity and importance of polybasic motif. J Biol Chem 1994; 269:29024 - 31; PMID: 7961867
- Bromberg Y, Shani E, Joseph G, Gorzalczany Y, Sperling O, Pick E. The GDP-bound form of the small G protein Rac1 p21 is a potent activator of the superoxide-forming NADPH oxidase of macrophages. J Biol Chem 1994; 269:7055 - 8; PMID: 8125910
- Joseph G, Pick E. “Peptide walking” is a novel method for mapping functional domains in proteins. Its application to the Rac1-dependent activation of NADPH oxidase. J Biol Chem 1995; 270:29079 - 82; http://dx.doi.org/10.1074/jbc.270.49.29079; PMID: 7493930
- Hancock JF, Paterson H, Marshall CJ. A polybasic domain or palmitoylation is required in addition to the CAAX motif to localize p21ras to the plasma membrane. Cell 1990; 63:133 - 9; http://dx.doi.org/10.1016/0092-8674(90)90294-O; PMID: 2208277
- Williams CL. The polybasic region of Ras and Rho family small GTPases: a regulator of protein interactions and membrane association and a site of nuclear localization signal sequences. Cell Signal 2003; 15:1071 - 80; http://dx.doi.org/10.1016/S0898-6568(03)00098-6; PMID: 14575862
- Michaelson D, Abidi W, Guardavaccaro D, Zhou M, Ahearn I, Pagano M, et al. Rac1 accumulates in the nucleus during the G2 phase of the cell cycle and promotes cell division. J Cell Biol 2008; 181:485 - 96; http://dx.doi.org/10.1083/jcb.200801047; PMID: 18443222
- Sandrock K, Bielek H, Schradi K, Schmidt G, Klugbauer N. The nuclear import of the small GTPase Rac1 is mediated by the direct interaction with karyopherin alpha2. Traffic 2010; 11:198 - 209; http://dx.doi.org/10.1111/j.1600-0854.2009.01015.x; PMID: 19961560
- Pop M, Aktories K, Schmidt G. Isotype-specific degradation of Rac activated by the cytotoxic necrotizing factor 1. J Biol Chem 2004; 279:35840 - 8; http://dx.doi.org/10.1074/jbc.M404346200; PMID: 15143066
- Worthylake DK, Rossman KL, Sondek J. Crystal structure of Rac1 in complex with the guanine nucleotide exchange region of Tiam1. Nature 2000; 408:682 - 8; http://dx.doi.org/10.1038/35047014; PMID: 11130063
- van Helden SF, Anthony EC, Dee R, Hordijk PL. Rho GTPase expression in human myeloid cells. PLoS One 2012; 7:e42563; http://dx.doi.org/10.1371/journal.pone.0042563; PMID: 22916134
- Vikis HG, Stewart S, Guan KL. SmgGDS displays differential binding and exchange activity towards different Ras isoforms. Oncogene 2002; 21:2425 - 32; http://dx.doi.org/10.1038/sj.onc.1205306; PMID: 11948427
- Hamel B, Monaghan-Benson E, Rojas RJ, Temple BR, Marston DJ, Burridge K, et al. SmgGDS is a guanine nucleotide exchange factor that specifically activates RhoA and RhoC. J Biol Chem 2011; 286:12141 - 8; http://dx.doi.org/10.1074/jbc.M110.191122; PMID: 21242305
- ten Klooster JP, Jaffer ZM, Chernoff J, Hordijk PL. Targeting and activation of Rac1 are mediated by the exchange factor beta-Pix. J Cell Biol 2006; 172:759 - 69; http://dx.doi.org/10.1083/jcb.200509096; PMID: 16492808
- Manser E, Loo TH, Koh CG, Zhao ZS, Chen XQ, Tan L, et al. PAK kinases are directly coupled to the PIX family of nucleotide exchange factors. Mol Cell 1998; 1:183 - 92; http://dx.doi.org/10.1016/S1097-2765(00)80019-2; PMID: 9659915
- Knaus UG, Wang Y, Reilly AM, Warnock D, Jackson JH. Structural requirements for PAK activation by Rac GTPases. J Biol Chem 1998; 273:21512 - 8; http://dx.doi.org/10.1074/jbc.273.34.21512; PMID: 9705280
- Chang F, Lemmon CA, Park D, Romer LH. FAK potentiates Rac1 activation and localization to matrix adhesion sites: a role for betaPIX. Mol Biol Cell 2007; 18:253 - 64; http://dx.doi.org/10.1091/mbc.E06-03-0207; PMID: 17093062
