Abstract
Rac1b is frequently expressed in a number of human cancer cells. It is still unclear, however, whether Rac1b causes morphological abnormalities in epithelial tissues. To investigate whether Rac1b induces morphological changes in 3-dimensional epithelial structures, we utilized an auxin-dependent protein expression system, which enabled us to rapidly induce and evaluate Rac1b function in MDCK (Madin-Darby Canine Kidney) cysts, a model for polarized epithelial structure. Cells carrying the wild-type Rac1, Rac1b and constitutively active Rac1V12 gene were morphologically indistinguishable from normal, when their coding proteins were not expressed. However, upon protein induction, Rac1V12, but not the wild-type Rac1 or Rac1b, significantly induced the luminal cell accumulation. Live cell imaging with cell cycle indicators showed that expression of Rac1V12, but not the wild-type Rac1 or Rac1b, promoted cell cycle progression. From these results, we concluded that the expression of Rac1b per se cannot induce cell proliferation. Rather, it is considered that Rac1b expression may participate in progression of malignancy.
Keywords: :
Introduction
Rac1 is a member of the Rho-family GTPases and is involved in the signal transduction pathways for actin reorganizationCitation1 and cell cycle progression.Citation2 Studies of clinical materials have suggested the involvement of Rac1 in carcinogenesis. For example, the Rac1 activator Tiam1 is overexpressed in colon adenoma,Citation3 and its active mutant has been found in renal cell carcinoma.Citation4 Rac1b, a splice variant of Rac1, was preferentially expressed in patients with colon, breast and lung cancer.Citation5-Citation7 The Rac1b transcript contains an additional 57 nucleotides, which encode a protein with an in-frame insertion of 19 amino acids between Rac1 residues 75 and 76 and that is positioned immediately carboxyl-terminal to the switch II domain.Citation5,Citation6 The switch II domain, along with the switch I domain, constitutes the regions that change in conformation during GDP/GTP cycling. Several reports have confirmed that Rac1b showed enhanced intrinsic guanine nucleotide exchange activity and impaired intrinsic GTPase activity;Citation8-Citation10 that is, Rac1b behaves in a constitutively active fashion in the signaling pathway for cell growth. Indeed, Rac1b overexpression in NIH3T3 fibroblasts has been shown to cause transformation.Citation10
A series of studies has revealed roles of Rac1 in epithelial cells. For example, in MDCK (Madin-Darby Canine Kidney) cells cultured on a 2-dimensional sheet, Rac1-mediated signaling was shown to be involved in the initiation of tight or adherence junctions,Citation11-Citation13 and Rac1 has been shown to be activated upon the establishment of cadherin-based cell junctions.Citation14 Rac1b induced breakdown of cell-cell adhesion and increased cell motility in SCp2 mouse mammary epithelial cells.Citation15 Similarly, in HCT116 colon cancer cells, Rac1b expression reduced endogenous E-cadherin protein, and changed E-cadherin distribution from the plasma membrane to the cytoplasm.Citation16 It is therefore naturally expected that Rac1b expression causes morphological changes in the epithelial tissue. However, it remains technically challenging to observe signaling events in vivo.
Numerous 3-dimensional cell cultures in gel have been developed to reconstitute the in vivo microenvironment.Citation17 One representative is the MDCK cyst system; a single MDCK cell seeded in an extracellular matrix-rich gel grows to form a cyst that comprises a monolayer of polarized cells surrounding a fluid-filled lumen, which is similar to the epithelial structure in our body. Using this system, it was shown that Rac1 mediated laminin accumulation on the basal surface of cysts to orientate apico-basal polarity.Citation18 We recently demonstrated that Rac1 is preferentially activated in the lateral membrane in MDCK mature cysts using imaging with a Förster (Fluorescent) Resonance Energy Transfer (FRET)-based biosensor.Citation19 In addition, a novel conditional protein expression system has been developed based on ubiquitin degradation mediated by the plant hormone auxin.Citation20 In this system, an AID (Auxin-Inducible Degron)-tagged protein is degraded in the presence of auxin, and is restored after the removal of auxin. Applying this method to MDCK cysts, we recently established an in vitro system to investigate the molecular mechanisms by which Ras activation causes aberrant epithelial structures in vivo.Citation21 In this study using the MDCK cyst and AID system, we sought to determine whether Rac1b expression induces transformation in the mature epithelial structure.