- Tolias KF, Cantley LC, Carpenter CL. Rho family GTPases bind to phosphoinositide kinases. J Biol Chem 1995; 270:17656 - 9; http://dx.doi.org/10.1074/jbc.270.30.17656; PMID: 7629060
- Tolias KF, Couvillon AD, Cantley LC, Carpenter CL. Characterization of a Rac1- and RhoGDI-associated lipid kinase signaling complex. Mol Cell Biol 1998; 18:762 - 70; PMID: 9447972
- Tolias KF, Hartwig JH, Ishihara H, Shibasaki Y, Cantley LC, Carpenter CL. Type Ialpha phosphatidylinositol-4-phosphate 5-kinase mediates Rac-dependent actin assembly. Curr Biol 2000; 10:153 - 6; http://dx.doi.org/10.1016/S0960-9822(00)00315-8; PMID: 10679324
- Di-Poï N, Fauré J, Grizot S, Molnár G, Pick E, Dagher MC. Mechanism of NADPH oxidase activation by the Rac/Rho-GDI complex. Biochemistry 2001; 40:10014 - 22; http://dx.doi.org/10.1021/bi010289c; PMID: 11513579
- Grizot S, Fauré J, Fieschi F, Vignais PV, Dagher MC, Pebay-Peyroula E. Crystal structure of the Rac1-RhoGDI complex involved in nadph oxidase activation. Biochemistry 2001; 40:10007 - 13; http://dx.doi.org/10.1021/bi010288k; PMID: 11513578
- Chae YC, Kim JH, Kim KL, Kim HW, Lee HY, Heo WD, et al. Phospholipase D activity regulates integrin-mediated cell spreading and migration by inducing GTP-Rac translocation to the plasma membrane. Mol Biol Cell 2008; 19:3111 - 23; http://dx.doi.org/10.1091/mbc.E07-04-0337; PMID: 18480413
- van Hennik PB, ten Klooster JP, Halstead JR, Voermans C, Anthony EC, Divecha N, et al. The C-terminal domain of Rac1 contains two motifs that control targeting and signaling specificity. J Biol Chem 2003; 278:39166 - 75; http://dx.doi.org/10.1074/jbc.M307001200; PMID: 12874273
- van Duijn TJ, Anthony EC, Hensbergen PJ, Deelder AM, Hordijk PL. Rac1 recruits the adapter protein CMS/CD2AP to cell-cell contacts. J Biol Chem 2010; 285:20137 - 46; http://dx.doi.org/10.1074/jbc.M109.099481; PMID: 20404345
- de Kreuk BJ, Nethe M, Fernandez-Borja M, Anthony EC, Hensbergen PJ, Deelder AM, et al. The F-BAR domain protein PACSIN2 associates with Rac1 and regulates cell spreading and migration. J Cell Sci 2011; 124:2375 - 88; http://dx.doi.org/10.1242/jcs.080630; PMID: 21693584
- Mizuki K, Takeya R, Kuribayashi F, Nobuhisa I, Kohda D, Nunoi H, et al. A region C-terminal to the proline-rich core of p47phox regulates activation of the phagocyte NADPH oxidase by interacting with the C-terminal SH3 domain of p67phox. Arch Biochem Biophys 2005; 444:185 - 94; http://dx.doi.org/10.1016/j.abb.2005.10.012; PMID: 16297854
- Rubini C, Ruzza P, Spaller MR, Siligardi G, Hussain R, Udugamasooriya DG, et al. Recognition of lysine-rich peptide ligands by murine cortactin SH3 domain: CD, ITC, and NMR studies. Biopolymers 2010; 94:298 - 306; http://dx.doi.org/10.1002/bip.21350; PMID: 19921743
- Watanabe G, Saito Y, Madaule P, Ishizaki T, Fujisawa K, Morii N, et al. Protein kinase N (PKN) and PKN-related protein rhophilin as targets of small GTPase Rho. Science 1996; 271:645 - 8; http://dx.doi.org/10.1126/science.271.5249.645; PMID: 8571126
- Vincent S, Settleman J. The PRK2 kinase is a potential effector target of both Rho and Rac GTPases and regulates actin cytoskeletal organization. Mol Cell Biol 1997; 17:2247 - 56; PMID: 9121475
- Zhao ZS, Manser E. PAK and other Rho-associated kinases--effectors with surprisingly diverse mechanisms of regulation. Biochem J 2005; 386:201 - 14; http://dx.doi.org/10.1042/BJ20041638; PMID: 15548136
- Owen D, Lowe PN, Nietlispach D, Brosnan CE, Chirgadze DY, Parker PJ, et al. Molecular dissection of the interaction between the small G proteins Rac1 and RhoA and protein kinase C-related kinase 1 (PRK1). J Biol Chem 2003; 278:50578 - 87; http://dx.doi.org/10.1074/jbc.M304313200; PMID: 14514689
- Wullschleger S, Loewith R, Hall MN. TOR signaling in growth and metabolism. Cell 2006; 124:471 - 84; http://dx.doi.org/10.1016/j.cell.2006.01.016; PMID: 16469695
- Jacinto E, Loewith R, Schmidt A, Lin S, Rüegg MA, Hall A, et al. Mammalian TOR complex 2 controls the actin cytoskeleton and is rapamycin insensitive. Nat Cell Biol 2004; 6:1122 - 8; http://dx.doi.org/10.1038/ncb1183; PMID: 15467718
- Saci A, Cantley LC, Carpenter CL. Rac1 regulates the activity of mTORC1 and mTORC2 and controls cellular size. Mol Cell 2011; 42:50 - 61; http://dx.doi.org/10.1016/j.molcel.2011.03.017; PMID: 21474067
- Durán RV, Hall MN. Regulation of TOR by small GTPases. EMBO Rep 2012; 13:121 - 8; http://dx.doi.org/10.1038/embor.2011.257; PMID: 22240970
- Aslan JE, McCarty OJ. Regulation of the mTOR-Rac1 axis in platelet function. Small GTPases 2012; 3:67 - 70; http://dx.doi.org/10.4161/sgtp.19137; PMID: 22714420
- Gulhati P, Bowen KA, Liu J, Stevens PD, Rychahou PG, Chen M, et al. mTORC1 and mTORC2 regulate EMT, motility, and metastasis of colorectal cancer via RhoA and Rac1 signaling pathways. Cancer Res 2011; 71:3246 - 56; http://dx.doi.org/10.1158/0008-5472.CAN-10-4058; PMID: 21430067
- Dustin ML, Olszowy MW, Holdorf AD, Li J, Bromley S, Desai N, et al. A novel adaptor protein orchestrates receptor patterning and cytoskeletal polarity in T-cell contacts. Cell 1998; 94:667 - 77; http://dx.doi.org/10.1016/S0092-8674(00)81608-6; PMID: 9741631
- Dikic I. CIN85/CMS family of adaptor molecules. FEBS Lett 2002; 529:110 - 5; http://dx.doi.org/10.1016/S0014-5793(02)03188-5; PMID: 12354621
- Lynch DK, Winata SC, Lyons RJ, Hughes WE, Lehrbach GM, Wasinger V, et al. A Cortactin-CD2-associated protein (CD2AP) complex provides a novel link between epidermal growth factor receptor endocytosis and the actin cytoskeleton. J Biol Chem 2003; 278:21805 - 13; http://dx.doi.org/10.1074/jbc.M211407200; PMID: 12672817
- Hutchings NJ, Clarkson N, Chalkley R, Barclay AN, Brown MH. Linking the T cell surface protein CD2 to the actin-capping protein CAPZ via CMS and CIN85. J Biol Chem 2003; 278:22396 - 403; http://dx.doi.org/10.1074/jbc.M302540200; PMID: 12690097
- Shih NY, Li J, Karpitskii V, Nguyen A, Dustin ML, Kanagawa O, et al. Congenital nephrotic syndrome in mice lacking CD2-associated protein. Science 1999; 286:312 - 5; http://dx.doi.org/10.1126/science.286.5438.312; PMID: 10514378
- Shih NY, Li J, Cotran R, Mundel P, Miner JH, Shaw AS. CD2AP localizes to the slit diaphragm and binds to nephrin via a novel C-terminal domain. Am J Pathol 2001; 159:2303 - 8; http://dx.doi.org/10.1016/S0002-9440(10)63080-5; PMID: 11733379
- Liu XL, Kilpeläinen P, Hellman U, Sun Y, Wartiovaara J, Morgunova E, et al. Characterization of the interactions of the nephrin intracellular domain. FEBS J 2005; 272:228 - 43; http://dx.doi.org/10.1111/j.1432-1033.2004.04408.x; PMID: 15634346
- Akilesh S, Suleiman H, Yu H, Stander MC, Lavin P, Gbadegesin R, et al. Arhgap24 inactivates Rac1 in mouse podocytes, and a mutant form is associated with familial focal segmental glomerulosclerosis. J Clin Invest 2011; 121:4127 - 37; http://dx.doi.org/10.1172/JCI46458; PMID: 21911940
- Johnson RI, Seppa MJ, Cagan RL. The Drosophila CD2AP/CIN85 orthologue Cindr regulates junctions and cytoskeleton dynamics during tissue patterning. J Cell Biol 2008; 180:1191 - 204; http://dx.doi.org/10.1083/jcb.200706108; PMID: 18362180
- Mustonen H, Lepistö A, Lehtonen S, Lehtonen E, Puolakkainen P, Kivilaakso E. CD2AP contributes to cell migration and adhesion in cultured gastric epithelium. Biochem Biophys Res Commun 2005; 332:426 - 32; http://dx.doi.org/10.1016/j.bbrc.2005.04.140; PMID: 15910750