Results
Induction of various AID-Rac1 proteins in MDCK cells
To induce various Rac1 proteins, we utilized the Auxin Inducible Degron (AID) system.Citation20 In the presence of 1-naphthaleneacetic acid (NAA), a synthetic auxin, the TIR1 plant F-box protein binds to the AID protein, leading to poly-ubiquitination of the AID protein and thereby degradation of the AID protein by the proteasome. Thus, an AID-tagged protein, such as AID-Rac1, should also be degraded in an auxin-dependent manner (). MDCK cells were first infected by a retrovirus carrying the TIR1 gene. After single-cell cloning, the clone with the highest level of TIR1 expression was designated as MDCK-TIR and used thereafter. The MDCK-TIR cells were super-infected with a retrovirus carrying the gene of AID-tagged Rac1, the wild-type of Rac1, Rac1b or the active form of Rac1 (Rac1G12V, denoted as Rac1V12 hereafter). The resulting cell lines were designated MDCK-Rac1WT, -Rac1b and -Rac1V12, and were passaged in the presence of NAA.Citation21
Figure 1. Inducible expression of various AID-tagged Rac1s in MDCK cells. (A) Schematic representation of the Auxin-Inducible Degron (AID) system. Auxin (indole-3-acetic acid) binding to TIR1, a plant F-box protein, promotes the interaction between TIR1 and the AID-tag, which is derived from Arabidopsis thaliana. The Skip1-Cul1-Rbx1 complex bound to TIR1 acts as an E3 ubiquitin ligase to effect polyubiquitylation of the AID-tagged proteins, followed by proteasome-mediated degradation. (B) MDCK-TIR (denoted as control) and various MDCK-Rac1-expressing Fucci cell cycle indicator cells were cultured with 50 μM NAA for 24 h, and further cultured for 24 h with or without NAA, followed by SDS-PAGE and western blotting analysis with anti-myc and anti-Rac1 antibodies to quantify TIR and AID-tagged and endogenous (denoted as Endo) Rac1, respectively. The result shown here is representative of two independent results. (C) Quantification of western blotting in B. After subtracting the background, the expression levels of AID-Rac1 (gray box) and endogenous Rac1 (white box) were normalized to that of TIR. These values were divided by the value of the endogenous Rac1/TIR without NAA. The averages from two independent experiments are shown. (D) MDCK-TIR cells expressing GFP-Syntaxin4 (denoted as Syntaxin4) and various AID-Rac1s (denoted as HA) were cultured on 35-mm glass-bottom dishes in medium without NAA for 24 h. The cells were fixed, immune-stained as described in Materials and Methods, and observed by a confocal microscope. The scale bar is 10 μm. (E) Measurement of guanine nucleotides bound to Rac1 and Rac1b. MDCK cells expressing GFP-Rac1WT and -Rac1b were subjected to quantification of the GTP/GDP ratio as described in the Materials and Methods. Shown in the graph is the average from two independent experiments.
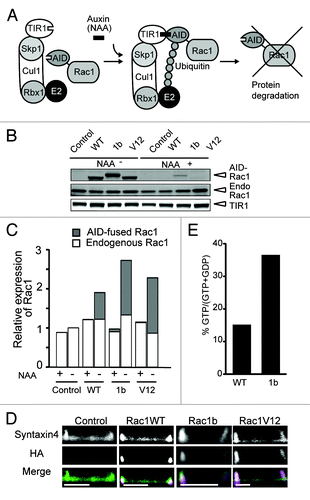
First, we compared the expression level of the AID-Rac1 protein to that of the endogenous Rac1 protein. MDCK-Rac1WT, -Rac1b and -Rac1V12 cells were further infected with retroviruses as a cell cycle indicator Fucci. Therefore, we examined AID-tagged Rac1 proteins in these cells. As a control, MDCK-TIR cells were used throughout this study. These cells were cultured in the presence or absence of NAA, and subjected to quantitative western blotting analysis with an anti-Rac1 antibody (). We found that the expression level of endogenous Rac1 was comparable among cells treated with or without NAA in MDCK-TIR cells expressing a Fucci indicator (, + and – NAA). In these cells, AID-Rac1WT, -1b and -V12 were conditionally induced to a level similar to that of endogenous Rac1. Importantly, the induced AID-tagged Rac1 proteins had little effect on the endogenous Rac1 protein level. Immunofluorescent staining showed that these AID-tagged Rac1 proteins were preferentially localized to the lateral membrane in the two-dimensional sheets (). We also confirmed that the GTP/GDP ratio of Rac1b is higher than that of Rac1-WT by thin layer chromatography ().
Perturbation of cystogenesis by Rac1V12 and Rac1b
A normal MDCK cell embedded in Matrigel grows to form a cyst, whose lumen faces the apical surface of polarized monolayer MDCK cells. Using the AID-inducible expression system, we next examined the effect of Rac1 protein expression on cystogenesis. To visualize the nucleus and the plasma membrane, the MDCK cell lines were dually infected with retroviruses for histone H1 fused to fluorescent protein dKeima (H1-Keima) and plasma membrane-targeted fluorescent protein GFP (GFP-CAAX). The cells were cultured in Matrigel with NAA for 10 d to form cysts, and further cultured with or without NAA. In the presence of NAA, the MDCK-Rac1WT, -Rac1b and -Rac1V12 cells formed a spherical cyst with a central lumen as did the control MDCK cells (, NAA+).Citation19 Upon deprivation of NAA to induce AID-tagged proteins, the morphology was not altered in the control MDCK, MDCK-Rac1WT or MDCK-Rac1b cysts (, NAA−). Upon deprivation of NAA in the MDCK-Rac1V12 cysts, however, a cyst-like cell mass was observed in the lumen.
Figure 2. Morphological change upon Rac1 expression. (A) MDCK-TIR (denoted as control) and various MDCK-Rac1 cells expressing H1-Keima (denoted as Nucleus) and GFP-CAAX (denoted as Plasma membrane) were cultured to form cysts in the presence of 50 μM NAA for 10 d. Cysts were washed and further cultured for 5 d with (+) and without (−) NAA and imaged by a confocal microscope. Shown are the representative images from at least two independent experiments. The scale bar is 20 μm. (B) Fluorescent images obtained in were quantified. The numbers of cells in the luminal space were counted from H1-Keima images. Averages with the standard deviation (sd) are shown. *p < 0.05 by Student’s t-test. The numbers of scored cysts were as follows: control (+) 152, control (−) 155; Rac1 WT (+) 663, Rac1WT (−) 554; Rac1b(+) 461, Rac1b (−) 538; Rac1V12 (+) 164, Rac1V12 (−) 172.
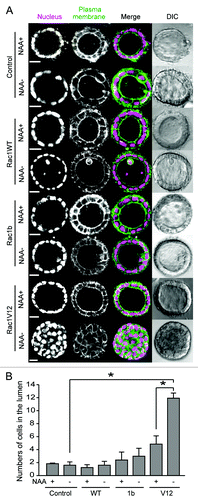
The numbers of cells in the lumen counted on the fluorescent images of H1-Keima are shown in . The average cell numbers were 1.79 in the MDCK-TIR cyst treated with NAA, 1.56 in MDCK-TIR cysts without NAA, 1.19 in the MDCK-Rac1WT cyst with NAA and 1.56 in the MDCK-Rac1WT cyst without NAA. These results indicated that Rac1WT did not significantly affect the number of cells in the lumen. The average cell numbers in the lumen were 2.36 in MDCK-Rac1b with NAA and 2.94 in MDCK-Rac1b without NAA, indicating that Rac1b induction increased the number of cells in the lumen slightly. In contrast, the numbers of cells in the lumen were 4.81 in MDCK-Rac1V12 with NAA and 11.9 in MDCK-Rac1V12 without NAA, clearly indicating that Rac1V12 induced accumulation of cells in the lumen. The increase in cells in the lumen of MDCK-Rac1V12 and MDCK-Rac1b in the presence of NAA was probably due to the leaky expression of Rac1V12 and Rac1b.
Accelerated cell cycle progression upon Rac1V12 expression
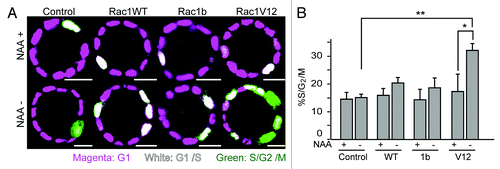
It has been reported that the recombinant Rac1 protein microinjected into quiescent fibroblasts stimulates cell cycle progression through G1 and subsequent DNA synthesis.Citation2,Citation22 It has also been reported that exogenously-expressed Rac1b promotes cell cycle progression in NIH3T3 cells,Citation23 and that depletion of Rac1b inhibits G1/S transition in CaCo2 cells.Citation24 Therefore, we next examined whether or not the expression of Rac1WT, Rac1b or Rac1V12 accelerated the cell cycle progression in MDCK cysts. To examine this, we utilized a cell-cycle indicator, Fucci.Citation25 Fucci consists of mCherry-hCdt1 and Venus-hGeminin, which are G1 and S/G2/M phase indicators, respectively. In the processed images (), magenta, white, and green indicate cells in the G1, G1/S, and S/G2/M phases. In the presence of NAA, MDCK-TIR, -Rac1WT, -Rac1b and -Rac1V12 cysts, most cells in the cyst wall were in G1 phase, and a small number of cells were in G1/S phase (, the upper panels). After NAA deprivation for two days (NAA−), the increase in S/G2/M cells was not significant in MDCK-TIR, -Rac1WT and -Rac1b cysts (). However, a marked increase in S/G2/M cells was observed in MDCK-Rac1V12 cysts. A similar increase in S/G2/M cells was observed in MDCK-Rac1V12 cysts cultured for four days (Fig. S1). Thus, the acceleration of cell cycle progression seems to contribute to the luminal cell accumulation in MDCK-Rac1V12 cysts.