- de Kreuk BJ, Hordijk PL. Control of Rho GTPase function by BAR-domains. Small GTPases 2012; 3:45 - 52; http://dx.doi.org/10.4161/sgtp.18960; PMID: 22714417
- Ritter B, Modregger J, Paulsson M, Plomann M. PACSIN 2, a novel member of the PACSIN family of cytoplasmic adapter proteins. FEBS Lett 1999; 454:356 - 62; http://dx.doi.org/10.1016/S0014-5793(99)00830-3; PMID: 10431838
- Qualmann B, Koch D, Kessels MM. Let’s go bananas: revisiting the endocytic BAR code. EMBO J 2011; 30:3501 - 15; http://dx.doi.org/10.1038/emboj.2011.266; PMID: 21878992
- Suetsugu S, Toyooka K, Senju Y. Subcellular membrane curvature mediated by the BAR domain superfamily proteins. Semin Cell Dev Biol 2010; 21:340 - 9; http://dx.doi.org/10.1016/j.semcdb.2009.12.002; PMID: 19963073
- Roberts-Galbraith RH, Gould KL. Setting the F-BAR: functions and regulation of the F-BAR protein family. Cell Cycle 2010; 9:4091 - 7; http://dx.doi.org/10.4161/cc.9.20.13587; PMID: 20948299
- Endris V, Haussmann L, Buss E, Bacon C, Bartsch D, Rappold G. SrGAP3 interacts with lamellipodin at the cell membrane and regulates Rac-dependent cellular protrusions. J Cell Sci 2011; 124:3941 - 55; http://dx.doi.org/10.1242/jcs.077081; PMID: 22159416
- Yang Y, Marcello M, Endris V, Saffrich R, Fischer R, Trendelenburg MF, et al. MEGAP impedes cell migration via regulating actin and microtubule dynamics and focal complex formation. Exp Cell Res 2006; 312:2379 - 93; http://dx.doi.org/10.1016/j.yexcr.2006.04.001; PMID: 16730001
- Vidal-Quadras M, Gelabert-Baldrich M, Soriano-Castell D, Lladó A, Rentero C, Calvo M, et al. Rac1 and calmodulin interactions modulate dynamics of ARF6-dependent endocytosis. Traffic 2011; 12:1879 - 96; http://dx.doi.org/10.1111/j.1600-0854.2011.01274.x; PMID: 21883766
- Abraham SJ, Nolet RP, Calvert RJ, Anderson LM, Gaponenko V. The hypervariable region of K-Ras4B is responsible for its specific interactions with calmodulin. Biochemistry 2009; 48:7575 - 83; http://dx.doi.org/10.1021/bi900769j; PMID: 19583261
- Nethe M, de Kreuk BJ, Tauriello DV, Anthony EC, Snoek B, Stumpel T, et al. Rac1 acts in conjunction with Nedd4 and dishevelled-1 to promote maturation of cell-cell contacts. J Cell Sci 2012; 125:3430 - 42; http://dx.doi.org/10.1242/jcs.100925; PMID: 22467858
- Williams TM, Lisanti MP. The Caveolin genes: from cell biology to medicine. Ann Med 2004; 36:584 - 95; http://dx.doi.org/10.1080/07853890410018899; PMID: 15768830
- Bailey KM, Liu J. Caveolin-1 up-regulation during epithelial to mesenchymal transition is mediated by focal adhesion kinase. J Biol Chem 2008; 283:13714 - 24; http://dx.doi.org/10.1074/jbc.M709329200; PMID: 18332144
- Nethe M, Hordijk PL. A model for phospho-caveolin-1-driven turnover of focal adhesions. Cell Adh Migr 2011; 5:59 - 64; http://dx.doi.org/10.4161/cam.5.1.13702; PMID: 20948305
- Pavlides S, Gutierrez-Pajares JL, Danilo C, Lisanti MP, Frank PG. Atherosclerosis, caveolae and caveolin-1. Adv Exp Med Biol 2012; 729:127 - 44; http://dx.doi.org/10.1007/978-1-4614-1222-9_9; PMID: 22411318
- Mercier I, Lisanti MP. Caveolin-1 and breast cancer: a new clinical perspective. Adv Exp Med Biol 2012; 729:83 - 94; http://dx.doi.org/10.1007/978-1-4614-1222-9_6; PMID: 22411315
- Grande-García A, Echarri A, Del Pozo MA. Integrin regulation of membrane domain trafficking and Rac targeting. Biochem Soc Trans 2005; 33:609 - 13; http://dx.doi.org/10.1042/BST0330609; PMID: 16042555