Figure 3. Accelerated cell cycle progression upon Rac1 induction. (A) MDCK-TIR and MDCK-Rac1 cells expressing mCherry-hCdt and Venus-hGem were cultured to form cysts in Matrigel containing 50 μM NAA for 10 d and transferred to a collagen gel with or without NAA. One day later, cells were imaged by a confocal microscope. Scale bars = 20 μm. (B) Quantification of (A). The percentage of S/G2/M cells, as indicated by green fluorescent cells, was scored. Data are the averages with s.d. *p < 0.05 by Student’s t-test. The numbers of scored cysts were as follows: control (+) Experiment (Exp)1 23, Exp2 24, Exp3 26, and Exp4 27, control (−), Exp1 23, Exp2 24, Exp3 10, and Exp4 27; Rac1 WT (+) Exp1 26, Exp2 38, Exp3 36, and Exp4 44; Rac1WT (−) Exp1 42, Exp2 40, Exp3 49, and Exp4 45; Rac1b (+) Exp1 23, Exp2 36, Exp3 28, Exp4 34, Exp5 65, and Exp6 65; Rac1b (−) Exp1 26, Exp2 35, Exp3 31, Exp4 38, Exp5 90, and Exp6 74; Rac1V12 (+) Exp1 28, Exp2 20 and Exp3 38; Rac1V12 (−) Exp1 25, Exp2 23 and Exp3 35.
Discussion
In this report, we have shown that the expression of constitutively active Rac1, but not expression of the wild-type or Rac1b, in MDCK cells within a mature cyst structure causes luminal cell accumulation through acceleration of the cell cycle progression ( and ).
Based on the previously reported biochemical properties of Rac1b,Citation10 we had expected that Rac1b would induce transformation/morphological changes in MDCK cysts. Since we confirmed that AID-tagged Rac1 proteins were expressed in similar amounts (), it appeared that the differences in quality among the various Rac1s influenced the cellular outputs. The most plausible difference between Rac1V12 and Rac1b would be a difference in their ability to interact with other molecules. For example, it has been well established that Rac1b does not interact with RhoGDI (GDP-dissociation inhibitor), a negative regulator for Rho family GTPases.Citation9,Citation10,Citation26 Whether or not Rac1b interacts with Tiam1, a guanine-nucleotide exchange factor for Rac1, is controversial.Citation8-Citation10 In a study using co-precipitation and proteomic methods, it was shown that Rac1b did not bind to common regulators of Rac1, such as GIT-1 and IQGAP.Citation26 Instead, Rac1b showed enhanced binding to the proteins SmgGDS, RACK1 and p120 catenin. It has been established that the cell cycle progression is regulated by the levels of cyclin D1 expression and cyclin-dependent kinase inhibitors,Citation27 and conflicting results have been reported with respect to Rac1b and cyclin D1 expression. In NIH3T3 cells, Rac1b is unable to stimulate or induce canonical downstream effectors, including NF-κB and cyclin D1.Citation10 In HEK293 cells, however, exogenous expression of Rac1b induces cyclin D1 expression.Citation16 This difference may be cell-type dependent. As shown in the present study, Rac1V12, but not Rac1b, could promote cell cycle progression in MDCK cysts (). Unlike in previous studies that employed overexpression systems, Rac1b was expressed at a level comparable to that of the endogenous Rac1 in MDCK cysts. In addition, the 3D condition, which seems to be more physiological, may also attenuate the effect of Rac1b on the cell cycle and morphology of MDCK cells. Thus, as is often the case with other oncogenes, Rac1b may contribute to the oncogenesis, but the expression of Rac1b alone may not be insufficient to induce either cell cycle progression or morphological alteration in a physiological context.
We found that Rac1V12 expression induced luminal cell accumulation (), while earlier studies showed that Rac1V12 expression did not alter the polarity of MDCK cells.Citation28,Citation29 In the present study, the expression level of Rac1V12 was approximately equal to the level of endogenous Rac1 (), whereas the expression level of Rac1V12 was not determined in the previous studies,Citation18 leaving the possibility that the difference was caused by the expression level. Another unexpected result in this study is that Rac1V12 stimulated cell cycle progression. We have shown that Rac1 activity is lower at the apical plasma membrane than the lateral plasma membrane, and that forced activation of Rac1 at the apical plasma membrane induces luminal cell accumulation by reorientation of the cell division axis, but not by accelerated cell cycle progression.Citation19 These data suggest that Rac1 activation by GTPase activity deficiency and that by the increased GEF activity cause different cell biological outcomes, even though the resulting morphological changes are very similar to each other.