- del Pozo MA, Alderson NB, Kiosses WB, Chiang HH, Anderson RG, Schwartz MA. Integrins regulate Rac targeting by internalization of membrane domains. Science 2004; 303:839 - 42; http://dx.doi.org/10.1126/science.1092571; PMID: 14764880
- Grande-García A, Echarri A, de Rooij J, Alderson NB, Waterman-Storer CM, Valdivielso JM, et al. Caveolin-1 regulates cell polarization and directional migration through Src kinase and Rho GTPases. J Cell Biol 2007; 177:683 - 94; http://dx.doi.org/10.1083/jcb.200701006; PMID: 17517963
- Kanters E, van Rijssel J, Hensbergen PJ, Hondius D, Mul FP, Deelder AM, et al. Filamin B mediates ICAM-1-driven leukocyte transendothelial migration. J Biol Chem 2008; 283:31830 - 9; http://dx.doi.org/10.1074/jbc.M804888200; PMID: 18809679
- Stahlhut M, van Deurs B. Identification of filamin as a novel ligand for caveolin-1: evidence for the organization of caveolin-1-associated membrane domains by the actin cytoskeleton. Mol Biol Cell 2000; 11:325 - 37; PMID: 10637311
- Sverdlov M, Shinin V, Place AT, Castellon M, Minshall RD. Filamin A regulates caveolae internalization and trafficking in endothelial cells. Mol Biol Cell 2009; 20:4531 - 40; http://dx.doi.org/10.1091/mbc.E08-10-0997; PMID: 19759182
- Muriel O, Echarri A, Hellriegel C, Pavón DM, Beccari L, Del Pozo MA. Phosphorylated filamin A regulates actin-linked caveolae dynamics. J Cell Sci 2011; 124:2763 - 76; http://dx.doi.org/10.1242/jcs.080804; PMID: 21807941
- Visvikis O, Lorès P, Boyer L, Chardin P, Lemichez E, Gacon G. Activated Rac1, but not the tumorigenic variant Rac1b, is ubiquitinated on Lys 147 through a JNK-regulated process. FEBS J 2008; 275:386 - 96; http://dx.doi.org/10.1111/j.1742-4658.2007.06209.x; PMID: 18093184
- Boulter E, Garcia-Mata R, Guilluy C, Dubash A, Rossi G, Brennwald PJ, et al. Regulation of Rho GTPase crosstalk, degradation and activity by RhoGDI1. Nat Cell Biol 2010; 12:477 - 83; http://dx.doi.org/10.1038/ncb2049; PMID: 20400958
- Garcia-Mata R, Boulter E, Burridge K. The ‘invisible hand’: regulation of RHO GTPases by RHOGDIs. Nat Rev Mol Cell Biol 2011; 12:493 - 504; http://dx.doi.org/10.1038/nrm3153; PMID: 21779026
- Oberoi TK, Dogan T, Hocking JC, Scholz RP, Mooz J, Anderson CL, et al. IAPs regulate the plasticity of cell migration by directly targeting Rac1 for degradation. EMBO J 2012; 31:14 - 28; http://dx.doi.org/10.1038/emboj.2011.423; PMID: 22117219
- Castillo-Lluva S, Tan CT, Daugaard M, Sorensen PH, Malliri A. The tumour suppressor HACE1 controls cell migration by regulating Rac1 degradation. Oncogene 2012; In press http://dx.doi.org/10.1038/onc.2012.189; PMID: 22614015
- Torrino S, Visvikis O, Doye A, Boyer L, Stefani C, Munro P, et al. The E3 ubiquitin-ligase HACE1 catalyzes the ubiquitylation of active Rac1. Dev Cell 2011; 21:959 - 65; http://dx.doi.org/10.1016/j.devcel.2011.08.015; PMID: 22036506
- Kim J, Park J, Choi S, Chi SG, Mowbray AL, Jo H, et al. X-linked inhibitor of apoptosis protein is an important regulator of vascular endothelial growth factor-dependent bovine aortic endothelial cell survival. Circ Res 2008; 102:896 - 904; http://dx.doi.org/10.1161/CIRCRESAHA.107.163667; PMID: 18309102
- Kim J, Ahn S, Ko YG, Boo YC, Chi SG, Ni CW, et al. X-linked inhibitor of apoptosis protein controls alpha5-integrin-mediated cell adhesion and migration. Am J Physiol Heart Circ Physiol 2010; 299:H300 - 9; http://dx.doi.org/10.1152/ajpheart.00180.2010; PMID: 20472758
- Evangelista M, Zigmond S, Boone C. Formins: signaling effectors for assembly and polarization of actin filaments. J Cell Sci 2003; 116:2603 - 11; http://dx.doi.org/10.1242/jcs.00611; PMID: 12775772