In summary, we found that Rac1b expression alone was insufficient to initiate morphological changes or cell cycle progression in MDCK cysts. Since Rac1b expression is regulated by ASF/SF2 or SRp20,Citation30 Dishevelled-3Citation16 and MMP-3,Citation15 it is plausible that these regulators may co-operatively work with Rac1b. This idea is supported by the recent report that Rac1b is dispensable for carcinogenesis but enhances active Ras-induced tumorigenesis.Citation7 A recent report also suggested a role of Rac1b in cell invasion by showing that Rac1b expression led to a reduction of membrane-bound Rac1 and promoted RhoA activation.Citation31 Considering that RhoA and Rac1 are both required for cell migration, but regulate different functions for motility,Citation32 and that the MMP-3-mediated epithelial-mesenchymal transition is Rac1-dependent,Citation15 it is tempting to speculate that Rac1b modulates the Rac1/RhoA balance in order to induce morphological change for invasion. Further experiments using the system established here will help to fully clarify the Rac1b-involved signaling in cancer progression.
Materials and Methods
Plasmids and antibodies
AID was amplified by PCR and cloned into a related retroviral expression vector to create pCX4bsr-3HA-AID-AID, which carries a blasticidin-resistance selectable marker. The cDNAs of wild-type Rac1, Rac1b and Rac1V12 were amplified and cloned into this vector. To express GFP-CAAX and Histone H1-Keima, the plasmids pCX4bsr-EGFP-KRasCT and pCX4neo-H1-dKeima were used. Plasmids for Fucci, CSII-EF-MCS-mCherry-hCdt, and CSII-EF-MCS-Venus-hGem were provided by Dr. Atsushi Miyawaki, RIKEN.Citation25
The primary antibodies used in this study were: anti-myc (Santa Cruz, sc-40 or sc-789), anti-HA (Roche, 11867423001) and anti-Rac1 (BD Biosciences, 610650). Secondary antibodies IRDye 800CW or IRDye 680 (LI-COR, 926-32210, 926-32221, Rockland Immunochemicals, Inc., 612–131–120) were used for western blotting, and anti-rat IgG antibody conjugated with Alexa647 (Life Technologies, A-21247) was used for immunostaining.
Cell culture
MDCK cells were purchased from RIKEN BioResource Center (No. RCB0995), and maintained in minimal essential medium (MEM) containing Earle’s balanced salt solution (Invitrogen,10370-021) supplemented with 10% fetal bovine serum (FBS; Equitech-Bio, SFBM-0500), 3% L-Gln, 0.1% non-essential amino acids, 1 mM sodium pyruvate, 100 units/ml penicillin and 100 μg/ml streptomycin, in a 5% CO2 humidified incubator at 37°C.
Establishment of cell lines
Viruses were produced in BOSC23 cells that were transfected with the expression vectors, the packaging plasmid pGP,Citation33 and the envelope plasmid pCMV-VSV-G-Rsv-Rev (provided by Hiroyuki Miyoshi and Atsushi Miyawaki, RIKEN). MDCK cells were first infected with the retrovirus for TIR-9myc expression, then cloned and designated as MDCK-TIR.Citation21 The MDCK-TIR cells were next infected with the retrovirus for various 3HA-AID tagged proteins, and selected by antibiotics. To establish the MDCK cells stably expressing the fluorescence markers or Fucci, MDCK-TIR cells were infected with retro- or lenti-viruses, respectively. To create lentiviruses, pCAG-HIVgp (provided by Hiroyuki Miyoshi, RIKEN) and pCMV-VSV-G-Rsv-Rev were utilized for virus production. For some experiments, virus-containing media was concentrated by RetroX (Clontech, 631456). After infection, cells were subjected to selection for two days with 8 mg/ml of G418 for pCX4neo vectors and 80 μg/ml of blasticidin for pCX4bsr vectors. For conditional degradation of AID-tagged proteins, the cells were treated with 50 μM synthetic auxin, 1-naphthaleneacetic acid (NAA), in culture medium.
Western blotting
Cells were lysed in 1x SDS Sample Buffer [62.5 mM TRIS-HCl (pH 6.8), 12% glycerol, 2% SDS, 0.004% BPB and 10% 2-mercaptoethanol]. After sonication, proteins were separated by SDS-PAGE on 5–20% Nagaiki precast gels (Intertechno Co., Ltd, GM-0520-3N, GM-0520-7N), and transferred to PVDF membranes (Immobilon FL-PVDF; Millipore,IPFL00010). After blocking with 5% skim milk for 90 min, membranes were blotted with various antibodies diluted 1:500 in a solution of Odyssey blocking buffer (LI-COR, 927-40000) and Tris-buffered saline, followed by incubation with secondary antibodies. Membranes were then scanned with an Odyssey IR scanner. Quantification was performed by MetaMorph Software (Molecular Devices).
Immunofluorescent staining
MDCK-TIR cells expressing GFP-Syntaxin4 and various AID-Rac1s were cultured on 35-mm glass-bottom dishes in medium with or without NAA for 24 h. The cells were fixed by 4% paraformaldehyde (PFA) for 20 min at room temperature and permeabilized with 0.1% TritonX-100 for 30 min at room temperature. After washing with phosphate buffered saline (PBS), the samples were treated with 0.7% fish skin gelatin (Sigma-Aldrich,G7765) and 0.025% saponin/PBS, followed by incubation with anti-HA antibody diluted to 1:200 overnight at 4°C. Samples were then treated with anti-rat IgG antibody conjugated with Alexa647. Images were obtained by FV1000 confocal microscopy (Olympus).
Measurement of guanine nucleotides bound to Rac1
Quantitation of guanine nucleotides bound to low-molecular-weight G proteins was performed as described previously.Citation34 Briefly, MDCK cells expressing GFP-Rac1WT and –Rac1b were cultured for 24 h. Cells were labeled with 0.05 mCi of 32Pi (NEN) in 0.5 ml of phosphate-free DMEM (Invitrogen, 11971–025) for 2 h and lysed in lysis buffer [20 mM TRIS-HCl (pH 7.5), 150 mM NaCl, 20 mM MgCl2, 1 mM Na3VO4, 0.5% Triton X-100] containing anti-GFP antibody (a gift from Dr Naoki Mochizuki). After centrifugation, the cleared cell lysates were incubated with protein-A agarose beads at 4°C for 30 min. After washing with lysis buffer, the immunoprecipitates were separated by thin layer chromatography, and developed by BAS2000 (Fuji Film Japan).
Cystogenesis
Cysts were generated as previously described, with modification.Citation19,Citation35,Citation36 Briefly, 8 x 103 MDCK cells were placed on an 8-chamber slide glass (Iwaki glass Co., Ltd., 3911–035) coated with 80 μl of polymerized Matrigel (BD Biosciences, 356231), and then supplied with culture medium containing 2% Matrigel for the indicated periods.
Confocal fluorescence imaging
For fluorescent imaging, an IX81 inverted microscope equipped with an FV1000 (Olympus), UPlanSApo 20X/0.75 (Olympus) was used. The confocal aperture size and image size were set at 80 μm and at 512 x 512 pixels with various zoom factors, respectively. The excitation laser and fluorescence filter settings were as follows: excitation laser, 440 nm for dKeima, 488 nm for GFP, 515 nm for Venus, 559 nm for mCherry, 635 nm for Alexa647; excitation dichroic mirror, DM405/488/559/635 for Alexa647 and GFP and BS20/80 for Venus, mCherry and dKeima; GFP and Venus channel PMT dichroic mirror, SDM 560; GFP channel PMT spectral setting, 500–545 nm; Venus channel PMT spectral setting, 530–545 nm; mCherry channel PMT spectral setting, 570–670 nm; dKeima channel PMT spectral setting, 575–675 nm; Alexa647 channel PMT spectral setting, 655–755 nm.
Additional material
Download Zip (791.6 KB)Acknowledgments
We are grateful to Ms. Y. Inaoka, K. Hirano, A. Kawagishi and K. Miyamoto for their technical assistance, and the staff of the Matsuda Laboratory for their technical advice and helpful input. This work was supported by grants from the Ministry of Education, Culture, Sports, Science, and Technology of Japan and the Mochida Memorial Foundation for Medical and Pharmaceutical Research. S.Y. and A.S. were supported by research fellowships from the Japan Society for the Promotion of Science for Young Scientists.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplemental Material
Supplemental materials may be found here: www.landesbioscience.com/journals/smallgtpases/article/23311
References
- Ridley AJ, Paterson HF, Johnston CL, Diekmann D, Hall A. The small GTP-binding protein rac regulates growth factor-induced membrane ruffling. Cell 1992; 70:401 - 10; http://dx.doi.org/10.1016/0092-8674(92)90164-8; PMID: 1643658
- Olson MF, Ashworth A, Hall A. An essential role for Rho, Rac, and Cdc42 GTPases in cell cycle progression through G1. Science 1995; 269:1270 - 2; http://dx.doi.org/10.1126/science.7652575; PMID: 7652575
- Malliri A, Rygiel TP, van der Kammen RA, Song JY, Engers R, Hurlstone AF, et al. The rac activator Tiam1 is a Wnt-responsive gene that modifies intestinal tumor development. J Biol Chem 2006; 281:543 - 8; http://dx.doi.org/10.1074/jbc.M507582200; PMID: 16249175
- Engers R, Zwaka TP, Gohr L, Weber A, Gerharz CD, Gabbert HE. Tiam1 mutations in human renal-cell carcinomas. Int J Cancer 2000; 88:369 - 76; http://dx.doi.org/10.1002/1097-0215(20001101)88:3<369::AID-IJC8>3.0.CO;2-K; PMID: 11054665