- Westendorf JJ. The formin/diaphanous-related protein, FHOS, interacts with Rac1 and activates transcription from the serum response element. J Biol Chem 2001; 276:46453 - 9; http://dx.doi.org/10.1074/jbc.M105162200; PMID: 11590143
- Koka S, Neudauer CL, Li X, Lewis RE, McCarthy JB, Westendorf JJ. The formin-homology-domain-containing protein FHOD1 enhances cell migration. J Cell Sci 2003; 116:1745 - 55; http://dx.doi.org/10.1242/jcs.00386; PMID: 12665555
- Gasteier JE, Madrid R, Krautkrämer E, Schröder S, Muranyi W, Benichou S, et al. Activation of the Rac-binding partner FHOD1 induces actin stress fibers via a ROCK-dependent mechanism. J Biol Chem 2003; 278:38902 - 12; http://dx.doi.org/10.1074/jbc.M306229200; PMID: 12857739
- Takeya R, Taniguchi K, Narumiya S, Sumimoto H. The mammalian formin FHOD1 is activated through phosphorylation by ROCK and mediates thrombin-induced stress fibre formation in endothelial cells. EMBO J 2008; 27:618 - 28; http://dx.doi.org/10.1038/emboj.2008.7; PMID: 18239683
- Fouladkou F, Lu C, Jiang C, Zhou L, She Y, Walls JR, et al. The ubiquitin ligase Nedd4-1 is required for heart development and is a suppressor of thrombospondin-1. J Biol Chem 2010; 285:6770 - 80; http://dx.doi.org/10.1074/jbc.M109.082347; PMID: 20026598
- Yang B, Gay DL, MacLeod MK, Cao X, Hala T, Sweezer EM, et al. Nedd4 augments the adaptive immune response by promoting ubiquitin-mediated degradation of Cbl-b in activated T cells. Nat Immunol 2008; 9:1356 - 63; http://dx.doi.org/10.1038/ni.1670; PMID: 18931680
- Liu Y, Oppenheim RW, Sugiura Y, Lin W. Abnormal development of the neuromuscular junction in Nedd4-deficient mice. Dev Biol 2009; 330:153 - 66; http://dx.doi.org/10.1016/j.ydbio.2009.03.023; PMID: 19345204
- Elbert M, Cohen D, Müsch A. PAR1b promotes cell-cell adhesion and inhibits dishevelled-mediated transformation of Madin-Darby canine kidney cells. Mol Biol Cell 2006; 17:3345 - 55; http://dx.doi.org/10.1091/mbc.E06-03-0193; PMID: 16707567
- Na J, Lykke-Andersen K, Torres Padilla ME, Zernicka-Goetz M. Dishevelled proteins regulate cell adhesion in mouse blastocyst and serve to monitor changes in Wnt signaling. Dev Biol 2007; 302:40 - 9; http://dx.doi.org/10.1016/j.ydbio.2006.08.036; PMID: 17005174
- Nishimura T, Honda H, Takeichi M. Planar cell polarity links axes of spatial dynamics in neural-tube closure. Cell 2012; 149:1084 - 97; http://dx.doi.org/10.1016/j.cell.2012.04.021; PMID: 22632972
- Debidda M, Wang L, Zang H, Poli V, Zheng Y. A role of STAT3 in Rho GTPase-regulated cell migration and proliferation. J Biol Chem 2005; 280:17275 - 85; http://dx.doi.org/10.1074/jbc.M413187200; PMID: 15705584
- Qiu RG, Chen J, Kirn D, McCormick F, Symons M. An essential role for Rac in Ras transformation. Nature 1995; 374:457 - 9; http://dx.doi.org/10.1038/374457a0; PMID: 7700355
- Qiu RG, Chen J, McCormick F, Symons M. A role for Rho in Ras transformation. Proc Natl Acad Sci U S A 1995; 92:11781 - 5; http://dx.doi.org/10.1073/pnas.92.25.11781; PMID: 8524848
- Olson MF, Ashworth A, Hall A. An essential role for Rho, Rac, and Cdc42 GTPases in cell cycle progression through G1. Science 1995; 269:1270 - 2; http://dx.doi.org/10.1126/science.7652575; PMID: 7652575
- Chook YM, Blobel G. Karyopherins and nuclear import. Curr Opin Struct Biol 2001; 11:703 - 15; http://dx.doi.org/10.1016/S0959-440X(01)00264-0; PMID: 11751052
- Riddick G, Macara IG. A systems analysis of importin-alpha-beta mediated nuclear protein import. J Cell Biol 2005; 168:1027 - 38; http://dx.doi.org/10.1083/jcb.200409024; PMID: 15795315
- Nagata K, Kawase H, Handa H, Yano K, Yamasaki M, Ishimi Y, et al. Replication factor encoded by a putative oncogene, set, associated with myeloid leukemogenesis. Proc Natl Acad Sci U S A 1995; 92:4279 - 83; http://dx.doi.org/10.1073/pnas.92.10.4279; PMID: 7753797
- Seo SB, McNamara P, Heo S, Turner A, Lane WS, Chakravarti D. Regulation of histone acetylation and transcription by INHAT, a human cellular complex containing the set oncoprotein. Cell 2001; 104:119 - 30; http://dx.doi.org/10.1016/S0092-8674(01)00196-9; PMID: 11163245
- Perrotti D, Neviani P. ReSETting PP2A tumour suppressor activity in blast crisis and imatinib-resistant chronic myelogenous leukaemia. Br J Cancer 2006; 95:775 - 81; http://dx.doi.org/10.1038/sj.bjc.6603317; PMID: 16953242
- Christensen DJ, Chen Y, Oddo J, Matta KM, Neil J, Davis ED, et al. SET oncoprotein overexpression in B-cell chronic lymphocytic leukemia and non-Hodgkin lymphoma: a predictor of aggressive disease and a new treatment target. Blood 2011; 118:4150 - 8; http://dx.doi.org/10.1182/blood-2011-04-351072; PMID: 21844565
- Trotta R, Ciarlariello D, Dal Col J, Mao H, Chen L, Briercheck E, et al. The PP2A inhibitor SET regulates granzyme B expression in human natural killer cells. Blood 2011; 117:2378 - 84; http://dx.doi.org/10.1182/blood-2010-05-285130; PMID: 21156847
- Fan Z, Beresford PJ, Oh DY, Zhang D, Lieberman J. Tumor suppressor NM23-H1 is a granzyme A-activated DNase during CTL-mediated apoptosis, and the nucleosome assembly protein SET is its inhibitor. Cell 2003; 112:659 - 72; http://dx.doi.org/10.1016/S0092-8674(03)00150-8; PMID: 12628186
- Vera J, Estanyol JM, Canela N, Llorens F, Agell N, Itarte E, et al. Proteomic analysis of SET-binding proteins. Proteomics 2007; 7:578 - 87; http://dx.doi.org/10.1002/pmic.200600458; PMID: 17309103
- Vasudevan NT, Mohan ML, Gupta MK, Hussain AK, Naga Prasad SV. Inhibition of protein phosphatase 2A activity by PI3Kγ regulates β-adrenergic receptor function. Mol Cell 2011; 41:636 - 48; http://dx.doi.org/10.1016/j.molcel.2011.02.025; PMID: 21419339
- ten Klooster JP, Leeuwen Iv, Scheres N, Anthony EC, Hordijk PL. Rac1-induced cell migration requires membrane recruitment of the nuclear oncogene SET. EMBO J 2007; 26:336 - 45; http://dx.doi.org/10.1038/sj.emboj.7601518; PMID: 17245428
- Qu D, Zhang Y, Ma J, Guo K, Li R, Yin Y, et al. The nuclear localization of SET mediated by impalpha3/impbeta attenuates its cytosolic toxicity in neurons. J Neurochem 2007; 103:408 - 22; PMID: 17608644
- Pullar CE, Chen J, Isseroff RR. PP2A activation by beta2-adrenergic receptor agonists: novel regulatory mechanism of keratinocyte migration. J Biol Chem 2003; 278:22555 - 62; http://dx.doi.org/10.1074/jbc.M300205200; PMID: 12697752
- Li G, Ji XD, Gao H, Zhao JS, Xu JF, Sun ZJ, et al. EphB3 suppresses non-small-cell lung cancer metastasis via a PP2A/RACK1/Akt signalling complex. Nat Commun 2012; 3:667; http://dx.doi.org/10.1038/ncomms1675; PMID: 22314363
- Mayhew MW, Webb DJ, Kovalenko M, Whitmore L, Fox JW, Horwitz AF. Identification of protein networks associated with the PAK1-betaPIX-GIT1-paxillin signaling complex by mass spectrometry. J Proteome Res 2006; 5:2417 - 23; http://dx.doi.org/10.1021/pr060140t; PMID: 16944954
- Humphries JD, Byron A, Bass MD, Craig SE, Pinney JW, Knight D, et al. Proteomic analysis of integrin-associated complexes identifies RCC2 as a dual regulator of Rac1 and Arf6. Sci Signal 2009; 2:ra51; http://dx.doi.org/10.1126/scisignal.2000396; PMID: 19738201
- Switzer CH, Cheng RY, Vitek TM, Christensen DJ, Wink DA, Vitek MP. Targeting SET/I(2)PP2A oncoprotein functions as a multi-pathway strategy for cancer therapy. Oncogene 2011; 30:2504 - 13; http://dx.doi.org/10.1038/onc.2010.622; PMID: 21297667