- Jordan P, Brazåo R, Boavida MG, Gespach C, Chastre E. Cloning of a novel human Rac1b splice variant with increased expression in colorectal tumors. Oncogene 1999; 18:6835 - 9; http://dx.doi.org/10.1038/sj.onc.1203233; PMID: 10597294
- Schnelzer A, Prechtel D, Knaus U, Dehne K, Gerhard M, Graeff H, et al. Rac1 in human breast cancer: overexpression, mutation analysis, and characterization of a new isoform, Rac1b. Oncogene 2000; 19:3013 - 20; http://dx.doi.org/10.1038/sj.onc.1203621; PMID: 10871853
- Zhou C, Licciulli S, Avila JL, Cho M, Troutman S, Jiang P, et al. The Rac1 splice form Rac1b promotes K-ras-induced lung tumorigenesis. Oncogene 2012; In Press http://dx.doi.org/10.1038/onc.2012.99; PMID: 22430205
- Matos P, Collard JG, Jordan P. Tumor-related alternatively spliced Rac1b is not regulated by Rho-GDP dissociation inhibitors and exhibits selective downstream signaling. J Biol Chem 2003; 278:50442 - 8; http://dx.doi.org/10.1074/jbc.M308215200; PMID: 14506233
- Fiegen D, Haeusler LC, Blumenstein L, Herbrand U, Dvorsky R, Vetter IR, et al. Alternative splicing of Rac1 generates Rac1b, a self-activating GTPase. J Biol Chem 2004; 279:4743 - 9; http://dx.doi.org/10.1074/jbc.M310281200; PMID: 14625275
- Singh A, Karnoub AE, Palmby TR, Lengyel E, Sondek J, Der CJ. Rac1b, a tumor associated, constitutively active Rac1 splice variant, promotes cellular transformation. Oncogene 2004; 23:9369 - 80; http://dx.doi.org/10.1038/sj.onc.1208182; PMID: 15516977
- Jou TS, Schneeberger EE, Nelson WJ. Structural and functional regulation of tight junctions by RhoA and Rac1 small GTPases. J Cell Biol 1998; 142:101 - 15; http://dx.doi.org/10.1083/jcb.142.1.101; PMID: 9660866
- Takaishi K, Sasaki T, Kotani H, Nishioka H, Takai Y. Regulation of cell-cell adhesion by rac and rho small G proteins in MDCK cells. J Cell Biol 1997; 139:1047 - 59; http://dx.doi.org/10.1083/jcb.139.4.1047; PMID: 9362522
- Jou TS, Nelson WJ. Effects of regulated expression of mutant RhoA and Rac1 small GTPases on the development of epithelial (MDCK) cell polarity. J Cell Biol 1998; 142:85 - 100; http://dx.doi.org/10.1083/jcb.142.1.85; PMID: 9660865
- Ehrlich JS, Hansen MD, Nelson WJ. Spatio-temporal regulation of Rac1 localization and lamellipodia dynamics during epithelial cell-cell adhesion. Dev Cell 2002; 3:259 - 70; http://dx.doi.org/10.1016/S1534-5807(02)00216-2; PMID: 12194856
- Radisky DC, Levy DD, Littlepage LE, Liu H, Nelson CM, Fata JE, et al. Rac1b and reactive oxygen species mediate MMP-3-induced EMT and genomic instability. Nature 2005; 436:123 - 7; http://dx.doi.org/10.1038/nature03688; PMID: 16001073
- Esufali S, Charames GS, Pethe VV, Buongiorno P, Bapat B. Activation of tumor-specific splice variant Rac1b by dishevelled promotes canonical Wnt signaling and decreased adhesion of colorectal cancer cells. Cancer Res 2007; 67:2469 - 79; http://dx.doi.org/10.1158/0008-5472.CAN-06-2843; PMID: 17363564
- O’Brien LE, Zegers MM, Mostov KE. Opinion: Building epithelial architecture: insights from three-dimensional culture models. Nat Rev Mol Cell Biol 2002; 3:531 - 7; http://dx.doi.org/10.1038/nrm859; PMID: 12094219
- O’Brien LE, Jou TS, Pollack AL, Zhang Q, Hansen SH, Yurchenco P, et al. Rac1 orientates epithelial apical polarity through effects on basolateral laminin assembly. Nat Cell Biol 2001; 3:831 - 8; http://dx.doi.org/10.1038/ncb0901-831; PMID: 11533663
- Yagi S, Matsuda M, Kiyokawa E. Suppression of Rac1 activity at the apical membrane of MDCK cells is essential for cyst structure maintenance. EMBO Rep 2012; 13:237 - 43; http://dx.doi.org/10.1038/embor.2011.249; PMID: 22261715
- Nishimura K, Fukagawa T, Takisawa H, Kakimoto T, Kanemaki M. An auxin-based degron system for the rapid depletion of proteins in nonplant cells. Nat Methods 2009; 6:917 - 22; http://dx.doi.org/10.1038/nmeth.1401; PMID: 19915560