- Perona R, Montaner S, Saniger L, Sánchez-Pérez I, Bravo R, Lacal JC. Activation of the nuclear factor-kappaB by Rho, CDC42, and Rac-1 proteins. Genes Dev 1997; 11:463 - 75; http://dx.doi.org/10.1101/gad.11.4.463; PMID: 9042860
- Sontag E, Sontag JM, Garcia A. Protein phosphatase 2A is a critical regulator of protein kinase C zeta signaling targeted by SV40 small t to promote cell growth and NF-kappaB activation. EMBO J 1997; 16:5662 - 71; http://dx.doi.org/10.1093/emboj/16.18.5662; PMID: 9312025
- Del Pozo MA, Kiosses WB, Alderson NB, Meller N, Hahn KM, Schwartz MA. Integrins regulate GTP-Rac localized effector interactions through dissociation of Rho-GDI. Nat Cell Biol 2002; 4:232 - 9; http://dx.doi.org/10.1038/ncb759; PMID: 11862216
- Furuhjelm J, Peränen J. The C-terminal end of R-Ras contains a focal adhesion targeting signal. J Cell Sci 2003; 116:3729 - 38; http://dx.doi.org/10.1242/jcs.00689; PMID: 12890755
- Wang B, Zou JX, Ek-Rylander B, Ruoslahti E. R-Ras contains a proline-rich site that binds to SH3 domains and is required for integrin activation by R-Ras. J Biol Chem 2000; 275:5222 - 7; http://dx.doi.org/10.1074/jbc.275.7.5222; PMID: 10671570
- Hajdo-Milasinović A, Ellenbroek SI, van Es S, van der Vaart B, Collard JG. Rac1 and Rac3 have opposing functions in cell adhesion and differentiation of neuronal cells. J Cell Sci 2007; 120:555 - 66; http://dx.doi.org/10.1242/jcs.03364; PMID: 17244648
- Filippi MD, Harris CE, Meller J, Gu Y, Zheng Y, Williams DA. Localization of Rac2 via the C terminus and aspartic acid 150 specifies superoxide generation, actin polarity and chemotaxis in neutrophils. Nat Immunol 2004; 5:744 - 51; http://dx.doi.org/10.1038/ni1081; PMID: 15170212
- Yamauchi A, Marchal CC, Molitoris J, Pech N, Knaus U, Towe J, et al. Rac GTPase isoform-specific regulation of NADPH oxidase and chemotaxis in murine neutrophils in vivo. Role of the C-terminal polybasic domain. J Biol Chem 2005; 280:953 - 64; http://dx.doi.org/10.1074/jbc.M408820200; PMID: 15504745
- Abankwa D, Gorfe AA, Inder K, Hancock JF. Ras membrane orientation and nanodomain localization generate isoform diversity. Proc Natl Acad Sci U S A 2010; 107:1130 - 5; http://dx.doi.org/10.1073/pnas.0903907107; PMID: 20080631
- Castillo-Lluva S, Tatham MH, Jones RC, Jaffray EG, Edmondson RD, Hay RT, et al. SUMOylation of the GTPase Rac1 is required for optimal cell migration. Nat Cell Biol 2010; 12:1078 - 85; http://dx.doi.org/10.1038/ncb2112; PMID: 20935639
- Navarro-Lérida I, Sánchez-Perales S, Calvo M, Rentero C, Zheng Y, Enrich C, et al. A palmitoylation switch mechanism regulates Rac1 function and membrane organization. EMBO J 2012; 31:534 - 51; http://dx.doi.org/10.1038/emboj.2011.446; PMID: 22157745
- Quilliam LA, Mueller H, Bohl BP, Prossnitz V, Sklar LA, Der CJ, et al. Rap1A is a substrate for cyclic AMP-dependent protein kinase in human neutrophils. J Immunol 1991; 147:1628 - 35; PMID: 1908879
- Bokoch GM, Quilliam LA, Bohl BP, Jesaitis AJ, Quinn MT. Inhibition of Rap1A binding to cytochrome b558 of NADPH oxidase by phosphorylation of Rap1A. Science 1991; 254:1794 - 6; http://dx.doi.org/10.1126/science.1763330; PMID: 1763330
- Nusser N, Gosmanova E, Makarova N, Fujiwara Y, Yang L, Guo F, et al. Serine phosphorylation differentially affects RhoA binding to effectors: implications to NGF-induced neurite outgrowth. Cell Signal 2006; 18:704 - 14; http://dx.doi.org/10.1016/j.cellsig.2005.06.010; PMID: 16109481
- Welch CM, Elliott H, Danuser G, Hahn KM. Imaging the coordination of multiple signalling activities in living cells. Nat Rev Mol Cell Biol 2011; 12:749 - 56; http://dx.doi.org/10.1038/nrm3212; PMID: 22016058