- Sakurai A, Matsuda M, Kiyokawa E. Activated Ras protein accelerates cell cycle progression to perturb Madin-Darby canine kidney cystogenesis. J Biol Chem 2012; 287:31703 - 11; http://dx.doi.org/10.1074/jbc.M112.377804; PMID: 22829590
- Lamarche N, Tapon N, Stowers L, Burbelo PD, Aspenström P, Bridges T, et al. Rac and Cdc42 induce actin polymerization and G1 cell cycle progression independently of p65PAK and the JNK/SAPK MAP kinase cascade. Cell 1996; 87:519 - 29; http://dx.doi.org/10.1016/S0092-8674(00)81371-9; PMID: 8898204
- Matos P, Jordan P. Expression of Rac1b stimulates NF-kappaB-mediated cell survival and G1/S progression. Exp Cell Res 2005; 305:292 - 9; http://dx.doi.org/10.1016/j.yexcr.2004.12.029; PMID: 15817154
- Vigil D, Cherfils J, Rossman KL, Der CJ. Ras superfamily GEFs and GAPs: validated and tractable targets for cancer therapy?. Nat Rev Cancer 2010; 10:842 - 57; http://dx.doi.org/10.1038/nrc2960; PMID: 21102635
- Sakaue-Sawano A, Kurokawa H, Morimura T, Hanyu A, Hama H, Osawa H, et al. Visualizing spatiotemporal dynamics of multicellular cell-cycle progression. Cell 2008; 132:487 - 98; http://dx.doi.org/10.1016/j.cell.2007.12.033; PMID: 18267078
- Orlichenko L, Geyer R, Yanagisawa M, Khauv D, Radisky ES, Anastasiadis PZ, et al. The 19-amino acid insertion in the tumor-associated splice isoform Rac1b confers specific binding to p120 catenin. J Biol Chem 2010; 285:19153 - 61; http://dx.doi.org/10.1074/jbc.M109.099382; PMID: 20395297
- Fu M, Wang C, Li Z, Sakamaki T, Pestell RG. Minireview: Cyclin D1: normal and abnormal functions. Endocrinology 2004; 145:5439 - 47; http://dx.doi.org/10.1210/en.2004-0959; PMID: 15331580
- Rogers KK, Jou TS, Guo W, Lipschutz JH. The Rho family of small GTPases is involved in epithelial cystogenesis and tubulogenesis. Kidney Int 2003; 63:1632 - 44; http://dx.doi.org/10.1046/j.1523-1755.2003.00902.x; PMID: 12675838
- Yu W, Datta A, Leroy P, O’Brien LE, Mak G, Jou TS, et al. β1-integrin orients epithelial polarity via Rac1 and laminin. Mol Biol Cell 2005; 16:433 - 45; http://dx.doi.org/10.1091/mbc.E04-05-0435; PMID: 15574881
- Gonçalves V, Matos P, Jordan P. Antagonistic SR proteins regulate alternative splicing of tumor-related Rac1b downstream of the PI3-kinase and Wnt pathways. Hum Mol Genet 2009; 18:3696 - 707; http://dx.doi.org/10.1093/hmg/ddp317; PMID: 19602482
- Nimnual AS, Taylor LJ, Nyako M, Jeng HH, Bar-Sagi D. Perturbation of cytoskeleton dynamics by the opposing effects of Rac1 and Rac1b. Small GTPases 2010; 1:89 - 97; http://dx.doi.org/10.4161/sgtp.1.2.14427; PMID: 21686260
- Sahai E, Marshall CJ. RHO-GTPases and cancer. Nat Rev Cancer 2002; 2:133 - 42; http://dx.doi.org/10.1038/nrc725; PMID: 12635176
- Akagi T, Shishido T, Murata K, Hanafusa H. v-Crk activates the phosphoinositide 3-kinase/AKT pathway in transformation. Proc Natl Acad Sci U S A 2000; 97:7290 - 5; http://dx.doi.org/10.1073/pnas.140210297; PMID: 10852971
- Kiyokawa E, Hashimoto Y, Kobayashi S, Sugimura H, Kurata T, Matsuda M. Activation of Rac1 by a Crk SH3-binding protein, DOCK180. Genes Dev 1998; 12:3331 - 6; http://dx.doi.org/10.1101/gad.12.21.3331; PMID: 9808620
- Martin-Belmonte F, Gassama A, Datta A, Yu W, Rescher U, Gerke V, et al. PTEN-mediated apical segregation of phosphoinositides controls epithelial morphogenesis through Cdc42. Cell 2007; 128:383 - 97; http://dx.doi.org/10.1016/j.cell.2006.11.051; PMID: 17254974
- Pollack AL, Runyan RB, Mostov KE. Morphogenetic mechanisms of epithelial tubulogenesis: MDCK cell polarity is transiently rearranged without loss of cell-cell contact during scatter factor/hepatocyte growth factor-induced tubulogenesis. Dev Biol 1998; 204:64 - 79; http://dx.doi.org/10.1006/dbio.1998.9091; PMID: 9851843